Abstract
Insulin-like growth factor-I (IGF-I) is a major anabolic hormone for skeletal muscle and a potent stimulus for protein synthesis and translation initiation. Recent studies suggest that translation can be inhibited by over expression of the mammalian target of rapamycin (mTOR) repressor REDD1. The purpose of the present study was to determine whether IGF-I alters the expression of REDD1 and whether this is associated with a concomitant change in protein synthesis in vitro. Subcutaneous injection of IGF-I or intravenous delivery of insulin for 3–4 h increased REDD1 mRNA in skeletal muscle 7- to10-fold. A 3-fold increase in REDD1 was observed when C2C12 myotubes were treated with IGF-I. REDD1 protein continued to be expressed for up to 24 h after addition of IGF-I to cells. Withdrawal of IGF-I from myotubes lead to a rapid loss of REDD1 protein content. IGF-I-induced REDD1 mRNA and protein expression were prevented by inhibitors of transcription and translation. IGF-I had an additive effect with dexamethasone (Dex) on REDD1 protein content in myotubes. The PI3K inhibitor LY294002 blocked IGF-I but not Dex induced REDD1. IGF-I also stimulated REDD1 promoter activity. Although REDD1 protein was elevated 5–6 h after addition of IGF-I to myotubes, protein synthesis measured during this 1 h window was paradoxically greater in myotubes expressing more REDD1. In contrast to the IGF-I induced increase in REDD1 mRNA, REDD2 mRNA was decreased by IGF-I. We conclude that IGF-I stimulates REDD1 expression in skeletal muscle and myotubes but under these conditions the REDD1 response is not sufficient to repress protein synthesis.
Keywords: Skeletal Muscle, IGF-I, C2C12 Myotubes, Protein Synthesis
INTRODUCTION
Muscle growth is exquisitely sensitive to nutrient availability, cellular and organismal stress, age, and the local production of cytokines and growth factors [Drummond and Rasmussen, 2008; Frost and Lang, 2008; Smith et al., 2008; Vary and Lynch, 2007]. The control of muscle mass is therefore balanced by the anabolic and catabolic response to these inputs [Frost and Lang, 2007; Guttridge, 2004; Lang et al., 2007a]. Foremost among the anabolic factors impinging on muscle is the peptide hormone insulin-like growth factor (IGF)-I [Velloso, 2008]. IGF-I is unique because it stimulates protein synthesis in skeletal and cardiac muscle but not in a variety of other tissues [Bark et al., 1998]. IGF-I is also a potent inhibitor of muscle atrophy via its ability to suppress the expression of atrophy genes although this is not a universal finding [Criswell et al., 1998; Dehoux et al., 2007; Glass, 2005].
The mammalian target of rapamycin (mTOR) is a serine (S)/threonine (T) kinase that plays a pivotal role in integrating positive input from nutrients and growth factors as well as negative input related to energy stress, such as hypoxia and free radicals [Dunlop and Tee, 2009]. mTOR functions as a central signaling molecule controlling the translation initiation machinery via phosphorylation of its substrates S6K1 and 4E-BP1. Together these substrates are thought to facilitate the recruitment of ribosomes for cap-dependent translation [Choo et al., 2008; Holz et al., 2005]. mTOR can exist in two complexes referred to as mTOR complex (mTORc)1 and -2 and it is the first complex that is thought to control translation initiation. An additional mTORc1 and -2 independent complex has also been proposed and is thought to enhance translational efficiency of mRNAs with highly structured 5′ untranslated regions [Patursky-Polischuk et al., 2009].
mTOR activity is tightly regulated by the upstream tumor suppressor proteins known as Tuberous Sclerosis Complex (TSC)-1 and -2. When present as a heterodimer the TSC1-TSC2 complex inhibits mTOR indirectly by converting the small GTPase Ras homolog enriched in brain (Rheb) to an inactive GDP bound form. Indeed, over expression of TSC1 in mouse skeletal muscle leads to a significant increase in the TSC1-TSC2 complex and a reduction in muscle mass [Wan et al., 2006]. In contrast, activation of the PI3K/Akt pathway by growth factors promotes the phosphorylation of TSC2 on S924 and T1518, inactivates the TSC complex, and restores Rheb and mTOR activity [Potter et al., 2002]. A second key mechanism by which Akt may alter the TSC1-TSC2 complex is by phosphorylating TSC2 on S939 and S981. Phosphorylation of these residues facilitates the movement of TSC2 away from Rheb and sequesters TSC2 with 14-3-3 proteins [Cai et al., 2006; Huang and Manning, 2009]. The association of TSC2 with 14-3-3 allows for strong stimulation of mTOR activity but this may only provide a simplistic explanation of the regulation of the TSC1-TSC2 complex by growth factors given the wide spectrum of potential binding partners to which TSC1 and -2 can associate [Rosner et al., 2008].
Over expression of REDD1 during hypoxic stress inhibits mTOR activity as evidenced by the decreased phosphorylation of its targets S6K1 and 4E-BP1 [Brugarolas et al., 2004; Corradetti et al., 2005]. REDD1, like TSC2, binds to 14-3-3 proteins and mutants of REDD1 that fail to bind 14-3-3 protein are defective at inhibiting mTOR activity. De Young et al have hypothesized that REDD1 inhibits mTOR activity by displacing endogenous TSC2 from 14-3-3, activating theTSC1-TSC2 complex and subsequent inhibition of Rheb [DeYoung et al., 2008]. The importance of REDD1 is underscored by the ability of REDD1 to dominantly suppress mTOR activity even in the presence of the strong growth signal elicited by a myristylated form of Akt [DeYoung et al., 2008].
Although much is known about how REDD1 can inhibit mTOR activity less is known about its regulation by factors other than hypoxia. We hypothesized that as a negative regulator of mTOR activity that REDD1 expression might be down regulated by growth factors. To test this hypothesis we examined REDD1 mRNA expression in skeletal muscle and myotubes in culture. We found a paradoxical increase in REDD1 in skeletal muscle of IGF-I treated rats and myotubes treated continuously with IGF-I. Although IGF-I treatment was associated with a 3-fold increase in endogenous REDD1, this change did not have a negative effect on global protein synthesis as measured by the incorporation of phenylalanine into muscle protein.
EXPERIMENTAL PROCEDURES
Experimental protocols for in vivo studies
The following experimental protocols were approved by the Institutional Animal Care and Use Committee of The Pennsylvania State University College of Medicine and adhered to the National Institutes of Health guidelines for the use of experimental animals. Pathogen-free male Sprague-Dawley rats (300–325 g; Charles River Breeding Laboratories, Cambridge, MA) were used in all of the experiments. Rats were housed in a controlled environment and provided commercial laboratory food (Harlan #2018; 18% protein rodent diet; Madison, WI) and water ad libitum.
For the IGF-I study freely fed rats were injected with recombinant human IGF-I (Genentech, South San Francisco, CA, 200 μg/kg body weight, n=5) subcutaneously with subsequent removal of the gastrocnemius 4 h afterward. Comparable doses of IGF-I have been reported to increase protein synthesis in skeletal muscle [Bark et al., 1998]. Time-matched control animals were injected with an equal volume of 0.9% (wt/vol) sterilesaline (n=6). For the insulin study rats were infused intravenously with recombinant human insulin (Humulin® R, Lilly, Indianapolis, IN, 4 mU/min/kg) and glucose infused IV to maintain euglycemia (100 ± 5 mg/dl). This insulin infusion rate provides plasma insulin concentrations in the high physiological range (~ 100 μU/ml). Time-matched control animals were infused with an equal volume of 0.9% (wt/vol) sterilesaline with the gastrocnemius isolated from both groups 3 h later (n=4 per group). Rats in both studies were anesthetized with pentobarbital sodium (75 mg/kg) prior to dissection of the gastrocnemius which was rapidly frozen between liquid nitrogen-cooled clamps. Frozen tissue was powdered with a mortar and pestleand subsequently stored at −70°C until analyzed. Muscle was isolated at time points during which we have previously demonstrated an IGF-I-induced fall in muscle atrophy genes and an increase in the phosphorylation of translation initiation factors [Lang et al., 2007b].
Cell culture
The C2C12 mouse myoblast cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA) and used for all studies. Cells were grown in 100 mm Petri dishes (Greiner Bio-One, Frickenhausen, Germany) and cultured in minimal essential media (MEM) containing 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml) and amphotericin B (25 μg/ml) (all from Mediatech, Herndon, VA). Cells were grown to near confluence and sub-cultured to 6-well plates for all experiments. Cells were differentiated to myotubes in 10% bovine calf serum (BCS). In most experiments cells were differentiated for ≥ 7 days to obtain cultures composed of greater than 90% myotubes. Just prior to the experiments myotubes were switched to fresh serum-free MEM. Experiments were performed with IGF-I obtained from Genentech (South San Francisco, CA) and used at 100 ng/ml, a dose previously shown to both increase and decrease gene expression [Adi et al., 2000; Dehoux et al., 2007]. Dexamethasone was obtained from Sigma and used at a dose (10 μM) previously shown to inhibit protein synthesis and increase REDD1 protein content [Shah et al., 2000; Wang et al., 2006]. Other chemicals including 5,6-dichloro-1-beta-ribofuranosyl benzimidazole (DRB), cycloheximide, LY294002, PD98059, were used at a concentrations previously shown to inhibit transcription, translation, and kinase activities and are noted in their respective figure legends. Protein was isolated in Laemelli sample buffer containing SDS, phosphatase, and protease inhibitors.
RNA isolation and ribonuclease protection assay (RPA)
Total RNA, DNA and protein were extracted from C2C12 cells in a mixture of phenol and guanidine thiocyanate (TRI Reagent, Molecular Research Center, Cincinnati, OH) using the manufacturer’s protocol. RNA was separated from protein and DNA by the addition of bromocholoropropane and precipitation in isopropanol. After a 75% ethanol wash and resuspension in formamide, RNA samples were quantified by spectrophotometry. Ten micrograms of RNA was used for each assay.
Riboprobes were synthesized from a custom multi-probe mouse template set containing a probe for mouse REDD1, REDD2 and L32 mRNA detection. Riboprobes for rat REDD1 and L32 were generated separately. Primer selection for the probes was determined with the help of Genefisher software [Giegerich et al., 1996]. Primers were synthesized (IDT, Coralville, IA) with restriction sites for EcoRI or KpnI at the 5′ end and with 3 extra bases at the extreme 5′ end as follows: mREDD1—Forward (5′-GCA GAA TTC GGC CGG AGG AAG ACT CCT CA-3′), Reverse (5′-GCA GGT ACC CTT CTT GAT GAC TCT GAA GCC GGT A -3′), mREDD2—Forward (5′-GCA GAA TTC CTG AGA AAC TGA CCC AGA GAA TTG C-3′), Reverse (5′-CCA GGT ACC GTC GTT CCA ATC AGG GAG TAC AGT -3′), mL32 —Forward (5′-GCA GAA TTC CGG CCT CTG GTG AAG CCC AA-3′), Reverse (5′-GCA GGT ACC CCT TCT CCG CAC CCT GTT GTC A-3′) rREDD1 —Forward (5′-GCA GAA TTC CCT CTT CGT CCT CGT CCC GAA-3′), Reverse (5′-GCA GGT ACC GGC ACT AGA CTG GGG TCC AGA-3′) and L32—Forward (5′-GCA GAA TTC CGG CCT CTG GTG AAG CCC AA-3′), Reverse (5′-GCAGGT ACC CCT TCT CCG CAC CCT GTT GTC A-3′). Briefly, protected RNAs were separated using a 5% acrylamide gel (19:1acrylamide / bisacrylamide). Gels were transferred to blotting paper and dried under vacuum on a gel dryer. Dried gels were exposed to a phosphorimager screen (Molecular Dynamics, Sunnyvale, CA) and the resulting data were quantified using ImageQuant™ software and normalized to the mouse or rat ribosomal protein L32 mRNA signal in each lane.
Quantitative real-time PCR (QT-RT-PCR)
Total RNA was extracted as described above and reversed transcribed with a Superscript ™ First Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) using random hexamers and 1.5 ug of RNA. No template and no amplification controls were included. Quantitative PCR was performed using Quantitect SYBR Green PCR kits (Qiagen) with final PCR primer concentrations of 300 nM in a reaction volume of 20 μl. Primers for actin were purchased from Qiagen and REDD2 (forward primer, 5′-CCAGCCTCAAGGACTTCTTC, 3′; reverse primer, 5′-TCCTCAATGACTGTCGTTCC-3 was obtained from Dr. Scot Kimball (Hershey, PA). The optimal concentration of cDNA and primers and the efficiency of amplification were obtained from five-point dilution curve analysis for each gene. Polymerase chain reaction analyses were performed using an ABI Prism 7700 Sequence Detection System (Applied Biosystems). Amplification and annealing temperatures were 60 °C. Melting curve analyses were performed to assess the specificity of the PCR products. After PCR was completed, baseline and threshold levels were adjusted using the ABI Prism 7700 software and CT values determined. Care was taken to ensure that threshold levels were within the log-linear range of all amplified plots. The relative standard curve method was used to analyze the data, with relative amounts of unknown samples being calculated using linear regression analysis. The relative value for REDD2 was divided by the actin value and the control group was set as the calibrator, a value of 1.0.
Western blot analysis
Cell extracts were electrophoresed on denaturing polyacrylamide gels and electrophoretically transferred to nitrocellulose with a semi-dry blotter (Bio-Rad Laboratories, Melville, NY). The resulting blots were blocked with 5% non-fat dry milk for 1.5 h and incubated with a polyclonal antibody against either REDD1 (Proteintech Group, Chicago IL, Cat # 10638-1AP), IGF-I receptor, IRS-1 (Santa Cruz Biotechnology, Santa Cruz, CA), MHC MF-20 (Developmental Studies Hybridoma Bank, Iowa City, Iowa) TSC-1, TSC2, S6K1, S6K1 phosphorylated on threonine 389 and GAPDH (Cell Signaling Technology, Danvers, MA). Primary antibody dilutions ranged from 1:1000 to 1:10,000 depending on the strength of the antibody. Unbound primary antibody was removed by washing with Tris-buffered saline containing 0.05% Tween-20 and blots were incubated with anti-rabbit or anti-mouse immunoglobulin conjugated with horseradish peroxidase. Blots were briefly incubated with the components of an enhanced chemiluminescent detection system (Amersham, Buckinghamshire, England). Dried blots were used to expose x-ray film for 1–25 minutes to achieve a signal within the linear range. Each film was then scanned with a Microtek Scanmaker 4 scanner (Microtek, Cerritos, CA) equipped with a transparency tray to generate a digital image that was then analyzed and quantified with Scion Image 3b software (Scion Corporation, Frederick MD). When necessary, comparisons were made across gels by normalizing all the data to the signal(s) of the SFMEM control which was set to 1.0. Results normalized in this manner for individual gels were then averaged with similarly normalized data from additional gels for each of the experimental conditions.
REDD1 promoter construct
A 744 bp fragment of mREDD1, containing 373 bp of the mREDD1 promoter region was cloned into pBSII-SK+ (Stratagene, La Jolla, CA) along with a 1979 bp fragment of pTAL-Luc (BD Pharmingen) containing a fully functional firefly luciferase gene with attached SV40 late polyadenylation sequence as follows. Mouse genomic clone RP24 (BACPAC Resources, Oakland, CA) along with primers mProF (in mREDD1 promoter) and mREDD1R (located in coding sequence) were used to PCR the mREDD1 promoter. Nested PCR followed using the previous reaction’s product and primers mREDD1-DF and mREDD1-ER. Primer mREDD1-ER has a HindIII site. A KpnI site exists within the PCR product. After digestion with HindIII and KpnI, the expected 750 bp band was identified, gel-purified, and ligated into pBSII-SK+ HindIII/KpnI. Clones were ligated with luciferase, linearized with HindIII and SacI overnight, and gel purified.
Firefly luciferase was cloned from pTAL-Luc (BD Pharmingen) with the following PCR primer set: Forward 5′GGCCACGGGGATGAAGCAGA and Reverse 5′GCTGAGCTCGCTCTCAAGGGCATCGGTCGA. The PCR product was digested with HindIII and SacI overnight. The gel-purified fragment was ligated to HindIII/SacI-cut pBS/mREDD1. A positive clone was identified and sequencing showed the 5′ and 3′ ends of the luciferase insert as well as the cloned mREDD1 to be intact with no mutations.
Transient transfection and promoter assays
C2C12 myoblasts at 80–90% confluence were trypsinized from multiple 100 mm plates and 4 × 106 cells resuspended in 100 μl of Nucleofector V solution containing the manufacturer’s supplement (Amaxa, Walkersville, MD, 4:1 vol/vol). Cells were nucleofected with 6 μg of either empty vector (pTal-Luc), or a 744 bp fragment of the mouse REDD1 promoter (pBS-mRL) and a plasmid expressing Renilla luciferase as previously described [Frost et al., 2006]. Plasmids were nucleofected with a brief electrical pulse generated in an Amaxa Biosystems Nucleofector II electroporator using a preprogrammed setting of B-032 as suggested by the manufacturer. Electroporated cells were resuspended and plated at 3.7 × 105 cells per well in MEM containing 10% FBS. Fresh serum containing media was added 2 h later and cells subsequently switched to serum-free MEM the following day. The transfected cells were either grown in SFMEM alone or treated with IGF-I (100 ng/ml) for 24 h. Cells were isolated in 300 μl of passive lysis buffer as suggested by the manufacturer (Promega, Madison, WI, Dual Luciferase assay system). Cell extracts were centrifuged to remove debris and 25 μl of the supernatant assayed for luciferase activity on a Luminoscan Ascent luminometer (Thermo Labsystems, Waltham, MA).
Statistics
Values are means ± SEM. Unless otherwise noted, each experimental condition was tested in triplicate and each experiment was repeated at least two times. For transfections, a single transfection yielded enough cells to compare luciferase activity in 12 wells and thus 6 wells per condition and this was repeated 3 times. Data were analyzed by ANOVA followed by the Bonferroni-Holm post-hoc test to determine treatment effect when ANOVA indicated a difference among the means (Daniel Kraus, open source add in for Excel ®, sorceforge.net). Statistical significance was set at P<0.05.
RESULTS
IGF-I and insulin increase REDD1 in skeletal muscle
Other than the regulation of REDD1 expression by cell stressors such as hypoxia and glucocorticoids little is known about the regulation of REDD1 in vivo [Jin et al., 2007; Wang et al., 2006]. In skeletal muscle, REDD1 mRNA is elevated after acute alcohol intoxication and decreased by low intensity resistance exercise [Drummond et al., 2008; Lang et al., 2008]. Yet, there is a paucity of data on the effect of anabolic peptides such as insulin and IGF-I on REDD1 expression. Therefore, Sprague-Dawley rats were injected with IGF-I and skeletal muscle obtained after 4 h. IGF-I increased REDD1 mRNA in gastrocnemius by 6-fold as measured by RPA (Figure 1, Panel A and C). The constant infusion of the structurally similar anabolic hormone insulin for 3 h in rats also increased REDD1 mRNA12-fold in the gastrocnemius (Figure 1, Panel B and C). The rise in REDD1 mRNA was accompanied by a 3-fold increase in REDD1 protein content in the muscle (Figure 1, Panel B, Saline 1.0 ± 0.12, Insulin 3.3 ± 0.28 AU/GAPDH, p<0.05). The change in REDD1 in skeletal muscle was independent of changes in housekeeping genes such as L32 (mRNA) and GAPDH (protein) that were used to normalize the steady-state levels of REDD1 mRNA and protein, respectively.
Figure 1. Effect of IGF-I and insulin on REDD1 mRNA and protein expression in rat skeletal muscle.
Rats were administered IGF-I subcutaneously (200 μg/kg body weight, Panel A) or insulin as a constant intravenous solution (4 mU/min/kg, Panel B) for 3 h with glucose to maintain euglycemia. Muscle was flash frozen in liquid nitrogen, powdered, and analyzed at the peak of rat REDD1 expression (3–4 h). RNA was isolated and hybridized with a ribonuclease protection assay (RPA) template as described in the Methods and run on a 5% acrylamide gel (Panel A and B, RPA). Some of the tissue homogenate from the insulin infused rats was analyzed for REDD1 protein by Western blotting (Panel B, Western). The dried gel was exposed to a phosphor imager screen and quantified with ImageQuant™ software (Panel C). All data were normalized to ribosomal protein (rp)L32 mRNA as described in the Methods and expressed as a fold-increase relative to time-matched animals injected or infused with saline alone. Values are means ± SEM. Bars with different lower case letters are significantly different from each other (P<0.05, 4–6 animals per group).
IGF-I time-dependently increases REDD1 mRNA and protein in C2C12 myotubes
To determine whether IGF-I induced REDD1 mRNA predominantly in myofibers rather than connective, vascular, or nervous tissue in skeletal muscle we treated differentiated C2C12 myotubes with IGF-I in vitro. IGF-I stimulated REDD1 mRNA in as little as 2 h and the 2–3 fold increase in REDD1 mRNA was sustained for at least 6 h (Figure 2, Panel A). REDD1 mRNA was translated into protein as detected by Western blotting of cell extracts with a REDD1 specific antibody. A significant increase in REDD1 protein occurred 3–5 h after IGF-I addition to the cells and is consistent with the earlier rise in REDD1 mRNA (Figure 2, Panel B). Both REDD1 mRNA and protein levels reached a new steady-state such that the amount of newly synthesized REDD1 protein was unchanged between 5 and 24 h. Although glucocorticoids often inhibit IGF-I signaling and conversely IGF-I can also independently inhibit glucocorticoid signaling [Schakman et al., 2005; Singleton et al., 2000], IGF-I and dexamethasone both increased REDD1 protein content in myotubes and the two together had an additive effect on REDD1 protein content (Figure 2, Panel C). All of the changes in REDD1 mRNA and protein in C2C12 myotubes were independent of changes in L32 mRNA and GAPDH protein. In addition, the total amount of a number of muscle IGF-I and mTOR signaling proteins were unchanged across the above conditions including: the IGF-I receptor (IGF-IR), IRS-1, TSC-1 and -2, S6K1, and myosin heavy chain (MHC) (Figure 2, Panel C). In contrast, the phosphorylation of S6K1 on T389 was increased by IGF-I and decreased by Dex.
Figure 2. IGF-I time-dependently increases REDD1 mRNA and protein in C2C12 myotubes.
C2C12 cells were subcultured into 6-well plates and allowed to differentiate into myotubes in the presence of 10% BCS for 7 days. Myotubes were switched to fresh serum-free MEM in the presence of absence of IGF-I (100 ng/ml). RNA was isolated and hybridized to a mouse REDD1 RPA template as described in the Methods. The dried gel was exposed to a phosphor imager screen and quantified (Panel A). A representative image of the 6 h samples is shown (Panel A, inset). Additional cells were cultured in either serum free media or IGF-I (100 ng/ml) for 1–24 h. Cell extracts were isolated and run on an SDS PAGE gel for Western blotting. A representative blot for duplicate samples from each time point is shown (Panel B). Finally, myotubes were cultured as above and treated with either IGF-I alone, dexamethasone (Dex; 10 μM) alone or the combination. Western blotting was used to detect REDD1 and a number of IGF-I, muscle, and mTOR relevant loading controls including: IGF-I receptor (IGF-IR), insulin receptor substrate-I (IRS-1), TSC-1 and -2, S6K1, myosin heavy chain (MHC) and GAPDH.
Accumulation of REDD1 mRNA and protein requires ongoing transcription and translation
The ability of C2C12 myotubes to respond to IGF-I was examined in the presence of the transcriptional inhibitor DRB. IGF-I increased REDD1 mRNA and this was completely inhibited by pretreatment with DRB (Figure 3, Panel A and B). Likewise, IGF-I stimulated the synthesis of REDD1 protein and this increase was completely inhibited by DRB (Figure 3, Panel C). Cycloheximide (CHX) also completely blocked the IGF-I-induced increase in REDD1 protein (Control 100 ± 27, IGF-I 252 ± 18, IGF-I + CHX 63 ± 13, AU/GAPDH, p<0.05).
Figure 3. IGF-I-induced REDD1 mRNA and protein is blocked by a transcriptional inhibitor.
C2C12 myoblasts were grown as described in Figure 2 for 8 h either in the presence or absence of the transcriptional inhibitor DRB (72 μM) with IGF-I (100 ng/ml). A representative RPA is shown (Panel A) and quantified (Panel B). Additional cell extracts were run on an SDS PAGE gel and probed for REDD1 protein by Western blotting (Panel C). All data were normalized for L32 mRNA or GAPDH protein as described in the Methods. Values are means ± SEM.
IGF-I withdrawal leads to a rapid loss of REDD1 protein
Preliminary data suggested that a constant exposure to IGF-I increased REDD1 protein but that REDD1 was not elevated in cells exposed to IGF-I episodically (data not shown). The aforementioned phenomena prompted us to determine whether a constant exposure to IGF-I was necessary for REDD1 protein expression. Myotubes were treated with IGF-I overnight to elevate REDD1 protein to a steady-state. The cells were then switched to fresh SFMEM alone or SFMEM containing IGF-I. Withdrawal of IGF-I decreased REDD1 protein to baseline levels in as little as 2 h (Figure 4, Panel A and C). In contrast, cells maintained either in the same media or media to which fresh IGF-I was added sustained the IGF-I induced level of REDD1 expression (Figure 4, Panel B and C). The level of GAPDH in cell extracts was unaltered by either IGF-I treatment or IGF-I withdrawal.
Figure 4. IGF-I withdrawal leads to a rapid loss of REDD1 protein.
C2C12 myotubes were grown overnight (O/N, 18 h) in the presence or absence of IGF-I (100 ng/ml) to induce REDD1 protein. IGF-I was then withdrawn from the cultures by switching the cells to fresh SFMEM for 1, 2, 4, or 8 h (Panel A, first two blots). Some cells were treated as above but they were not switched to fresh SFMEM (Panel A, second two blots). Withdrawal of IGF-I led to a rapid decline in REDD1protein as quantified in Panel B whereas REDD1 protein levels remained elevated in cells grown in the continuous presence of IGF-I. All data were normalized to GAPDH as described in the Methods and expressed as a fold increase above that of cells grown in SFMEM alone. Values are means ± SEM (*, p<0.05, compared to control cells).
Glucocorticoids maintain REDD1 in the absence of IGF-I
Because the withdrawal of IGF-I led to a rapid loss of REDD1 we determined whether glucocorticoids could maintain REDD1 protein levels after removal of IGF-I from the media. To test this idea myotubes were stimulated with IGF-I overnight to elevate REDD1 and then switched to either fresh SFMEM alone or SFMEM containing IGF-I or dexamethasone for an additional 5 h. Myotubes grown in the continued presence of IGF-I expressed 3-fold more REDD1 than cells that were switched back to SFMEM (Figure 5, Panel A and B). In addition, switching myotubes to fresh media containing dexamethasone enhanced the REDD1 content of the extracts even further.
Figure 5. IGF-I but not Dex-induced REDD1 is PI3K-dependent.
C2C12 myoblasts were grown as described above with IGF-I and then switched to either SFMEM, fresh IGF-I, IGF-I and LY294002 (LY, 10 μM), Dex (1 μM) or Dex and LY. REDD1 content was determined by Western blotting (Panel A) and quantified and normalized to GAPDH (Panel B). Values are means ± SEM of triplicate dishes. Bars with different lower case letters are significantly different from each other (P<0.05).
Because IGF-I strongly stimulates PI3 kinase (PI3K) signaling in skeletal muscle we determined whether PI3K activity was necessary for the continued expression of REDD1 protein. Switching cells to fresh IGF-I stimulated REDD1 protein content 3-fold and this was completely blocked in the presence of the PI3K inhibitor LY294002 (LY). In contrast, myotubes grown in the presence of IGF-I overnight followed by dexamethasone for the last 5 h of culture expressed 6-fold more REDD1 protein than cells switched to SFMEM, and the dexamethasone-induced REDD1 was only minimally affected by LY (Figure 5, Panels A and B). IGF-I may also stimulate the extracellular-signal related kinases in muscle and therefore we determined whether inhibition of the upstream activating kinase MAP kinase kinase with PD98059 altered REDD1 protein content. IGF-I stimulated REDD1 protein 3-fold (p<0.05) and neither IGF-I or dexamethasone-induced REDD1 was inhibited by PD98059 (Control 100 ± 1, IGF-I followed by: IGF-I 263 ± 5, IGF-I + PD 369 ± 30, Dex 523 ± 26, Dex + PD 450 ± 32, p>0.05 ).
IGF-I stimulates mREDD1 promoter activity
We cloned a 373 bp fragment of the mouse REDD1 promoter to determine if the IGF-I induced increase in REDD1 mRNA and protein was due to a corresponding increase in transcription of the REDD1 gene. IGF-I stimulated REDD1 promoter activity 3-fold as measured by the expression of firefly luciferase activity in cells transfected with a plasmid containing the REDD1 promoter but not with the same plasmid lacking the promoter sequence (empty vector) (Figure 6).
Figure 6. IGF-I stimulates REDD1 promoter activity.
C2C12 cells were grown as myoblasts and electroporated with 8 ug of either an empty vector (pTal-Luc), the mouse REDD1 promoter (pBS-mRL), or the REDD1 promoter in which the HRE was mutated (pBS-mRL/mtHRE). All plasmids expressed firefly luciferase. Electroporated cells were resuspended and plated in MEM containing 10% FBS. Fresh serum containing media was added 2 h later and subsequently switched to serum-free MEM the following day. Each transfection yielded 12 wells containing cells transfected with a particular vector. Half of the cells (6-wells) were grown in SFMEM alone with the other half (6-wellls) treated with IGF-I (100 ng/ml) for 24 h. Cells were isolated in 300 μl of passive lysis buffer, centrifuged to remove debris, and 25 μl of the supernatant assayed for luciferase activity. Values are means ± SE and are expressed as a fold increase compared with control cells isolated at the same time. Values with different letters are significantly different from each other (P< 0.05).
IGF-I stimulates protein synthesis in the presence of elevated endogenous levels of REDD1
Because transfection of cells with excess REDD1 inhibits mTOR activity and decreases cell size we examined whether the IGF-I-induced increase in REDD1 we observed in myotubes altered protein synthesis. IGF-I was added to myotubes and protein synthesis measured during a 1 h interval 5–6 h later, a time point during which REDD1 protein was elevated 3-fold. IGF-I increased protein synthesis by15%, compared to control cells grown in SFMEM (Figure 7, Panel A). Given that IGF-I withdrawal decreases REDD1 to levels seen in control cells we also grew cells in the presence of IGF-I 5 h and then either withdrew IGF-I for an additional 5 h or grew cells in the continued presence of IGF-I to maintain elevated REDD1 levels. Protein synthesis was elevated by 20–50% in cells continuously treated with IGF-I or cells to which fresh media and IGF-I was added (Figure 7, Panel B). In contrast, withdrawal of IGF-I decreased protein synthesis relative to cells grown in the continued presence of IGF-I (Figure 7, Panel B). Therefore, although IGF-I increased REDD1, this change was not associated with depressed protein synthesis. Conversely, although IGF-I withdrawal decreased REDD1, this change was not associated with a concomitant rise but rather a relative decrease in protein synthesis. Likewise, IGF-I strongly stimulated protein synthesis in the presence of Dex (40%) during a period when the REDD1 protein level was elevated (Figure 7, Panels B and C).
Figure 7. IGF-I increases protein synthesis despite elevated REDD1.

C2C12 myotubes were grown as described above in either SFMEM or IGF-I (100 ng/ml) for 5 h. Cells were then labeled with [3H]-phenylalanine for 1 h and phenylalanine incorporation into protein determined by TCA precipitation as described in the Methods (Panel A). Additional cells were grown in the presence of IGF-I for 5 h and then either left in the continuous presence of that media for an additional 5 h or switched to fresh SFMEM ± IGF-I. Cells were labeled with [3H]-phenylalanine for the final 5 h of the experiment. Some cells were also switched to Dex or Dex and IGF-I for the final 5 h. The amount of radioactively labeled protein was determined as described in the Methods (Panel B). Cells grown under the same conditions as described for Panel B were also isolated in sample buffer, run on SDS-PAGE gels and probed for REDD1, pAkt (S473), pS6K1 (T389), total S6K1, pS6 (S240/244), total rpS6 or MHC (Panel C). Values are means ± SEM and are expressed as a percentage increase in the incorporation of phenylalanine above that in control cells isolated at the same time point. Bars with different letters are statistically different (P<0.05).
In general, REDD1 was elevated under the same conditions during which both the mTORc1 substrate S6K1 and the mTORc2 substrate Akt were phosphorylated (Figure 7, Panel C). The S6K1 substrate rpS6 also followed a similar pattern of phosphorylation and the phosphorylation of all three substrates was reduced upon withdrawal of IGF-I or Dex treatment (Figure 7, Panel C). Yet, even in the presence of Dex, IGF-I restored mTOR signaling as assessed by an increase in protein synthesis (Figure 7B) and the phosphorylation of mTOR substrates (Figure 7, Panel C).
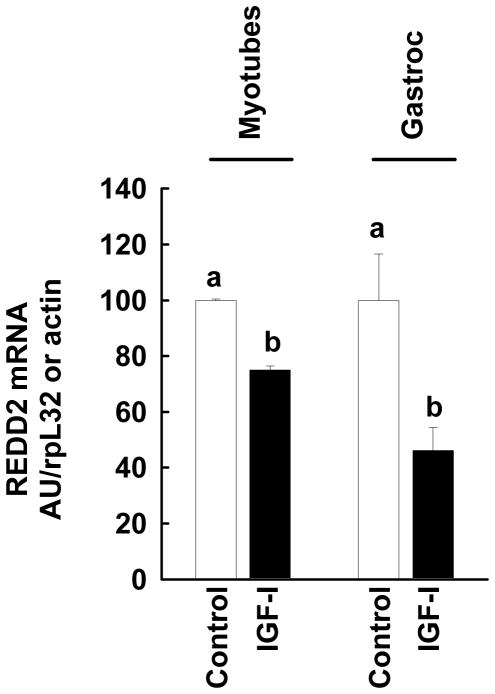
IGF-I decreases REDD2 mRNA in vivo and in vitro
Because the IGF-I induced increase in REDD1 was associated with an unexpected increase in protein synthesis in myotubes we examined whether the REDD1 homolog REDD2 might also be altered by IGF-I treatment and perhaps compensate for the rise in REDD1. Incubation of C2C12 myotubes for 5 h with IGF-I decreased REDD2 mRNA by 25% (Figure 8). Similarly, subcutaneous injection of IGF-I produced a 60% reduction in REDD2 mRNA in gastrocnemius after 4 h, compared to time-matched control values (Figure 8). Attempts to detect REDD2 protein by Western analysis using commercially available antibodies proved unsuccessful (ie. Abcam H00115265-B01P and Proteintech Group 12094-1-AP, data not shown).
Figure 8. IGF-I decreases REDD2 mRNA in vivo and in vitro.
C2C12 myotubes were grown as described above in either SFMEM or IGF-I (100 ng/ml) for 5 h. RNA was isolated and hybridized to a mouse REDD2 RPA template and quantified as described in the Methods (Myotubes). Rats were administered IGF-I subcutaneously and the gastrocnemius analyzed at the peak of rat REDD1 expression (3–4 h). RNA was isolated and the REDD2 mRNA determined by QT-RT-PCR as described in the Methods. All data were normalized to actin mRNA and expressed as a fold-increase relative to time-matched animals injected with saline alone. Values are means ± SEM. Bars with different lower case letters are significantly different from each other (P<0.05, 4–6 animals per group).
DISCUSSION
Our understanding of the molecular mechanism(s) regulating mTOR activity has expanded greatly in recent years with the identification of a series of steps known to activate the serine/threonine kinase. The role of TSC1-TSC2 as a gatekeeper of Rheb and therefore mTOR activity has also generated great interest in how mTOR is regulated and how it chooses its substrates. The binding of TSC2, raptor, PRAS40, and REDD1 to 14-3-3 proteins suggests that this family of proteins may play a key role in regulating not only mTOR in general but specifically muscle mass. For example, knockdown of raptor or over expression of TSC-1 in skeletal muscle induces muscle atrophy [Bentzinger et al., 2008; Wan et al., 2006]. Likewise, over expression of PRAS40, REDD1, or its homolog REDD2 strongly inhibits the phosphorylation of mTOR substrates and by association may alter the accretion of muscle mass [Corradetti et al., 2005; DeYoung et al., 2008; Miyazaki and Esser, 2009; Vander Haar et al., 2007; Wang et al., 2007].
Because REDD1 has been shown to inhibit mTOR activity and decrease cell size in transfected cells [Sofer et al., 2005], we examined whether IGF-I alters endogenous expression of REDD1 in skeletal muscle and myotubes. We hypothesized that an anabolic peptide, like IGF-I, would decrease REDD1 expression and therefore enhance mTOR activity. IGF-I was injected for a period of time over which we have previously demonstrated an IGF-I-induced fall in muscle atrophy genes and an increase in the phosphorylation of translation initiation factors [Lang et al., 2007b]. Contrary to our original hypothesis, IGF-I increased REDD1 mRNA expression in the gastrocnemius 3–4 h after a subcutaneous injection. A similar increase in REDD1 mRNA was seen in rats infused with insulin during a hyperinsulinemic euglycemic clamp and this was accompanied with a concomitant increase in REDD1 protein. The increase in REDD1 mRNA expression was surprising given REDD1 is normally elevated by stress and stress hormones including: hypoxia, DNA damage, energy stress, endoplasmic reticulum stress, acute alcohol intoxication, and glucocorticoids [DeYoung et al., 2008; Lang et al., 2008; Lin et al., 2005; Sofer et al., 2005; Wang et al., 2006; Whitney et al., 2009]. These results suggested that regulation of the REDD1 gene by IGF-I might be atypical when contrasted to the above stresses.
REDD1 mRNA and protein were elevated by IGF-I in the gastrocnemius and differentiated C2C12 myotubes in a comparable time frame suggesting that the regulation in vivo and in vitro is similar. We and others have previously shown that the synthetic glucocorticoid Dex can increase REDD1 in vivo and in L6 myoblasts and we confirmed that finding here in C2C12 myotubes [Lang et al., 2008; Wang et al., 2006]. Although IGF-I often inhibits Dex signaling, and vice versa [Dardevet et al., 1998; Waddell et al., 2008], the combination of Dex and IGF-I had an additive effect on REDD1 protein content in myotubes. The IGF-I-induced increase in REDD1 mRNA and protein requires both new transcription and translation of the REDD1 gene and mRNA because it was blocked by DRB and cycloheximide, respectively. IGF-I also stimulates a minimal REDD1 promoter to the same extent that it stimulates the accumulation of REDD1 mRNA and protein.
REDD1 protein expression requires ongoing stimulation of the IGF-I receptor as evidenced by a decrease in REDD1 protein in cells that are first grown in the presence of IGF-I and then have the media replaced with fresh media. The rapid decrease in REDD1 protein that we observed upon IGF-I removal is consistent with the short half-life of REDD1 reported by Kimball et al in response to cycloheximide [Kimball et al., 2008]. Interestingly, if Dex is added to myotubes upon IGF-I removal REDD1 protein levels are maintained or slightly augmented suggesting that Dex continues to promote the synthesis of REDD1 and/or prevents the loss of the protein upon IGF-I withdrawal.
The IGF-I-induced expression of REDD1 in myotubes is PI3K-dependent based on its inhibition by the PI3K inhibitor LY294002. Jin et al have also recently shown that both hypoxia and cell density increase REDD1 via a PI3K-dependent pathway [Jin et al., 2007]. Yet, because myotubes are grown at a maximal cell density and they have fused and differentiated we do not believe that IGF-I changes the density of the cultures over the short exposure time that is required to increase REDD1 mRNA and protein.
The specificity of the IGF-I induction of PI3K signaling as it pertains to REDD1 expression was ascertained in two experiments. First, although both IGF-I and Dex induced REDD1, only IGF-I-induced REDD1 was inhibited by LY294002. These results suggest Dex induces REDD1 by a PI3K-independent mechanism. Secondly, because IGF-I stimulates both the PI3K and MAP kinase pathways in most cells we determined whether the MEK inhibitor PD98059 blunted IGF-I-induced REDD1 expression. IGF-I and Dex-induced REDD1 expression were both MEK/MAP kinase-independent.
Other investigators have found that the JNK kinase inhibitor SP600125 stimulates REDD1 expression but that this relates to the capacity of this compound to induce endoplasmic reticulum (ER) stress and enhance expression of the basic leucine zipper family transcription factor ATF4 rather than its activity as a JNK inhibitor [Jin et al., 2009]. ATF4 is both necessary and sufficient for the induction of REDD1 in response to ER stress. Although it is not presently known how IGF-I stimulates REDD1, insulin increases the expression of ATF4 in mouse L cells and C2C12 myotubes and the induction is rapamycin sensitive [Adams, 2007]. Our preliminary data suggest that IGF-I-induced REDD1 is also blunted by rapamycin suggesting that IGF-I, like insulin, may also induce ATF4 in myotubes and consequently stimulate REDD1 (unpublished observation)[Malmberg and Adams, 2008].
REDD1 is normally increased by stress conditions and may make cells more prone to cell death induced by reactive oxygen species (ROS). Ellisen showed that epithelial cells transfected with REDD1 have a dramatic increase in ROS-induced death when treated with H2O2 [Ellisen et al., 2002]. Growth hormone and IGF-I increase ROS in some insulin sensitive tissues by increasing the activity of the NADPH oxidase subunit Nox4 and decreasing antioxidant defenses [Brown-Borg and Rakoczy, 2003; Goldstein et al., 2005]. It is not known whether IGF-I induced REDD1 expression alters the sensitivity of myotubes to ROS but this may explain some of the paradoxical effects of IGF-I in aging skeletal muscle [Fulle et al., 2005]
REDD1 over expression has universally been found to suppress mTOR signaling and decrease cell size but few studies have directly determined its effect on protein synthesis [Sofer et al., 2005]. We determined whether endogenous REDD1 expression might alter protein synthesis as measured by the incorporation of [3H] phenylalanine into protein during a 1 h window between 5–6 h post IGF-I addition to myotubes. The rationale for the timing of this study was that by 5–6 h the transient IGF-I induced increase in mTOR activity would have returned to baseline and REDD1 expression would be maximal. In contrast to our prediction, protein synthesis was 15 % higher in cells treated with IGF-I despite their elevated level of REDD1. Likewise, when IGF-I was removed from the media protein synthesis was reduced to half of that seen in the presence of IGF-I despite the reduced REDD1 content.
One potential explanation of this paradox is that at later time points IGF-I may enhance translation elongation but not translation initiation and therefore the overall measurement of protein synthesis (phenylalanine incorporation) remains high. This would be consistent with IGF-I inhibiting both eEF2 kinase and the subsequent phosphorylation of eEF2 on T54 to foster translation elongation [Wang et al., 2001]. In contrast, protein synthesis may decrease upon IGF-I withdrawal because translation elongation is restrained in the absence of IGF-I.
A second potential confounding factor on REDD1 activity may be brought about by IGF-I induced changes in REDD2. REDD2 also represses mTOR activity and we report herein that REDD2 mRNA is decreased by IGF-I in muscle under both in vivo and in vitro conditions. Because REDD2 is highly expressed in skeletal muscle a relatively small decrease in REDD2 might compensate for the relatively large change observed in REDD1 (3-fold). In contrast, over expression of REDD1 or -2 in cell lines may overwhelm the endogenous balance of the factors that control TSC1-TSC2 and have a dominant negative effect on mTOR activity.
In conclusion, the present work demonstrates that IGF-I rapidly increases REDD1 mRNA and protein in the gastrocnemius and myotubes in culture. The increase in REDD1 is PI3K dependent and requires ongoing transcription and translation. IGF-I also increases REDD1 promoter activity. Chronic IGF-I exposure is necessary to induce REDD1 and IGF-I and dexamethasone have an additive effect on REDD1 expression. Finally, although there are experimental conditions where over expression of REDD1 is associated with a reduction in cell size and mTOR activity the IGF-I-induced increase in REDD1 does not reduce protein synthesis in myotubes. As a consequence, the physiological relevance of the IGF-I induced increase in REDD1 we observe remains to be elucidated.
Acknowledgments
We thank Dr. Scot Kimball for providing the REDD2 QT-RT-PCR primers, Alex Tuckow for help in interpreting the QT-RT-PCR results and Gerald Nystrom for his excellent technical contributions. This work was supported in part by the National Institutes of Health Grants GM-38032 and AA-11290.
This work was supported in part by the National Institutes of Health Grants GM-38032 and AA-11290.
References
- Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282:16744–53. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- Adi S, Cheng ZQ, Zhang PL, Wu NY, Mellon SH, Rosenthal SM. Opposing early inhibitory and late stimulatory effects of insulin-like growth factor-I on myogenin gene transcription. J Cell Biochem. 2000;78:617–26. [PubMed] [Google Scholar]
- Bark TH, McNurlan MA, Lang CH, Garlick PJ. Increased protein synthesis after acute IGF-I or insulin infusion is localized to muscle in mice. Am J Physiol. 1998;275:E118–23. doi: 10.1152/ajpendo.1998.275.1.E118. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Ruegg MA. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–24. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Rakoczy SG. Growth hormone administration to long-living dwarf mice alters multiple components of the antioxidative defense system. Mech Ageing Dev. 2003;124:1013–24. doi: 10.1016/j.mad.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–89. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–9. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280:9769–72. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- Criswell DS, Booth FW, DeMayo F, Schwartz RJ, Gordon SE, Fiorotto ML. Overexpression of IGF-I in skeletal muscle of transgenic mice does not prevent unloading-induced atrophy. Am J Physiol. 1998;275:E373–9. doi: 10.1152/ajpendo.1998.275.3.e373. [DOI] [PubMed] [Google Scholar]
- Dardevet D, Sornet C, Savary I, Debras E, Patureau-Mirand P, Grizard J. Glucocorticoid effects on insulin- and IGF-I-regulated muscle protein metabolism during aging. J Endocrinol. 1998;156:83–9. doi: 10.1677/joe.0.1560083. [DOI] [PubMed] [Google Scholar]
- Dehoux M, Gobier C, Lause P, Bertrand L, Ketelslegers JM, Thissen JP. IGF-I does not prevent myotube atrophy caused by proinflammatory cytokines despite activation of Akt/Foxo and GSK-3beta pathways and inhibition of atrogin-1 mRNA. Am J Physiol Endocrinol Metab. 2007;292:E145–50. doi: 10.1152/ajpendo.00085.2006. [DOI] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–51. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Fujita S, Abe T, Dreyer HC, Volpi E, Rasmussen BB. Human muscle gene expression following resistance exercise and blood flow restriction. Med Sci Sports Exerc. 2008;40:691–8. doi: 10.1249/MSS.0b013e318160ff84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Rasmussen BB. Leucine-enriched nutrients and the regulation of mammalian target of rapamycin signalling and human skeletal muscle protein synthesis. Curr Opin Clin Nutr Metab Care. 2008;11:222–6. doi: 10.1097/MCO.0b013e3282fa17fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop EA, Tee AR. Mammalian target of rapamycin complex 1: signalling inputs, substrates and feedback mechanisms. Cell Signal. 2009;21:827–35. doi: 10.1016/j.cellsig.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- Frost RA, Lang CH. Protein kinase B/Akt: a nexus of growth factor and cytokine signaling in determining muscle mass. J Appl Physiol. 2007;103:378–87. doi: 10.1152/japplphysiol.00089.2007. [DOI] [PubMed] [Google Scholar]
- Frost RA, Lang CH. Regulation of muscle growth by pathogen-associated molecules. J Anim Sci. 2008;86:E84–93. doi: 10.2527/jas.2007-0483. [DOI] [PubMed] [Google Scholar]
- Frost RA, Nystrom GJ, Lang CH. Multiple Toll-like receptor ligands induce an IL-6 transcriptional response in skeletal myocytes. Am J Physiol Regul Integr Comp Physiol. 2006;290:R773–84. doi: 10.1152/ajpregu.00490.2005. [DOI] [PubMed] [Google Scholar]
- Fulle S, Belia S, Di Tano G. Sarcopenia is more than a muscular deficit. Arch Ital Biol. 2005;143:229–34. [PubMed] [Google Scholar]
- Giegerich R, Meyer F, Schleiermacher C. GeneFisher--software support for the detection of postulated genes. Proc Int Conf Intell Syst Mol Biol. 1996;4:68–77. [PubMed] [Google Scholar]
- Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37:1974–84. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Mahadev K, Wu X. Redox paradox: insulin action is facilitated by insulin-stimulated reactive oxygen species with multiple potential signaling targets. Diabetes. 2005;54:311–21. doi: 10.2337/diabetes.54.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge DC. Signaling pathways weigh in on decisions to make or break skeletal muscle. Curr Opin Clin Nutr Metab Care. 2004;7:443–50. doi: 10.1097/01.mco.0000134364.61406.26. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–80. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–22. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HO, An S, Lee HC, Woo SH, Seo SK, Choe TB, Yoo DH, Lee SB, Um HD, Lee SJ, Park MJ, Kim JI, Hong SI, Rhee CH, Park IC. Hypoxic condition- and high cell density-induced expression of Redd1 is regulated by activation of hypoxia-inducible factor-1alpha and Sp1 through the phosphatidylinositol 3-kinase/Akt signaling pathway. Cell Signal. 2007;19:1393–403. doi: 10.1016/j.cellsig.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Jin HO, Seo SK, Woo SH, Kim ES, Lee HC, Yoo DH, Choe TB, Hong SI, Kim JI, Park IC. SP600125 negatively regulates the mammalian target of rapamycin via ATF4-induced Redd1 expression. FEBS Lett. 2009;583:123–7. doi: 10.1016/j.febslet.2008.11.035. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Do AN, Kutzler L, Cavener DR, Jefferson LS. Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. J Biol Chem. 2008;283:3465–75. doi: 10.1074/jbc.M706643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang CH, Frost RA, Vary TC. Regulation of muscle protein synthesis during sepsis and inflammation. Am J Physiol Endocrinol Metab. 2007a;293:E453–9. doi: 10.1152/ajpendo.00204.2007. [DOI] [PubMed] [Google Scholar]
- Lang CH, Frost RA, Vary TC. Acute alcohol intoxication increases REDD1 in skeletal muscle. Alcohol Clin Exp Res. 2008;32:796–805. doi: 10.1111/j.1530-0277.2008.00637.x. [DOI] [PubMed] [Google Scholar]
- Lang CH, Huber D, Frost RA. Burn-induced increase in atrogin-1 and MuRF-1 in skeletal muscle is glucocorticoid independent but downregulated by IGF-I. Am J Physiol Regul Integr Comp Physiol. 2007b;292:R328–36. doi: 10.1152/ajpregu.00561.2006. [DOI] [PubMed] [Google Scholar]
- Lin L, Qian Y, Shi X, Chen Y. Induction of a cell stress response gene RTP801 by DNA damaging agent methyl methanesulfonate through CCAAT/enhancer binding protein. Biochemistry. 2005;44:3909–14. doi: 10.1021/bi047574r. [DOI] [PubMed] [Google Scholar]
- Malmberg SE, Adams CM. Insulin signaling and the general amino acid control response. Two distinct pathways to amino acid synthesis and uptake. J Biol Chem. 2008;283:19229–34. doi: 10.1074/jbc.M801331200. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Esser KA. REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch. Am J Physiol Cell Physiol. 2009b;296:C583–92. doi: 10.1152/ajpcell.00464.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J, Ruegg MA, Hall MN, Meyuhas O. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol. 2009;29:640–9. doi: 10.1128/MCB.00980-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–65. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- Rosner M, Hanneder M, Siegel N, Valli A, Hengstschlager M. The tuberous sclerosis gene products hamartin and tuberin are multifunctional proteins with a wide spectrum of interacting partners. Mutat Res. 2008;658:234–46. doi: 10.1016/j.mrrev.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Schakman O, Gilson H, de Coninck V, Lause P, Verniers J, Havaux X, Ketelslegers JM, Thissen JP. Insulin-like growth factor-I gene transfer by electroporation prevents skeletal muscle atrophy in glucocorticoid-treated rats. Endocrinology. 2005;146:1789–97. doi: 10.1210/en.2004-1594. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Kimball SR, Jefferson LS. Among translational effectors, p70S6k is uniquely sensitive to inhibition by glucocorticoids. Biochem J. 2000;347:389–97. doi: 10.1042/0264-6021:3470389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton JR, Baker BL, Thorburn A. Dexamethasone inhibits insulin-like growth factor signaling and potentiates myoblast apoptosis. Endocrinology. 2000;141:2945–50. doi: 10.1210/endo.141.8.7621. [DOI] [PubMed] [Google Scholar]
- Smith GI, Atherton P, Villareal DT, Frimel TN, Rankin D, Rennie MJ, Mittendorfer B. Differences in muscle protein synthesis and anabolic signaling in the postabsorptive state and in response to food in 65–80 year old men and women. PLoS ONE. 2008;3:e1875. doi: 10.1371/journal.pone.0001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–45. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Vary TC, Lynch CJ. Nutrient signaling components controlling protein synthesis in striated muscle. J Nutr. 2007;137:1835–43. doi: 10.1093/jn/137.8.1835. [DOI] [PubMed] [Google Scholar]
- Velloso CP. Regulation of muscle mass by growth hormone and IGF-I. Br J Pharmacol. 2008;154:557–68. doi: 10.1038/bjp.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab. 2008;295:E785–97. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Wu X, Guan KL, Han M, Zhuang Y, Xu T. Muscle atrophy in transgenic mice expressing a human TSC1 transgene. FEBS Lett. 2006;580:5621–7. doi: 10.1016/j.febslet.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem. 2006;281:39128–34. doi: 10.1074/jbc.M610023200. [DOI] [PubMed] [Google Scholar]
- Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–44. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. Embo J. 2001;20:4370–9. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem Biophys Res Commun. 2009;379:451–5. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]