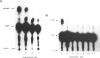Abstract
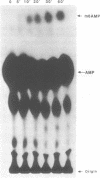
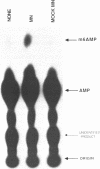
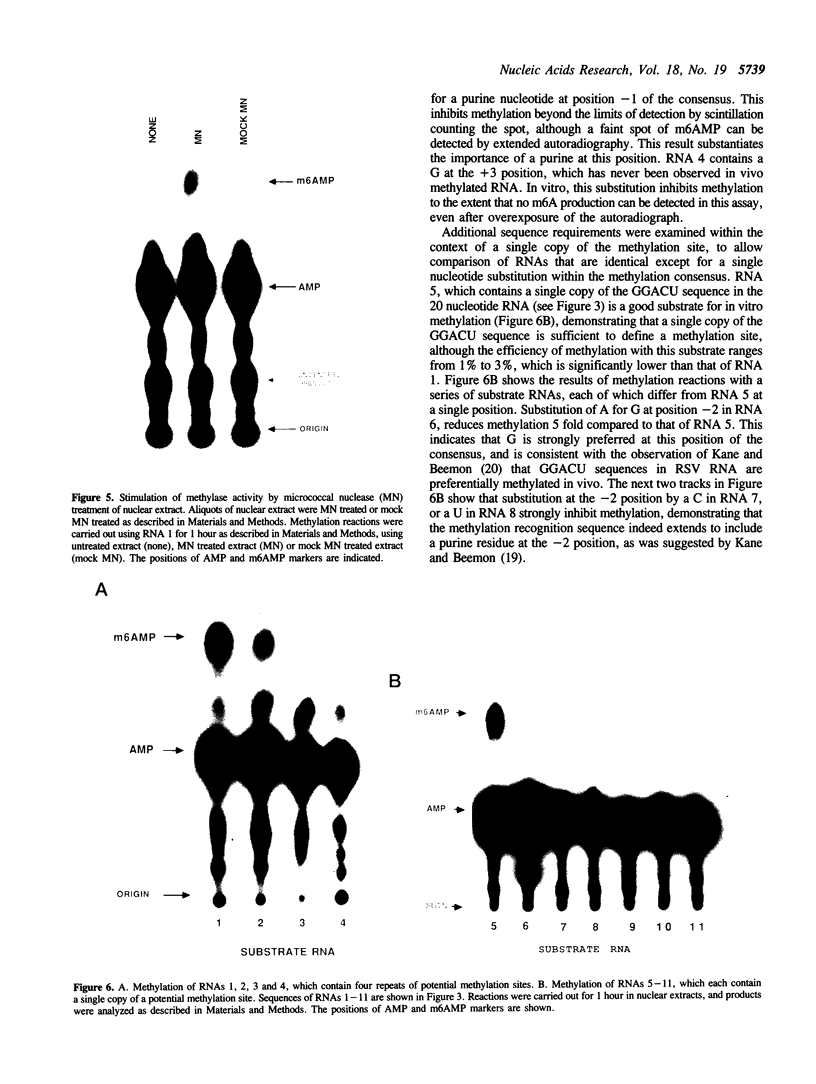
N6-adenosine methylation is a frequent modification of mRNAs and their precursors, but little is known about the mechanism of the reaction or the function of the modification. To explore these questions, we developed conditions to examine N6-adenosine methylase activity in HeLa cell nuclear extracts. Transfer of the methyl group from S-[3H methyl]-adenosylmethionine to unlabeled random copolymer RNA substrates of varying ribonucleotide composition revealed a substrate specificity consistent with a previously deduced consensus sequence, Pu[G greater than A]AC[A/C/U]. 32-P labeled RNA substrates of defined sequence were used to examine the minimum sequence requirements for methylation. Each RNA was 20 nucleotides long, and contained either the core consensus sequence GGACU, or some variation of this sequence. RNAs containing GGACU, either in single or multiple copies, were good substrates for methylation, whereas RNAs containing single base substitutions within the GGACU sequence gave dramatically reduced methylation. These results demonstrate that the N6-adenosine methylase has a strict sequence specificity, and that there is no requirement for extended sequences or secondary structures for methylation. Recognition of this sequence does not require an RNA component, as micrococcal nuclease pretreatment of nuclear extracts actually increased methylation efficiency.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Cory S. Modified nucleosides and bizarre 5'-termini in mouse myeloma mRNA. Nature. 1975 May 1;255(5503):28–33. doi: 10.1038/255028a0. [DOI] [PubMed] [Google Scholar]
- Aloni Y., Dhar R., Khoury G. Methylation of nuclear simian virus 40 RNAs. J Virol. 1979 Oct;32(1):52–60. doi: 10.1128/jvi.32.1.52-60.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemon K., Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977 Jun 15;113(1):165–179. doi: 10.1016/0022-2836(77)90047-x. [DOI] [PubMed] [Google Scholar]
- Camper S. A., Albers R. J., Coward J. K., Rottman F. M. Effect of undermethylation on mRNA cytoplasmic appearance and half-life. Mol Cell Biol. 1984 Mar;4(3):538–543. doi: 10.1128/mcb.4.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani D., Kahana C., Lavi S., Groner Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979 Jun 25;6(8):2879–2899. doi: 10.1093/nar/6.8.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Kiang S., Nevins J. R., Darnell J. E., Jr N-6-methyl-adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J Mol Biol. 1979 Dec 15;135(3):733–752. doi: 10.1016/0022-2836(79)90174-8. [DOI] [PubMed] [Google Scholar]
- Christofori G., Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988 Sep 9;54(6):875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimock K., Stoltzfus C. M. Processing and function of undermethylated chicken embryo fibroblast mRNA. J Biol Chem. 1979 Jul 10;254(13):5591–5594. [PubMed] [Google Scholar]
- Dimock K., Stoltzfus C. M. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977 Feb 8;16(3):471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Taylor R. H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975 Oct;2(10):1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel D., Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983 Dec;131(2):409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Morgan M., Shatkin A. J., Jelinek W., Salditt-Georgieff M., Darnell J. E. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci U S A. 1975 May;72(5):1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A. J., Stavnezer E., Bishop J. M. Blocked, methylated 5'-terminal sequence in avian sarcoma virus RNA. Nature. 1975 Oct 16;257(5527):618–620. doi: 10.1038/257618a0. [DOI] [PubMed] [Google Scholar]
- Gilmartin G. M., McDevitt M. A., Nevins J. R. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988 May;2(5):578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Horowitz A., Nilsen T. W., Munns T. W., Rottman F. M. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. E., Beemon K. Inhibition of methylation at two internal N6-methyladenosine sites caused by GAC to GAU mutations. J Biol Chem. 1987 Mar 5;262(7):3422–3427. [PubMed] [Google Scholar]
- Kane S. E., Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985 Sep;5(9):2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANE B. G. The separation of adenosine, guanosine, cytidine and uridine by one-dimensional filter-paper chromatography. Biochim Biophys Acta. 1963 May 28;72:110–112. [PubMed] [Google Scholar]
- MAGASANIK B., VISCHER E., DONIGER R., ELSON D., CHARGAFF E. The separation and estimation of ribonucleotides in minute quantities. J Biol Chem. 1950 Sep;186(1):37–50. [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Gershowitz A., Weber L. A., Baglioni C. Histone mRNAs contain blocked and methylated 5' terminal sequences but lack methylated nucleosides at internal positions. Cell. 1977 Jan;10(1):113–120. doi: 10.1016/0092-8674(77)90145-3. [DOI] [PubMed] [Google Scholar]
- Narayan P., Rottman F. M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science. 1988 Nov 25;242(4882):1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Friderici K., Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5' terminus. Cell. 1975 Apr;4(4):387–394. doi: 10.1016/0092-8674(75)90159-2. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E. Methylated constituents of heterogeneous nuclear RNA: presence in blocked 5' terminal structures. Cell. 1975 Sep;6(1):13–19. doi: 10.1016/0092-8674(75)90068-9. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Scherrer K. The methylated constituents of globin mRNA. FEBS Lett. 1975 Sep 1;57(1):73–78. doi: 10.1016/0014-5793(75)80155-4. [DOI] [PubMed] [Google Scholar]
- Salditt-Georgieff M., Jelinek W., Darnell J. E., Furuichi Y., Morgan M., Shatkin A. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell. 1976 Feb;7(2):227–237. doi: 10.1016/0092-8674(76)90022-2. [DOI] [PubMed] [Google Scholar]
- Schibler U., Kelley D. E., Perry R. P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977 Oct 5;115(4):695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976 Dec;9(4 Pt 2):645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Dane R. W. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J Virol. 1982 Jun;42(3):918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y., Ryner L. C., Manley J. L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988 Mar 11;52(5):731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- Wei C. M., Gershowitz A., Moss B. 5'-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976 Jan 27;15(2):397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- Wei C. M., Gershowitz A., Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975 Apr;4(4):379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- Wei C. M., Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977 Apr 19;16(8):1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- Wei C., Gershowitz A., Moss B. N6, O2'-dimethyladenosine a novel methylated ribonucleoside next to the 5' terminal of animal cell and virus mRNAs. Nature. 1975 Sep 18;257(5523):251–253. doi: 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]