Abstract
The SCF (Skp1, Cullins, F-box proteins) multisubunit E3 ubiquitin ligase, also known as CRL (Cullin-RING ubiquitin Ligase) is the largest E3 ubiquitin ligase family that promotes the ubiquitination of various regulatory proteins for targeted degradation, thus regulating many biological processes, including cell cycle progression, signal transduction, and DNA replication. The efforts to discover small molecule inhibitors of a SCF-type ligase or its components were expedited by the FDA approval of Bortezomib (also known as Velcade or PS-341), the first (and only) class of general proteasome inhibitor, for the treatment of relapsed/refractory multiple myeloma and mantle cell lymphoma. Although Bortezomib has demonstrated a certain degree of cancer cell selectivity with measurable therapeutic index, the drug is, in general, cytotoxic due to its inhibition of overall protein degradation. An alternative and ideal approach is to target a specific E3 ligase, known to be activated in human cancer, for a high level of specificity and selectivity with less associated toxicity, since such inhibitors would selectively stabilize a specific set of cellular proteins regulated by this E3. Here, we review recent advances in validation of SCF E3 ubiquitin ligase as an attractive anti-cancer target and discuss how MLN4924, a small molecule inhibitor of NEDD8-activating enzyme, can be developed as a novel class of anticancer agents by inhibiting SCF E3 ligase via removal of cullin neddylation. Finally, we discuss under future perspective how basic research on SCF biology will direct the drug discovery efforts surrounding this target.
Keywords: Ubiquitin-proteasome system, SCF E3 ubiquitin ligase, anticancer target, drug discovery, neddylation, cullins, F-box proteins, RING ligases
INTRODUCTION
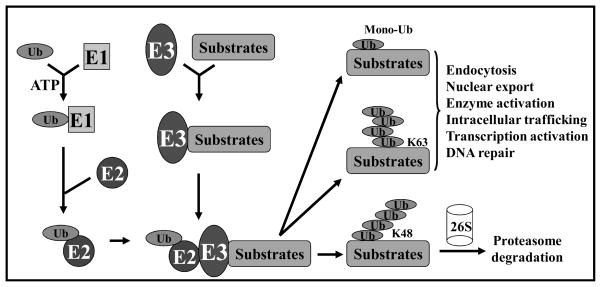
The ubiquitin-proteasome system (UPS) regulates many biological processes through timely degradation of diverse cellular proteins. It, therefore, plays an essential role in maintaining homeostasis and in response to environmental stimuli [1,2]. UPS-targeted protein degradation requires substrate ubiquitination, which is a multi-step enzymatic process catalyzed by a cascade of enzymes, including ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin ligase E3. While E1 and E2 activate and transfer ubiquitin in the reaction, E3 recognizes the substrate and catalyzes the covalent attachment of ubiquitin to the substrate [3]. Multiple runs of this reaction result in polyubiquitination of substrate. The fate of ubiquitinated proteins is determined, however, by the nature of ubiquitin attachment and the type of isopeptide linkage of the polyubiquitin chain. While the K48-linked polyubiquitination predominantly targets protein for degradation after being recognized by the proteasome, the K63-linked polyubiquitination and mono-ubiquitination normally alters protein function, cellular localization, enzyme activity, DNA repair, or interaction with other proteins [4–6]. Fig. (1) illustrates the ubiquitination cascade reactions and three potential fates of ubiquitinated proteins.
Fig. 1. Ubiquitination cascade in protein degradation and functional regulation.
Ubiquitin is first activated by ubiquitin activating enzyme, E1 and transferred to ubiquitin conjugating enzyme E2, and finally transferred to substrates by ubiquitin ligase E3, which recognizes the substrate and catalyzes the ubiquitin transfer. A single run of this ubiquitination reaction results in monoubiquitination of substrate protein with potential functional changes. Multiple runs of such a reaction lead to polyubiquitinatin of a substrate, which is either recognized by 26S proteasome for targeted degradation, if ubiquitin tag is in a K48 linkage, or subjected to activation, if it is in a K63 linkage.
The SCF multisubunit E3 ligase complex, consisting of Skp-1, Cullins, F-Box proteins and RBX/ROC RING finger proteins [7,8], is the largest family of ubiquitin ligases that promote the degradation of about 20% of UPS-regulated proteins [9], including cell cycle regulatory proteins, transcription factors, oncoproteins and tumor suppressors among others [10–15] (Table 1). The crystal structure of SCF-RBX complex revealed that Cul-1 acts as a scaffold that binds at its N-terminus the Skp-1 and F-box protein and at its C-terminus the RING protein RBX1 [16]. It is well established that the substrate specificity of SCF complex is determined by the F box proteins that bind to Skp1 and Cullins through its F-box domain and to substrates through its WD40 or leucine rich domains [16], whereas the core SCF E3 ubiquitin ligase is a complex of Cullins-RBX/ROC, in which RBX binds to E2 and facilitates ubiquitin transfer from E2 to substrates [17]. Furthermore, the activity of SCF E3 ubiquitin ligases requires cullin neddylation, which disrupts inhibitory binding of cullin by CAND1 [18–21]. In the human genome, there are 69 F-box proteins, including WD40 domain containing FBXWs, leucine-rich repeats-containing FBXLs and other diverse domains-containing FBXOs, seven cullins (Cul-1, -2, 3, 4A, 4B, -5, and -7) and two RING proteins, RBX1/ROC1 and RBX2/ROC2, also known as SAG (Sensitive to Apoptosis Gene) [22–26]. Cullin-based assembly of SCF E3 ligase subunits can be classified into four categories: Cul1-Skp1-F-box, Cul2/5-Elongins-B/C-VHL/SOCS box, Cul3-BTB, and Cul4A/B-DDB1-DWD [27], making SCF/CRL the largest family of E3 ubiqutin ligases, responsible for the degradation of ~20% of all proteins subjected to proteasomal degradation [9].
Table 1.
SCF E3 Ubiquitin Ligases and their Substrates
| Name | Substrates | References |
|---|---|---|
| 1) RBX1/Cullin-1/SKP1/F-Box proteins | Many, for near complete list, see cited references | [11,14,15] |
| a) Skp2 | p21, p27, p57, p130, Cyclins A/D1/E, E2F1, Orc1, Cdt1, c-Myc, B-Myb, Foxo1, Rassf1, Smad4, Brca2, Cdk9 | [10,15] |
| b) β-TrCP | IκB, β-Catenin, Emi1, Cdc25A/B, Weel, BimEL, Mcl1, p100, p105, p53, p63, Pdcd4, Per1/2, procaspase-3, Rest, Smad3, Snail, Stat-1, Weel, ATF4, Claspin, Cyclin D1, Dlg, H-ras | [10,15,52] |
| c) Fbxw7 | Aurora-A, c-Jun, c-Myb, c-Myc, Cyclin E1/2, mTOR, Notch1/4, Pgc1, Src3, Presenilin, SREBP | [10,15] |
| 2) RBX1/Cullin-2/Elongin BC/VHL | HIF-α, TEL-JAK2 | [115,116] |
| 3) RBX1/Cullin-3/BTB-domain proteins | MEI-1, Dishevelled (Dsh), Nrf2, RhoBTB2, topoisomerase I-DNA complex, caspase 8, DAPK | [117–125] |
| 4) RBX1/Cullin-4A/DDB1 | Cdt1, p21, Smug, Histone H2A/H3/H4, XPC, DDB2, Hoxa9, Chk1, β-Catenin, p53, TSC2, c-Jun, Merlin, Stat1/2/3, UNG2. | [27,126–134] |
| 5) RBX1/Cullin-5/elongin BC/BC-box proteins/SOCS | Disabled-1 (Dab1) | [135] |
| 6) RBX1/Cullin-7/SKP1/Fbw8 | Insulin receptor substrate 1 (IRS-1) | [136] |
SCF COMPLEX E3 UBIQUITIN LIGASES IN CANCER AND AS ANTICANCER TARGETS
The majority of SCF E3 ligase substrates are involved in regulation of cell cycle progression, gene transcription, signal transduction and DNA replication among others [11,12,14]. Through targeted degradation of these substrates, SCF E3 ligases regulate many biological processes. Accumulated evidence strongly suggests that abnormal regulation of SCF E3 ubiquitin ligases contributes to uncontrolled proliferation, genomic instability, and cancer [11]. Among the components of SCF, some are oncogenes (e.g. Skp2) that promote degradation of tumor suppressors and are amplified and/or overexpressed in human cancers, whereas others are tumor-suppressors (e.g. Fbxw7) that target the degradation of oncoproteins and are mutated in human cancers [28–30]. Some deregulated components of SCF E3 ligase in human cancer are listed in Table 2 and discussed below.
Table 2.
Components of the SCF E3 Ubiqutin Ligase with Oncogenic or Tumor Suppressive Activity
| Name | Alterations in cancer | Activity | Major functions | References |
|---|---|---|---|---|
| ROC1/RBX1 | Overexpression | Oncogenic | Cell cycle progression, DNA damage response | [37] |
| SAG/RBX2 | Overexpression | Oncogenic | Anti-apoptosis | [22,55] |
| Skp2 | Amplification, Overexpression | Oncogenic | Cell cycle progression | [28,29] |
| β-TrCP | Overexpression | Oncogenic | Anti-apoptosis, protein translation | [28,29] |
| Fbxw7 | Mutation, loss of expression | Tumor suppressive | Anti-proliferation, Signaling transduction | [30] |
| Cul-1 | Overexpression | Oncogenic | Cell cycle progression | [85] |
| Cul4A | Amplification, Overexpression | Oncogenic | DNA repair; Anti-apoptosis | [89,92,93] |
| Cul5 | Loss of expression | Tumor suppressive | Blockage of Src activity, Anti-proliferation | [87,88] |
RING-Finger Proteins
Two family members of the RING component of SCF E3 ligase in human and mouse are RBX1/ROC1 and RBX2/ROC2/SAG [22–25]. Both members contain a functional RING domain at the carboxyl terminus and are evolutionally conserved with a similar tissue expression pattern [8]. Under overexpressed conditions, both proteins bind to any one of six members of the cullin family (Cul 1-3, Cul4A, B and Cul-5) [24] and have an in vitro E3 ubiquitin ligase activity [31,32]. Either family member can fully rescue yeast death phenotype caused by deletion of Hrt1, a yeast orthologue of Rbx1/Roc1 [24,31,33]. A potential difference between the two members is that RBX1 is constitutively expressed and prefers to bind with Cul2/VHL, whereas RBX2/ROC2/SAG is stress-inducible and preferably binds to Cul-5/SOCS [34,35]. Our recent mouse knockout study revealed that these two members are functionally non-redundant. Under wild type Rbx2 background, Rbx1 deletion caused early embryonic lethality at E7.5 as the result of proliferation defects [36], whereas Sag knockout in the wild type Rbx1 background also caused embryonic lethality at the later stage (E11.5-12.5), associated with cardiovascular defects (manuscript submitted for publication).
RBX1/ROC1
Our recently study showed that compared to normal tissues, RBX1/ROC1 is overexpressed in diverse human primary cancers, particularly in lung cancer. SiRNA silencing of ROC1 triggered the DNA damage response and sequentially induced G2/M cell cycle arrest, senescence and apoptosis in a p53-independent manner, leading to suppression of cancer cell growth [37] (Fig. 2). The underlying mechanism for ROC1 silencing-induced senescence is likely attributable to accumulation of DNA replication licensing proteins (such as Cdt-1 and Orc1), known to be SCF E3 ligase substrates [15,38–41], which trigger the DNA damage response and senescence [42–44]. Thus, RBX1 is a cancer cell survival protein whose inhibition triggers various cell death pathways, eventually leading to cancer cell killing.
Fig. 2. Targeting the SCF E3 ubiquitin ligase to trigger multiple cell killing pathways.
SCF E3 ubiquitin ligase can be inactivated by targeting its oncogenic components, including RBX1/RBX2, Cul-4A, or Skp2 via siRNA silencing approach or by pharmaceutical inhibition of cullin neddylation with a NAE inhibitor, MLN4924. Inactivation of SCF E3 ligase causes the accumulation of its substrates which suppress cancer cell growth by triggering multiple cancer cell killing pathways, including apoptosis, senescence and autophagy with the mechanisms subjected to future investigation.
RBX2/ROC2/SAG
RBX2/ROC2/SAG is the second member of the RING component of SCF E3 ligases, which was originally cloned as a redox inducible antioxidant protein in our laboratory [22]. As an antioxidant, SAG suppresses apoptosis induced by many stimuli, including redox [22,45], tumor promoter, TPA [34], nitric oxide [46], ischemia/reoxygenation [47], neurotoxins [48], heat-shock [49] and UV-irradiation [50]. When complexed with other components of SCF, SAG exerts E3 ubiquitin ligase activity [31] and promotes the degradation of p27, c-Jun, procaspase-3, IκBα, HIF-1α, and Noxa, thus regulating cell proliferation, apoptosis, and skin carcinogenesis [34,51–55]. Significantly, SAG is overexpressed in multiple human tumor tissues, and patients with SAG overexpression have a poor prognosis [55–57]. SAG siRNA silencing selectively inhibited cancer cell proliferation via apoptosis induction, suppressed in vivo tumor growth and sensitized cancer cells to chemotherapeutic drugs and radiation [52,55], suggesting its potential as an anti-cancer target (Fig. 2).
F-Box Proteins
F-box proteins are the substrate-recognizing subunits of SCF E3 ligase which determine the substrate specificity of SCF. A single F-box protein can recognize and target multiple substrates (e.g. Skp2 targets p27, p21, p57), whereas the same substrate can be recognized and targeted by different F-box proteins (e.g. cyclin E targeted by both Skp2 and Fbxw7). More interestingly, a single F-box protein can target the degradation of several substrates with opposite biological functions (e.g. Skp2 targets p21/p27 as well as cyclin A/D1/E) [15]. Thus, when or whether a particular substrate is targeted for degradation by a given F-box protein will likely be cell context dependent, leading to different biological consequences. Among ~70 F-box proteins in the human genome, only three are well studied: oncogenic Skp2, tumor suppressive Fbxw7, and β-TrCP, which could be tumor suppressive as well as oncogenic in a substrate dependent manner [28,30]. Here we will only focus on Skp2 and β-TrCP as potential cancer targets.
Skp2
Skp2 recognizes and promotes the degradation of several negative cell cycle regulators, including p27, p21, p130 and p57 [11,12,14]. Skp2 is overexpressed in many human cancer types [58] with associated p27 decrease and poor prognosis, seen in gastric cancer [59,60], colon cancer [61,62], and breast cancer [63–65]. Tissue specific expression of Skp2 in mouse prostate gland caused hyperplasia, dysplasia and low-grade carcinoma [66], whereas targeted expression of Skp2 in the T-lymphoid lineage co-operated with activated N-Ras to induce T cell lymphomas with a short latent period and high penetrance [67]. Furthermore, a knock-in mouse model showed a crucial role of Skp2 dependent degradation of p27 for the progress of colon adenomas to carcinoma [68]. Interestingly, in a mouse knockout model, although Skp2 disruption on its own does not induce cellular senescence, Skp2-null environment facilitates tumor-suppressive senescence response upon inactivation of tumor suppressor genes or aberrant proto-oncogenic signals [69]. Consistently, down-regulation of Skp2 using an anti-sense oligonucleotide or siRNA silencing inhibited growth of melanoma [70], oral cancer cells [71], glioblastoma cells [72], and lung cancer cells [73–75]. Thus, pharmacological inhibitors of the Skp2 pathway would be of therapeutic value for cancer treatment.
β-TrCP
The F-box protein, β-TrCP1/2, is a substrate recognizing component of SCF E3 ubiquitin ligases that promote ubiquitination and degradation of various proteins. In some tissues, β-TrCP is characterized as an oncoprotein for targeted degradation of tumor suppressors, such as IκB [14,76] a negative regulator of NFκB [77], PDCD4 [78], a protein translation suppressor via inhibiting eIF4A [28] and BimEL1 [79], a proapoptotic protein. In a transgenic mouse model, targeted β-TrCP1 expression in mammary gland promotes mammary tumor formation via activating NFκB [80], whereas targeted β-TrCP1 expression in intestine, liver and kidney caused an increased incidence of tumor formation in these organs [81]. Consistently, β-TrCP is overexpressed in human breast cancer cell lines and primary tumors, and β-TrCP inhibition sensitizes breast cancer cells to chemotherapies [82]. Similarly, overexpression of β-TrCP increased NFκB activity and chemo-resistance, whereas siRNA silencing of β-TrCP reduced NFκB activation and chemo-resistance in pancreatic cancer cells [83] and sensitized cervical cancer cells to apoptosis induced by etoposide or TRAIL [52]. Thus, disruption of the binding between β-TrCP and its tumor suppressor substrates would be an effective and selective targeting approach.
Cullins
Cullins are scaffold proteins which assembly with other components of SCF into four functionally distinct E3 ubiqutin ligases [27]. So far, seven cullin family members (Cullin 1, 2, 3, 4A, 4B, 5, and 7) have been identified [84]. Among all cullins, Cul-1 is overexpressed in 40% of lung cancers, with active neddylated forms specifically expressed in high-grade neuroendocrine lung tumor tissues [85], whereas Cul-2 frameshift mutations were detected in two out of 41 colon cancers [86]. Cul-5 is a putative tumor suppressor that blocks Src activity [87] and inhibits breast cancer cell growth upon overexpression [88]. Most cullin-related studies focused on Cul-4A, which is overexpressed in a number of human cancers, including breast cancers [89,90] with poor prognosis [91], hepatocellular carcinomas [92], and mesotheliomas [93]. While Cul-4A overexpression was associated with tumor proliferation and cell cycle progression in breast cancer cells [90], knockdown of Cul4A by siRNA caused accumulation of p21 and p27 and G1 cell cycle arrest, leading to growth suppression of mesothelioma cells [93]. In a recent conditional knockout model, skin specific Cul4A disruption dramatically increased resistance to UV-induced skin carcinogenesis [94], which highlights a potential protection from skin cancer caused by UV exposure using pharmacological Cul4A inhibitor. Taken together, these findings indicate that Cul4A amplification and overexpression plays an oncogenic role in carcinogenesis, and that Cul4A could be an attractive target for anticancer therapies. However, since Cul-4A has many protein substrates [27] that regulate a variety of cellular functions, the challenge will be how to selectively target its oncogenic substrates.
TARGETING SCF E3 UBIQUITIN LIGASES FOR ANTICANCER THERAPY
Bortezomib (also known as Velcade or PS-341) is the first (and only) in class of general proteasome inhibitor, approved by the FDA for the treatment of relapsed/refractory multiple myeloma and mantle cell lymphoma [95,96]. Although it has demonstrated a certain degree of tumor cell selectivity that provides a therapeutic window, the drug is, in general, cytotoxic due to overall inhibition of proteolysis of a wide array of cellular proteins [96,97]. In contrast to general proteasome inhibitors, a specific E3 ligase inhibitor would selectively stabilize a specific set of cellular proteins regulated by this E3, thus avoiding some undesired effects on other cellular proteins. This would, therefore, achieve a high level of specificity with less associated toxicity [29,98,99]. Since the SCF E3 ubiquitin ligase is abnormally activated in many human cancers, contributing to uncontrolled proliferation and genomic instability [11], this E3 ubiquitin ligase or some of its components are being considered and validated as promising anticancer targets [98,99].
In theory, the following approaches can be used to screen for the inhibitors of SCF E3 ubiquitin ligase or its components. The first is to disrupt the interaction between RBX1/2 with E2 ubiquitin conjugation enzyme to prevent ubiquitin transfer to substrates. So far no such inhibitor has been reported. The second approach is to disrupt interaction between SCF components, such as disruption of Cks1-Skp2 interaction [100,101] for p27 accumulation, or to disrupt interaction between F-box proteins and their tumor suppressive substrates (e.g. β-TrCP vs. IκB) [102]. The third approach, which is not for ligase inhibition per se, but does prevent substrate degradation, is to inhibit a kinase that phosphorylates a particular SCF substrate on a degron/destruction motif to prevent its binding to a corresponding F-box protein. It has been well-established that substrate phosphorylation is a prerequisite for F-box protein binding and subsequent degradation [11,12,15]. This approach, however, is currently being used only as a research option for understanding how the substrate degradation by a particular SCF E3 ligase is regulated. The fourth is to use Protacs (protein-targeting chimeric molecule 1), the artificial chimeric molecules that recruit selected protein substrates to SCF complex for ubiquitination and degradation [103]. A high through-put screen could be set up to identify potential inhibitors. The fifth approach is to screen for small molecule inhibitors of SCF E3 ligase. Although a number of high-throughput screening methods have been established and optimized to screen for small molecule inhibitors of single peptide E3 ligase [104], none of them can be readily converted to screen for inhibitors of multi-component SCF E3 ligases. Recently, a small molecule inhibitor, CpdA, was identified from a biochemical-based screening using in vitro transcribed/translated and 35S-labeled p27, and an in vitro–reconstituted system incorporating purified cyclin E/Cdk2, Skp2, Skp1, Cul1, Roc1, and HeLa cell extract. CpdA was found to prevent incorporation of Skp2 into the SCF E3 ligase and to induce G1 arrest as well as p27-dependent cell killing via induction of autophagy. Furthermore, CpdA sensitized multiple myeloma to a number of anticancer drugs, including dexamethasone, doxorubicin, melphalan, and bortezomib [105].
TARGETING CULLIN NEDDYLATION AS AN ALTERNATIVE APPROACH FOR INACTIVATION OF SCF E3 LIGASES
Neddylation, a process of addition of ubiquitin-like protein NEDD8 to target proteins, is a novel type of post-translational modifications [106]. Neddylation is mediated by NEDD8 activating enzyme E1 (NAE), NEDD8 conjugating enzyme E2 (Ubc12) and NEDD8-E3 ligase, which consecutively activate and transfer NEDD8 to substrates. The cullin family of proteins has been well characterized as the major substrates for neddylation [107,108], and activity of SCF E3 ubiquitin ligases requires cullin neddylation, which disrupts inhibitory binding by CAND1 [18–21]. Thus, a clever approach, employed by Soucy et al. [9], is to screen for NAE inhibitors with expected inhibitory activity against SCF E3 ubiquitin ligases. Indeed, MLN4924, the first class of such inhibitors was identified via a HTS screen and did exactly what was expected: selective inhibition of NAE and SCF E3, leading to accumulation of SCF E3 ligase substrates to trigger cell death [9,109].
Mechanistically, MLN4924 binds to NAE at the active site to create a covalent Nedd8-MLN4924 adduct, which can resemble NEDD8 adenylate, but cannot be further utilized in subsequent intraenzyme reactions. This modification inhibits NAE activity with subsequent abrogation of cullin neddylation [110]. By doing so, MLN4924 inhibits activity of SCF E3 ligases and causes accumulation of a number of SCF E3 substrates, resulting in an abnormal cell cycle profile with accumulation of anueploid cell populations and induction of apoptosis [9]. In vivo xenograft assays also showed that MLN4924 slows tumor growth and was well tolerated in mice at various doses and treatment regimens [9], demonstrating a cancer cell selective killing. Most recently, MLN4924 showed potent activity against acute myeloid leukemia by inducing apoptosis [111,112] and against prostate cancer cell in vivo growth by triggering cellular senescence [69]. With all these promises, MLN4924 has advanced to several Phase I clinical trials for solid tumors and hematological malignancies [109].
FUTURE PROSPECTIVES
Compared to the general proteasome inhibitor Bortezomib, which blocks the entire UPS-mediated protein degradation, drugs that target a particular E3 ligase are expected to have better selectivity with less associated toxicity. Indeed, SCF complex E3 ligase inhibitor MLN4924, which blocks ~20% of all cellular proteins subjected to proteasomal degradation, is well-tolerated in mice [9]. Newly released Phase I clinical data showed that patients do develop some symptoms, such as fatigue, nausea, myalgia with increased transaminases1, which could be associated with an overall inhibition of SCF E3 ligase as well as of other proteins whose function is subjected to neddylation regulation [106]. The following areas of basic research on SCF E3 ligase would further our mechanistic understanding of this multi-functional ligase with the ultimate goal to develop highly selective and specific inhibitors against a subset of pathways regulated by this E3 ligase.
Characterization of F-Box Proteins and their Corresponding Substrates
Among a total of ~70 F-box proteins in the human genome, which serve as the receptors for substrate recognition [26], only few are well characterized [28,30]. A recent global protein stability profiling study has identified over 350 potential SCF E3 substrates which are involved in regulation of cell cycle progression, apoptosis and cell signaling [113,114]. A complete characterization of the F-box proteins and identification of their corresponding substrates with elucidation of associated biological functions will provide an opportunity to screen for drugs that specifically target a given receptor-substrate interaction for potential activation or inactivation of a particular pathway.
Mechanistic Understanding of the Death Pathway Triggered by Inactivation of the SCF E3 Ligase Complex
Our recent work and unpublished data showed that inactivation of SCF E3 ubiquitin ligase complex via siRNA silencing of ROC1/RBX1 or treatment with MLN4924 caused accumulation of SCF E3 substrates, followed by cancer cell growth arrest and killing via apoptosis, senescence, and autophagy [37] (Fig. 2). How these death pathways are mediated, possibly by a subset of accumulated substrates, and whether different types of cell death occur sequentially or in parallel are subjects for future study. Mechanistic understanding of how these death pathways are triggered will help to design screening strategies to target a particular pathway with an aim to reduce normal cell toxicity.
Functional Differentiation of RBX1-SCF or RBX2-SCF E3 Ligase
SCF E3 ubiquitin ligases contain only two RING family members, RBX1/ROC1 or RBX2/ROC2/SAG. Our recent knockout study showed that they are functionally non-redundant. During mouse embryogenesis, Rbx1 controls cell proliferation at the early stage [36], whereas Rbx2/Sag regulates vascular development at the later stage (manuscript submitted). Our cell culture work also showed that siRNA silencing of RBX1 induced cell death via apoptosis, senescence, and autophagy [37], whereas siRNA silencing of RBX2/SAG only triggered apoptosis [55]. These studies suggest that RBX1-SCF and RBX2-SCF E3 ligases may selectively target non-overlapping as well as overlapping sets of protein substrates for degradation. Functional characterization of these two types of E3 ligases and their associated substrates would help to design a unique screening strategy for selectively targeting either one of them.
In summary, SCF E3 ubiquitin ligase appears to be a promising anti-cancer target. MLN4924, a small molecule inhibitor of SCF, holds promise for further clinical development as a novel class of anticancer agents. As our understanding of this ligase progresses, more specific drugs targeting a particular subset of SCF controlled pathways will be discovered with expectation of highly cancer cell selectivity with minimal normal cell toxicity.
Acknowledgments
We apologize for not being able to cite all related original articles, but instead, citing few excellent review articles, due to the space limitation. We thank two anonymous reviewers for their thoughtful suggestions. This work is supported by NCI grants (CA111554 and CA118762) to Yi Sun and startup funding from Fudan University in China and the Chinese National Nature Science Foundation (31071204) to Lijun Jia.
ABBREVIATIONS
- BTB
Bric-a-brac/Tramtrack/Broad domain
- β-TrCP
β-Transducin Repeat-Containing Protein
- CAND1
Cullin-Associated and Neddylation-Dissociate-1
- Cks1
Cdc kinase subunit 1
- CpdA
Compound A
- CRL
Cullin-RING ubiquitin Ligases
- DDB
Damaged DNA-Binding Protein
- Fbxw7
F-box and WD repeat domain–containing 7
- HTS
High-Throughput Screen
- NAE
Nedd8-Activating Enzyme
- NEDD
Neural precursor Cell-expressed Developmentally Down-regulated
- RBX1
Ring box protein-1
- ROC
Regulator of Cullins
- SAG
Sensitive to Apoptosis
- SCF
Skp1, Cullins, F-box proteins
- siRNA
small interfering RNA
- SOCS
Suppressors of cytokine signalling
- Skp1
S-phase kinase associated protein 1
- Skp2
S-phase kinase associated protein 2
- TRAIL
TNF-Related Apoptosis-Inducing Ligand
- UPS
Ubiquitin-proteasome system
- VHL
Von Hippel-Lindau
Footnotes
Smith, P. G. Inhibition of NEDD8-activating enzyme for cancer treatment: preclinal validation to clinic application of MLN4924. AACR 101st Annual Meeting 2010. 2010, Pg 258.
References
- 1.Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickart CM. Ubiquitin in chains. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 6.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635–650. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- 9.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 13.Jia L, Sun Y. RBX1/ROC1-SCF E3 ubiquitin ligase is required for mouse embryogenesis and cancer cell survival. Cell Div. 2009;4:16. doi: 10.1186/1747-1028-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 15.Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F box proteins II. Cell. 2009;137:1358, 1358, e1351. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 16.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 17.Wu K, Fuchs SY, Chen A, Tan P, Gomez C, Ronai Z, Pan ZQ. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 24.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 25.Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IkBa. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 26.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 30.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 31.Swaroop M, Wang Y, Miller P, Duan H, Jatkoe T, Madore S, Sun Y. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: Chip profiling implicates its role in cell cycle regulation. Oncogene. 2000;19:2855–2866. doi: 10.1038/sj.onc.1203635. [DOI] [PubMed] [Google Scholar]
- 32.Furukawa M, Ohta T, Xiong Y. Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J Biol Chem. 2002;277:15758–15765. doi: 10.1074/jbc.M108565200. [DOI] [PubMed] [Google Scholar]
- 33.Seol JH, Feldman RMR, Zachariae WZ, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Shevchenko A, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67:3616–3625. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- 35.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan M, Davis SW, Saunders TL, Zhu Y, Sun Y. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc Natl Acad Sci U S A. 2009;106:6203–6208. doi: 10.1073/pnas.0812425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854–30858. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- 39.Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem. 2006;281:3753–3756. doi: 10.1074/jbc.C500464200. [DOI] [PubMed] [Google Scholar]
- 41.Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, Lygerou Z, Nishimoto T. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 43.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d’Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 44.Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y. Alteration of SAG mRNA in human cancer cell lines: Requirement for the RING finger domain for apoptosis protection. Carcinogenesis. 1999;20:1899–1903. doi: 10.1093/carcin/20.10.1899. [DOI] [PubMed] [Google Scholar]
- 46.Yang ES, Park JW. Regulation of nitric oxide-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Res. 2006;40:279–284. doi: 10.1080/10715760500511500. [DOI] [PubMed] [Google Scholar]
- 47.Chanalaris A, Sun Y, Latchman DS, Stephanou A. SAG attenuates apoptotic cell death caused by simulated ischaemia/reoxygenation in rat cardiomyocytes. J Mol Cell Cardiol. 2003;35:257–264. doi: 10.1016/s0022-2828(03)00003-8. [DOI] [PubMed] [Google Scholar]
- 48.Kim SY, Kim MY, Mo JS, Park JW, Park HS. SAG protects human neuroblastoma SH-SY5Y cells against 1-methyl-4-phenylpyridinium ion (MPP+)-induced cytotoxicity via the down-regulation of ROS generation and JNK signaling. Neurosci Lett. 2007;413:132–136. doi: 10.1016/j.neulet.2006.11.074. [DOI] [PubMed] [Google Scholar]
- 49.Lee SJ, Yang ES, Kim SY, Shin SW, Park JW. Regulation of heat shock-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Biol Med. 2008;45:167–176. doi: 10.1016/j.freeradbiomed.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 50.He H, Gu Q, Zheng M, Normolle D, Sun Y. SAG/ROC2/RBX2 E3 ligase promotes UVB-induced skin hyperplasia, but not skin tumors, by simultaneously targeting c-Jun/AP-1 and p27. Carcinogenesis. 2008;29:858–865. doi: 10.1093/carcin/bgn021. [DOI] [PubMed] [Google Scholar]
- 51.Duan H, Tsvetkov LM, Liu Y, Song Y, Swaroop M, Wen R, Kung HF, Zhang H, Sun Y. Promotion of S-phase entry and cell growth under serum starvation by SAG/ROC2/Rbx2/Hrt2, an E3 ubiquitin ligase component: association with inhibition of p27 accumulation. Mol Carcinog. 2001;30:37–46. doi: 10.1002/1098-2744(200101)30:1<37::aid-mc1011>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 52.Tan M, Gallegos JR, Gu Q, Huang Y, Li J, Jin Y, Lu H, Sun Y. SAG/ROC-SCFbeta-TrCP E3 ubiquitin ligase promotes procaspase-3 degradation as a mechanism of apoptosis protection. Neoplasia. 2006;8:1042–1054. doi: 10.1593/neo.06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1alpha ubiquitination and degradation. Oncogene. 2008;27:1404–1411. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- 54.Gu Q, Bowden TG, Normolle D, Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage dependent targeting of c-Jun/AP1 and IkB/NF-kB. J Cell Biol. 2007;178:1009–1023. doi: 10.1083/jcb.200612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y, Duan H, Sun Y. Elevated expression of SAG/ROC2/Rbx2/Hrt2 in human colon carcinomas: SAG does not induce neoplastic transformation, but its antisense transfection inhibits tumor cell growth. Mol Carcinog. 2001;30:62–70. [PubMed] [Google Scholar]
- 57.Sasaki H, Yukiue H, Kobayashi Y, Moriyama S, Nakashima Y, Kaji M, Fukai I, Kiriyama M, Yamakawa Y, Fujii Y. Expression of the sensitive to apoptosis gene, SAG, as a prognostic marker in nonsmall cell lung cancer. Int J Cancer. 2001;95:375–377. doi: 10.1002/1097-0215(20011120)95:6<375::aid-ijc1066>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 58.Nalepa G, Wade Harper J. Therapeutic anti-cancer targets upstream of the proteasome. Cancer Treat Rev. 2003;29(Suppl 1):49–57. doi: 10.1016/s0305-7372(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 59.Masuda TA, Inoue H, Sonoda H, Mine S, Yoshikawa Y, Nakayama K, Mori M. Clinical and biological significance of S-phase kinase-associated protein 2 (Skp2) gene expression in gastric carcinoma: modulation of malignant phenotype by Skp2 overexpression, possibly via p27 proteolysis. Cancer Res. 2002;62:3819–3825. [PubMed] [Google Scholar]
- 60.Masuda TA, Inoue H, Nishida K, Sonoda H, Yoshikawa Y, Kakeji Y, Utsunomiya T, Mori M. Cyclin-dependent kinase 1 gene expression is associated with poor prognosis in gastric carcinoma. Clin Cancer Res. 2003;9:5693–5698. [PubMed] [Google Scholar]
- 61.Hershko DD, Shapira M. Prognostic role of p27Kip1 deregulation in colorectal cancer. Cancer. 2006;107:668–675. doi: 10.1002/cncr.22073. [DOI] [PubMed] [Google Scholar]
- 62.Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–1346. doi: 10.1002/cncr.20917. [DOI] [PubMed] [Google Scholar]
- 63.Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Invest. 2002;110:633–641. doi: 10.1172/JCI15795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Forozan F, Mahlamaki EH, Monni O, Chen Y, Veldman R, Jiang Y, Gooden GC, Ethier SP, Kallioniemi A, Kallioniemi OP. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res. 2000;60:4519–4525. [PubMed] [Google Scholar]
- 65.Sonoda H, Inoue H, Ogawa K, Utsunomiya T, Masuda TA, Mori M. Significance of skp2 expression in primary breast cancer. Clin Cancer Res. 2006;12:1215–1220. doi: 10.1158/1078-0432.CCR-05-1709. [DOI] [PubMed] [Google Scholar]
- 66.Shim EH, Johnson L, Noh HL, Kim YJ, Sun H, Zeiss C, Zhang H. Expression of the F-Box Protein SKP2 Induces Hyperplasia, Dysplasia, and Low-Grade Carcinoma in the Mouse Prostate. Cancer Res. 2003;63:1583–1588. [PubMed] [Google Scholar]
- 67.Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M. Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci U S A. 2001;98:2515–2520. doi: 10.1073/pnas.041475098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Timmerbeul I, Garrett-Engele CM, Kossatz U, Chen X, Firpo E, Grunwald V, Kamino K, Wilkens L, Lehmann U, Buer J, Geffers R, Kubicka S, Manns MP, Porter PL, Roberts JM, Malek NP. Testing the importance of p27 degradation by the SCFskp2 pathway in murine models of lung and colon cancer. Proc Natl Acad Sci U S A. 2006;103:14009–14014. doi: 10.1073/pnas.0606316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katagiri Y, Hozumi Y, Kondo S. Knockdown of Skp2 by siRNA inhibits melanoma cell growth in vitro and in vivo. J Dermatol Sci. 2006;42(3):215–224. doi: 10.1016/j.jdermsci.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 71.Kudo Y, Kitajima S, Ogawa I, Kitagawa M, Miyauchi M, Takata T. Small interfering RNA targeting of S phase kinase-interacting protein 2 inhibits cell growth of oral cancer cells by inhibiting p27 degradation. Mol Cancer Ther. 2005;4:471–476. doi: 10.1158/1535-7163.MCT-04-0232. [DOI] [PubMed] [Google Scholar]
- 72.Lee SH, McCormick F. Downregulation of Skp2 and p27/Kip1 synergistically induces apoptosis in T98G glioblastoma cells. J Mol Med. 2005;83:296–307. doi: 10.1007/s00109-004-0611-7. [DOI] [PubMed] [Google Scholar]
- 73.Sumimoto H, Yamagata S, Shimizu A, Miyoshi H, Mizuguchi H, Hayakawa T, Miyagishi M, Taira K, Kawakami Y. Gene therapy for human small-cell lung carcinoma by inactivation of Skp-2 with virally mediated RNA interference. Gene Ther. 2005;12:95–100. doi: 10.1038/sj.gt.3302391. [DOI] [PubMed] [Google Scholar]
- 74.Jiang F, Caraway NP, Li R, Katz RL. RNA silencing of S-phase kinase-interacting protein 2 inhibits proliferation and centrosome amplification in lung cancer cells. Oncogene. 2005;24:3409–3418. doi: 10.1038/sj.onc.1208459. [DOI] [PubMed] [Google Scholar]
- 75.Yokoi S, Yasui K, Saito-Ohara F, Koshikawa K, Iizasa T, Fujisawa T, Terasaki T, Horii A, Takahashi T, Hirohashi S, Inazawa J. A novel target gene, SKP2, within the 5p13 amplicon that is frequently detected in small cell lung cancers. Am J Pathol. 2002;161:207–216. doi: 10.1016/S0002-9440(10)64172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 78.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 79.Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton J, Pagano M. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kudo Y, Guardavaccaro D, Santamaria PG, Koyama-Nasu R, Latres E, Bronson R, Yamasaki L, Pagano M. Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Mol Cell Biol. 2004;24:8184–8194. doi: 10.1128/MCB.24.18.8184-8194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Belaidouni N, Peuchmaur M, Perret C, Florentin A, Benarous R, Besnard-Guerin C. Overexpression of human beta TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene. 2005;24:2271–2276. doi: 10.1038/sj.onc.1208418. [DOI] [PubMed] [Google Scholar]
- 82.Tang W, Li Y, Yu D, Thomas-Tikhonenko A, Spiegelman VS, Fuchs SY. Targeting beta-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 2005;65:1904–1908. doi: 10.1158/0008-5472.CAN-04-2597. [DOI] [PubMed] [Google Scholar]
- 83.Muerkoster S, Arlt A, Sipos B, Witt M, Grossmann M, Kloppel G, Kalthoff H, Folsch UR, Schafer H. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65:1316–1324. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- 84.Marin I. Diversification of the cullin family. BMC Evol Biol. 2009;9:267. doi: 10.1186/1471-2148-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salon C, Brambilla E, Brambilla C, Lantuejoul S, Gazzeri S, Eymin B. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J Pathol. 2007;213:303–310. doi: 10.1002/path.2223. [DOI] [PubMed] [Google Scholar]
- 86.Park SW, Chung NG, Hur SY, Kim HS, Yoo NJ, Lee SH. Mutational analysis of hypoxia-related genes HIF1alpha and CUL2 in common human cancers. APMIS. 2009;117:880–885. doi: 10.1111/j.1600-0463.2009.02550.x. [DOI] [PubMed] [Google Scholar]
- 87.Laszlo GS, Cooper JA. Restriction of Src activity by Cullin-5. Curr Biol. 2009;19:157–162. doi: 10.1016/j.cub.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson AE, Le IP, Buchwalter A, Burnatowska-Hledin MA. Estrogen-dependent growth and estrogen receptor (ER)-alpha concentration in T47D breast cancer cells are inhibited by VACM-1, a cul 5 gene. Mol Cell Biochem. 2007;301:13–20. doi: 10.1007/s11010-006-9392-3. [DOI] [PubMed] [Google Scholar]
- 89.Chen LC, Manjeshwar S, Lu Y, Moore D, Ljung BM, Kuo WL, Dairkee SH, Wernick M, Collins C, Smith HS. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- 90.Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, Palacios J, Nathanson KL, Garcia MJ, Benitez J. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009;11:R86. doi: 10.1186/bcr2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schindl M, Gnant M, Schoppmann SF, Horvat R, Birner P. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27:949–952. [PubMed] [Google Scholar]
- 92.Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, Emi M, Inazawa J. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476–1484. doi: 10.1053/jhep.2002.33683. [DOI] [PubMed] [Google Scholar]
- 93.Hung MS, Mao JH, Xu Z, Yang CT, Yu JS, Harvard C, Lin YC, Bravo DT, Jablons DM, You L. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8:508–513. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- 96.Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 97.Colson K, Doss DS, Swift R, Tariman J, Thomas TE. Bortezomib, a newly approved proteasome inhibitor for the treatment of multiple myeloma: nursing implications. Clin J Oncol Nurs. 2004;8:473–480. doi: 10.1188/04.CJON.473-480. [DOI] [PubMed] [Google Scholar]
- 98.Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Therapy. 2003;2:623–629. [PubMed] [Google Scholar]
- 99.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 101.Huang J, Sheung J, Dong G, Coquilla C, Daniel-Issakani S, Payan DG. High-throughput screening for inhibitors of the E3 ubiquitin ligase APC. Methods Enzymol. 2005;399:740–754. doi: 10.1016/S0076-6879(05)99049-6. [DOI] [PubMed] [Google Scholar]
- 102.Xu S, Patel P, Abbasian M, Giegel D, Xie W, Mercurio F, Cox S. In vitro SCFbeta-Trcp1-mediated IkappaBalpha ubiquitination assay for high-throughput screen. Methods Enzymol. 2005;399:729–740. doi: 10.1016/S0076-6879(05)99048-4. [DOI] [PubMed] [Google Scholar]
- 103.Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun Y. Overview of approaches for screening for ubiquitin ligase inhibitors. Methods Enzymol. 2005;399:654–663. doi: 10.1016/S0076-6879(05)99043-5. [DOI] [PubMed] [Google Scholar]
- 105.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouhde Parseval L, Webb DR, Mercurio F, Nakayama KI, Nakayama K, Orlowski RZ. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 107.Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009;66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 109.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 110.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 111.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O’Dwyer M, Nawrocki ST, Giles FJ, Carew JS. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 112.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, Smith PG. MLN4924; a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-κB-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 113.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 114.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 115.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 116.Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, Kitamura T, Kato H, Nakayama K, Yoshimura A. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem. 2001;276:12530–12538. doi: 10.1074/jbc.M010074200. [DOI] [PubMed] [Google Scholar]
- 117.Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 118.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, Peter M. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 120.Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 121.Niture SK, Jaiswal AK. Prothymosin-alpha mediates nuclear import of the INrf2/Cul3 Rbx1 complex to degrade nuclear Nrf2. J Biol Chem. 2009;284:13856–13868. doi: 10.1074/jbc.M808084200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 122.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 123.Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 2004;18:856–861. doi: 10.1101/gad.1177904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang HF, Tomida A, Koshimizu R, Ogiso Y, Lei S, Tsuruo T. Cullin 3 promotes proteasomal degradation of the topoisomerase I-DNA covalent complex. Cancer Res. 2004;64:1114–1121. doi: 10.1158/0008-5472.can-03-2858. [DOI] [PubMed] [Google Scholar]
- 125.Lee YR, Yuan WC, Ho HC, Chen CH, Shih HM, Chen RH. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 2010 Apr 13;2010 doi: 10.1038/emboj.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nag A, Bagchi S, Raychaudhuri P. Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res. 2004;64:8152–8155. doi: 10.1158/0008-5472.CAN-04-2598. [DOI] [PubMed] [Google Scholar]
- 127.Banks D, Wu M, Higa LA, Gavrilova N, Quan J, Ye T, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle. 2006;5:1719–1729. doi: 10.4161/cc.5.15.3150. [DOI] [PubMed] [Google Scholar]
- 128.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 129.Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ, Xiong Y. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22:866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang J, Chen J. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene. 2008;27:4056–4064. doi: 10.1038/onc.2008.44. [DOI] [PubMed] [Google Scholar]
- 131.Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 132.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 133.Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- 134.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 135.Feng L, Allen NS, Simo S, Cooper JA. Cullin 5 regulates Dab1 protein levels and neuron positioning during cortical development. Genes Dev. 2007;21:2717–2730. doi: 10.1101/gad.1604207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu X, Sarikas A, Dias-Santagata DC, Dolios G, Lafontant PJ, Tsai SC, Zhu W, Nakajima H, Nakajima HO, Field LJ, Wang R, Pan ZQ. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol Cell. 2008;30:403–414. doi: 10.1016/j.molcel.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]