Abstract
Recent findings suggest that nuclear IGF1R binds to enhancer regions and functions as a transcriptional cofactor. However, the downstream transcriptional regulators of this pathway remain to be defined. Here, we show that nuclear IGF1R associates with the transcription factor LEF1 and increases promoter activity of LEF1 downstream target genes cyclin D1 and axin2. Furthermore, nuclear IGF1R augments protein levels of cyclin D1 and axin2. Our findings suggest a novel function for IGF1R, thus further emphasizing the important role of this receptor in cancer biology.
Keywords: gene activation, LEF1, nuclear IGF1R
Introduction
The Wnt and insulin-like growth factor-1 receptor (IGF1R) signalling pathways function in numerous developmental processes, and alterations of both signalling pathways are associated with common pathological conditions, including cancer [1, 2].
Activation of the Wnt signalling pathway leads to the stabilization and nuclear translocation of β-catenin. Complex formation of β-catenin with the T cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors activates the transcription of target genes, several of which are known to be important for human tumourigenesis such as axin2 [3] and cyclin D1 [4]. LEF1 is unable to activate transcription on its own, but it can activate transcription in collaboration with other factors, such as β-catenin in response to Wnt signalling or cofactor ALY independent of Wnt signalling [5, 6]. In unstimulated cells, LEF1 can associate with the corepressor Groucho to repress Wnt-responsive genes [7].
IGF1R is overexpressed in many human cancers [8, 9], and its activation leads to downstream activation of the phosphatidylinositol 3′-kinase (PI3K)–Akt pathway [10] and the mitogen-activated protein kinase–Erk pathway [11, 12].
There is accumulating evidence for the interaction between IGF1R and Wnt oncogenic pathways. IGFs are known to cause translocation of β-catenin to the nucleus, where it activates the target genes [13, 14, 15, 16]. Insulin receptor substrate-1 (IRS1), a docking protein important for the activation of IGF1R-mediated signalling pathways, can upon IGF1 stimulation be translocated to the nucleus [17]. IRS1 is required for IGF1-mediated nuclear translocation of β-catenin [18]. Furthermore, nuclear IRS1 activates the rDNA and TCF/LEF promoters [19]. This promoter activation is PI3K-independent but is IGF1-dependent [19]. We recently demonstrated that IGF1 stimulates the SUMOylation of IGF1R at three evolutionarily conserved lysine residues—Lys1025, Lys1100 and Lys1120—in the β-subunit of the receptor. Mutation of these lysine residues blocks SUMOylation of the receptor and prevents its accumulation in the nucleus but does not interfere with its endocytosis or activation of the PI3K or mitogen-activated protein kinase pathways [20]. SUMOylation is a prerequisite for the nuclear translocation of IGF1R; however, SUMOylated IGF1R is predominantly localized perinuclearly and at the nuclear membrane. The SUMO-modified IGF1R is deSUMOylated after passage across the nuclear membrane [20]. Aleksic et al [21] reported high levels of nuclear IGF1R (nIGF1R) in primary renal cancer cells, preinvasive lesions in the breast and in proliferative nonmalignant tissues. In renal cancer, presence of nIGF1R was associated with poor prognosis [21]. The molecular mechanism by which nIGF1R mediates its function remains unknown. In our recent work we also demonstrated that nIGF1R binds to putative enhancer sites in genomic DNA and drives transcription of target genes [20]. On the basis of our findings and the mentioned reports, we asked whether nIGF1R could bind to the TCF–LEF complex and affect the expression of its targeted genes.
In this report, we demonstrate that IGF1R associates with LEF1 in the cell nucleus. Downstream target genes of TCF/LEF transcription factors, axin2 and cyclin D1 reporter promoters, responded to the addition of wild type (wt)-IGF1R with an increase in transcription. Elevated protein levels of axin2 and cyclin D1 were also detectable after wt-IGF1R overexpression. These promoter activations and protein inductions are absent when cells are transfected with the mutated form of IGF1R that cannot be SUMOylated and translocated to the cell nucleus.
Results And Discussion
IGF1R colocalizes with LEF1 in the cell nucleus
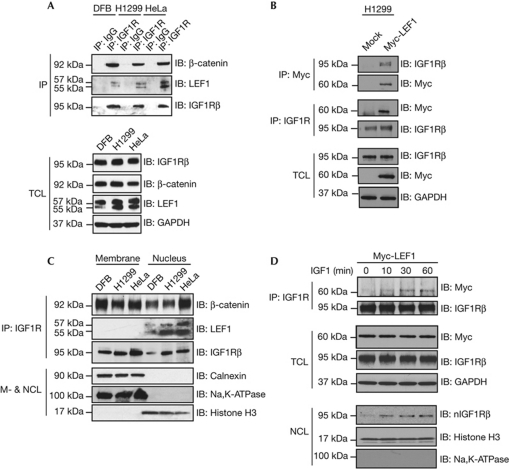
Interaction between β-catenin and IGF1R has been demonstrated in vitro [22] and in vivo [15]. We confirmed IGF1R–β-catenin association in human melanoma (DFB), nonsmall lung carcinoma (H1299) and human cervical carcinoma (HeLa) cell lines by co-immunoprecipitation (co-IP) (Fig 1A). On the basis of the recent discovery of nuclear translocation of IGF1R [20] and the fact that β-catenin forms a transcriptional complex with the TCF/LEF family of transcription factors, we investigated whether IGF1R could also associate with LEF1. As shown in Fig 1A, IGF1R co-IPs with endogenous LEF1. The specificity of the co-IP bands was confirmed by IP with IgG (Fig 1A). The expression levels of IGF1R, β-catenin and LEF1 in total cell lysate (TCL) are shown in Fig 1A, lower panel. To confirm IGF1R–LEF1 association, we next performed co-IP experiments using transiently transfected H1299 cells with Myc-LEF1. IGF1R was found to co-precipitate with Myc-LEF1. The reciprocal co-IP experiment also showed that Myc-LEF1 can be found in the IGF1R precipitate (Fig 1B).
Figure 1.
IGF1R colocalizes with β-catenin and LEF1. (A) IGF1R was immunoprecipitated from total cell lyaste and subjected to immunoblotting with anti-β-catenin and anti-LEF1 in DFB, H1299 and HeLa cells. Blots were stripped and incubated with anti-IGF1R to confirm equal loading. Rabbit IgG was used as a negative control. Expression levels of IGF1R, β-catenin and LEF1 in total cell lysate of abovementioned samples were analysed by IB. GAPDH was used as loading control. (B) Reciprocal IP of total cell lysate from H1299 cells, transiently transfected with Myc-LEF1, using anti-IGF1R and anti-Myc, was conducted followed by IB as indicated. Transfection efficiency of Myc-LEF1 and expression levels of IGF1R was analysed by IB (bottom). GAPDH was used as loading control. (C) Membrane and nuclear cell lysates (M- & NCL) of cells used in A were IPd with anti-IGF1R and were analysed by anti-β-catenin and anti-LEF1. Blots were stripped and incubated with anti-IGF1R to confirm equal loading. Fractions were analysed for the presence of markers of the plasma membrane (Na, K-ATPase), the ER (calnexin) and nucleus (histone H3). (D) Serum-starved H1299 cells transiently transfected with Myc-LEF1 were stimulated with IGF1 for the indicated times, IPd with anti-IGF1R and analysed by IB with anti-Myc, after which the blots were stripped and then incubated with anti-IGF1R to confirm equal loading. Transfection efficiency of Myc-LEF1 and expression levels of IGF1R were analysed by IB. Serum-starved H1299 cells were stimulated with IGF1 for the indicated times, after which they were fractionated and the nuclear fraction was analysed by IB for the presence of IGF1R. ER, endoplasmic reticulum; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IGF1R, insulin-like growth factor-1 receptor; IgG, immunoglobulin; Ipd, immunoprecipitated; LEF, lymphoid enhancer factor; TCL, total cell lysate.
To determine the subcellular localization of the complexes, IP was performed after fractionation of the cells. Although IGF1R colocalizes with β-catenin both in the membrane and nuclear fractions, LEF1 colocalization with IGF1R occurs exclusively in the nucleus (Fig 1C). Through the use of markers for plasma membrane (Na+- and K+-dependent adenosine triphosphatase, Na+, K+-ATPase), endoplasmic reticulum (calnexin) and nucleus (histone H3) we could confirm adequate purity of the fractions (Fig 1C). SUMOylation and nuclear localization of IGF1R under basal conditions in all the three cell lines were analysed confirming our previous publication (supplementary Fig S1A,B online).
To investigate the ligand-dependency of LEF1–IGF1R association, serum-starved H1299 cells transiently transfected with Myc-LEF1 were stimulated with IGF1 for indicated times. IGF1R was immunoprecipitated from TCLs and analysed by blotting for anti-Myc (Fig 1D). The IGF1R–Myc-LEF1 co-IP band appeared already after 10 min stimulation and increased at later time points (30–60 min). As previously shown, nuclear translocation of IGF1R is ligand-dependent [20, 21]. The kinetics of nIGF1R accumulation in H1299 cells was determined. Serum-starved cells, which were devoid of nIGF1R, accumulated nIGF1R 10–60 min after addition of ligand (Fig 1D, lower panel) correlating with IGF1R–LEF1 association and demonstrating the spatiotemporal nature of this interaction.
IGF1R–LEF1 association does not involve IRS1 or β-catenin
Previous studies have established that IGF1 stimulation enhances tyrosine phosphorylation of β-catenin and IRS1, and that this phosphorylation rapidly dissociates β-catenin from E-cadherin at the plasma membrane followed by relocation to the cellular cytoplasm [14]. Furthermore, it is shown that the IGF1 stimulation translocates IRS1 and β-catenin to the nucleus and activates the TCF/LEF reporter [18]. Therefore, the next question we addressed was whether β-catenin and IRS1 are involved and/or required for the IGF1R–LEF1 association.
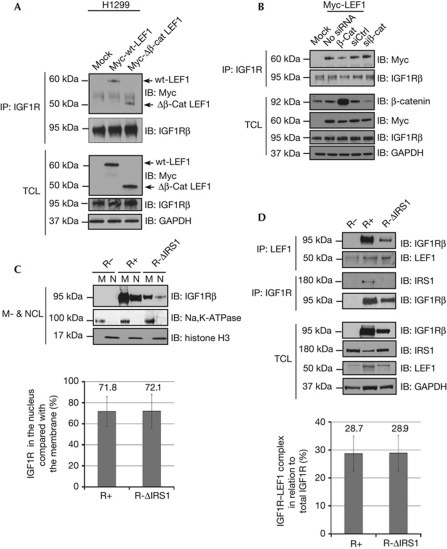
To determine whether β-catenin is required for IGF1R–LEF1 association, co-IP experiments were carried out using H1299 cells transfected with Mock, Myc-wt-LEF1 and LEF1 mutant lacking the β-catenin binding site (Myc-Δβ-cat-LEF1). Surprisingly, Myc-Δβ-cat-LEF1 did not augment the IGF1R–LEF1 association (Fig 2A), indicating that the IGF1R binding site on LEF1 is distinct from the β-catenin binding domain. Furthermore, the IGF1R–LEF1 association was neither compromised upon small interfering RNA-mediated β-catenin knockdown nor was it affected by β-catenin overexpression (Fig 2B), indicating that β-catenin is not required for IGF1R–LEF1 interaction. Fig 2A,B, lower panels, show levels of β-catenin, IGF1R and Myc-LEF1 proteins in TCL analysed by western blot.
Figure 2.
IGF1R colocalizes with LEF1 independent of β-catenin and IRS1. (A) Total cell lysate from H1299 cells transiently transfected with wt and mutant LEF1, which lacks the β-catenin binding domain, were subjected to IP with anti-IGF1R and were analysed by IB with anti-Myc. Blots were stripped and incubated with anti-IGF1R to confirm equal loading. Expression levels of IGF1R and Myc-LEF1 constructs in TCL were analysed by IB. GAPDH was used as loading control. (B) H1299 cells co-transfected Myc-LEF1 and siRNA targeting β-catenin, control siRNA or plasmids encoding β-catenin were IPd with anti-IGF1R and were analysed by IB using anti-Myc. Transfection efficacy was confirmed by anti-β-catenin and anti-Myc blotting. Expression levels of IGF1R and GAPDH were analysed by IB. (C) IGF1R null cells stably expressing either wt-IGF1R or Y950F-IGF1R, unable to bind IRS1, (R-ΔIRS1) were fractionated and membrane and nuclear fractions were analysed by anti-IGF1R. R− cells were used as negative control. Histone H3 and Na+/K+-ATPase were used as a nuclear and membrane markers, respectively. Percentage densitometric quantification of nuclear IGF1R similar to those shown in C normalized to membrane IGF1R is shown (bottom). Means and s.d.'s (n=3 experiments) are indicated. (D) Total cell lysate from cells in C were co-IPd with anti-LEF1, detected with anti-IGF1R or co-IPd with anti-IGF1R and detected with anti-IRS1. Blots were stripped and incubated with anti-LEF1 and anti-IGF1R, respectively, to confirm equal loading. Expression levels of IGF1R, IRS1 and LEF1 in TCL were analysed by IB. GAPDH was used as loading control. The graph shows percentage of densitometric quantification of IGF1R–LEF1 co-IP similar to those shown in D normalized against IPd LEF1 and compared with total IGF1R, normalized to GAPDH. Means and s.d.s (n=3 experiments) are indicated. β-cat, plasmid encoding β-catenin; Δβ-cat-LEF1, β-catenin binding domain; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IGF1R, insulin-like growth factor-1 receptor; IP, immunoprecipitation; IRS1, insulin receptor substrate-1; LEF1, lymphoid enhancer factor 1; M, membrane fraction; NCL, nuclear cell lysate; R−, IGF1R null cells; R+, IGF1R null cells expressing wt-IGF1R; siβ-cat, small interfering RNA (siRNA) targeting β-catenin; siCtrl, control siRNA; wt, wild type.
To assess whether IRS1 functions as a scaffold for IGF1R–LEF1 association, we used fibroblasts derived from IGF1R-null mice (R− cells), which were stably transfected with wt-IGF1R (R+) and a mutant IGF1R in which the putative IRS1 binding site had been mutated (R-ΔIRS1). R_ cells were used as negative control. In R-ΔIRS1 cells, the IGF1R tyrosine at amino acid 950, which is the major binding site for IRS1 [23, 24], has been mutated to phenylalanine (Y950F). The inability of Y950F to bind to IRS1 was confirmed by co-IP experiments using IGF1Rβ antibody followed by a western blot analysis with anti-IRS1 (Fig 2D, middle panel). As nuclear localization of IGF1R is essential for IGFR1–LEF1 association (Fig 1), R+ and R-ΔIRS1 cells were fractionated and analysed by western blotting to determine the presence of nIGF1Rβ in these cells (Fig 2C). The association of Y950F-IGF1R with LEF1 was also assessed (Fig 2D, upper panel). Altogether, our data show that Y950F-IGF1R, which is unable to bind IRS1, translocates to the nucleus and binds LEF1, demonstrating that IRS1 is not required for IGF1R nuclear translocation and LEF1 association. Levels of IGF1R, IRS1 and LEF1 in TCL derived from R_, R+ and R-ΔIRS1 cells were determined (Fig 2D, lower panel). The graphs show the quantified and normalized signals of nIGF1R and IGF1R–LEF1 association in R+ and R-ΔIRS1 cells based on three independent experiments.
nIGF1R increases axin2 and cyclin D1 promoter activity
To distinguish the nuclear and non-nuclear activities of IGF1R, we used the tri-SUMO-site mutant (TSM) IGF1R construct. As previously reported, TSM-IGF1R has the same internalization and signalling properties as wt-IGF1R but it does not translocate to the nucleus because of the omitted receptor SUMOylation sites [20]. Signalling and nuclear translocation of wt- and TSM-IGF1R were reconfirmed by transfecting IGF1R-deficient leiomyosarcoma SKUT1 cells [25] with wt- and TSM-IGF1R plasmids, as shown in supplementary Fig S1C online and Fig 3A. Supplementary Fig S1C online shows the comparable wt- and TSM-IGF1R signalling as measured by Akt and Erk phosphorylation in response to IGF1 stimulation and Fig 3A shows that despite equal expression levels in TCL, the nuclear fraction is abundant with wt- but not with TSM-IGF1R. This provides us with a system where we can investigate the effects that are specific to nuclear localization of IGF1R.
Figure 3.
IGF1R overexpression increases TCF/LEF activity. (A) Membrane and nuclear cell lysates of SKUT1 cells, transiently transfected with mock, wt- and TSM-IGF1R, were analysed for IGF1R by immunoblotting. Histone H3 was used as a nuclear marker. (B) IGF1R and LEF1 complexes were detected by in situ proximity ligation assay in cells described in A. Cells were counterstained with Hoechst (blue) to visualize nuclei and the IGF1R–LEF1 interaction was visualized as red dots. (C) Total cell lysate from cells described in A and B were subjected to IP with anti-LEF1 and analysed by IB with anti-IGF1R. Transfection efficacy of wt- and TSM-IGF1R was confirmed by analysing TCL with anti-IGF1R. Expression levels of LEF1 and GAPDH in TCL were analysed by IB. (D) Cells described in panels A–C were co-transfected with either cyclin D1 or axin2 promoter reporters and a Renilla luciferase as an internal transfection control. Promoter activity was defined as firefly luciferase activity/Renilla luciferase activity. The data shown represent averages of four independent experiments. Analysis of variance was performed using the normalized raw data, P<0.05. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IGF1R, insulin-like growth factor-1 receptor; IP, immunoprecipitation; LEF, lymphoid enhancer factor; M, membrane fraction; NCL, nuclear cell lysate; PLA, proximity ligation assay; TCF, T cell factor; TCL, total cell lysate; TSM, tri-SUMO-site mutant; wt, wild type.
IGF1R–LEF1 association and its subcellular localization were analysed by in situ proximity ligation assays (PLAs; Fig 3B). The PLA assay allows visualization of the interaction between two endogenous proteins in fixed cells strictly dependent on the simultaneous recognition of the target by two antibodies (see Methods). As shown in Fig 3B, wt-IGF1R binds to LEF1 exclusively in the cell nucleus but TSM-IGF1R does not bind to LEF1. LEF1–wt-IGF1R interaction and the lack of LEF1–TSM-IGF1R interaction was confirmed by co-IP experiments as shown in Fig 3C. Altogether, these data emphasize the importance of nuclear localization of IGF1R for IGF1R–LEF1.
Considering that cyclin D1 [26] and axin2 [27] genes are direct targets for transactivation by LEF1, we next investigated the activity of cyclin D1 and axin2 promoters using promoter-luciferase reporter constructs in the wt- and TSM-IGF1R transfected cells. Upon wt-IGF1R overexpression, the activity of cyclin D1 and axin2 promoters increased by 17% and 22%, respectively. In cells expressing TSM-IGF1R, the cyclin D1 and axin2 promoter activities decreased with 7% and 20%, respectively, compared with the control (Fig 3D). Taken together, the cyclin D1 and axin2 reporter data suggest that nIGF1R increases LEF1 downstream activity.
nIGF1R binds to genomic cyclin D1 promoter
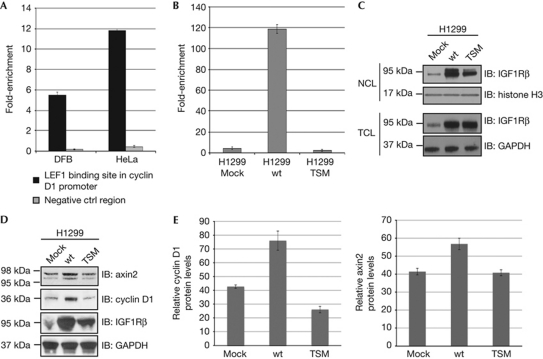
As a proof-of-concept the proposed assembly of IGF1R with LEF1 on the endogenous promoter for cyclin D1 was tested by chromatin IP (ChIP)–qPCR. On the basis of the possibility that the IGF1R-deficient SKUT1 cells might not contain the enhancer regions to which nIGF1R binds, we used cell lines with endogenous IGF1R expression. Chromatin fragments from DFB and HeLa cell lysates, which co-IPd with IGF1R antibody, were analysed by qPCR to detect the genomic regions containing the TCF/LEF binding element in the cyclin D1 promoter. Fig 4A shows that the binding of IGF1R to the LEF1 binding site in the cyclin D1 promoter region is nearly sixfold higher in DFB cells and about 12-fold higher in HeLa cells compared with a nonspecific control region.
Figure 4.
Nuclear IGF1R increases protein levels of axin2 and cyclin D1. (A) Increase in binding of IGF1R to the LEF1 binding site in the cyclin D1 promoter measured as fold-enrichment after ChIP followed by qPCR analysis in DFB and HeLa cells compared with its binding to an unspecific DNA control region. (B) Increase in binding of IGF1R to the LEF1 binding site in the cyclin D1 promoter measured as fold-enrichment after ChIP followed by qPCR analysis in H1299 cells transiently transfected with mock, wt- and TSM-IGF1R. (C) Cells as described in B were fractionated and the total cell lysate and nuclear cell lysate were analysed for the content of IGF1R by immunoblotting. Histone H3 was used as loading control for NCL and GAPDH was used as loading control for TCL. (D) Expression levels of axin2 and cyclin D1 in TCL of cells described in panels B and C were analysed by IB. GAPDH was used as loading control. (E) Densitometric ratio of cyclin D1/GAPDH (left) and axin2/GAPDH (right) are shown in D. Means and s.d.s (n=3 experiments) are shown. Analysis of variance was performed using the normalized raw data, P<0.05. ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IGF1R, insulin-like growth factor-1 receptor; IP, immunoprecipitation; LEF1, lymphoid enhancer factor 1; M, membrane fraction; NCL, nuclear cell lysate; TSM, tri-SUMO-site mutant; TCL, total cell lysate; wt, wild type.
Owing to high transfectability of the H1299 cells we were able to compare the binding of IGF1R with genomic cyclin D1 in H1299 cells transfected with mock, wt- and TSM-IGF1R. The abundance of nIGF1R was greater in cells transfected with wt-IGF1R than in cells transfected with either the empty vector or TSM-IGF1R (Fig 4C, upper panel). The levels of IGF1R in TCL are shown in Fig 4C, lower panel. We detected a nearly 120-fold increase in IGF1R binding to the LEF1 binding site of the cyclin D1 promoter in cells overexpressing wt-IGF1R compared with cells overexpressing TSM-IGF1R, suggesting that nIGF1R binds to the LEF1 binding site in the cyclin D1 promoter region (Fig 4B). TSM-IGF1R does not translocate to the nucleus in SKUT1 cells that are IGF1R deficient (Fig 3A). In contrast, H1299 cells transfected with TSM-IGF1R have increased levels of nIGF1R (Fig 4C). One possibility could be that TSM-IGF1R hetrodimerizes with endogenous receptor and therefore can be translocated to the nucleus. However, TSM-IGF1R transfection decreased the level of endogenous IGF1R binding to genomic cyclin D1 promoter, suggesting that although increased level of nIGF1R is detected, the function of endogenous IGF1R is impaired upon TSM-IGF1R transfection. One could speculate that the hetrodimerization of endogenous IGF1R with TSM-IGF1R might change the DNA binding capabilities of the endogenous IGF1R.
nIGF1R increases axin2 and cyclin D1 protein levels
To investigate whether the observed effects of nIGF1R on reporter assays, as shown in Fig 3D, could affect the expression of the TCF/LEF complex gene targets, H1299 cells were transfected with wt- and TSM-IGF1R and subsequently analysed by western blotting for changes in cyclin D1 and axin2 protein levels. Consistent with reporter assay experiments, wt-IGF1R increased the protein levels of axin2 and cyclin D1 (Fig 4D). Protein levels of cyclin D1 and axin2 are somewhat more significantly increased than the effects seen by reporter experiments. TSM-IGF1R decreased protein expression of cyclin D1 but had no significant effect on axin2 protein expression, which could be due to other mechanisms involved in regulating protein levels. Quantified data of axin2 and cyclin D1 protein levels, normalized to glyceraldehyde 3-phosphate dehydrogenase, are shown in Fig 3E.
Taken together, our data suggest that, apart from its classical tyrosine kinase activity, IGF1R binds to the LEF1 transcription factor in the nucleus, leading to elevated protein levels of cyclin D1 and axin2. This might be an additional molecular mechanism by which IGF1R promotes uncontrolled cell proliferation and thus contributes to the neoplastic transformation of cells. Future studies will be directed at further detailing the molecular mechanisms by which nIGF1R affects gene expression.
Methods
Duolink in situ proximity ligation assay. SKUT1 cultured on coverslips were fixed with 4% paraformaldehyde. Fixed cells were permeabilized with 0.2% Triton for 30 min followed by 30 min blocking with blocking buffer (5% bovine serum albumin, 5% Donkey serum, 0.3% Triton in phosphate-buffered saline). The cells were incubated with primary antibodies overnight at room temperature in a humidity chamber. Duolink in situ proximity ligation assay was conducted according to the manufacturer's protocol (OLINK Bioscience, Uppsala, Sweden) using PLA probe anti-mouse minus and PLA probe anti-rabbit plus.
Chromatin IP. ChIP assay was performed using ChIP assay kit (Upstate Biotechnology) according to the manufacturer's protocol. Briefly, chromatin was sonicated in a BioRupter to an average size of 300 bp. Rabbit IGF1R or IgG control antibodies were used for IP. Quantification of immunoprecipitated DNA was performed by qPCR using ABI PRISM 7500 Sequence Detection System and ChampionChip qPCR Primers and RT2 SYBR Green Master Mix assay (SABiosciences, Frederick, MD, USA). The queried site primer detects the LEF1 binding site in the CYCLIN D1 promoter (Human CCND1, NM_053056.2 (-)01 kb) and the negative primers measure the relative amount of nonspecific DNA sequences that co-precipitates during the ChIP procedure (ChIP–qPCR Human IGX1A Negative Control). The cycling parameters used were one cycle at 95 °C for 10 min; 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The fold-change in occupancy (also known as fold-enrichment) was calculated by determining the IP efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and normalized to the level observed at a control region.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported by the Swedish Cancer Foundation, Swedish Research Council, Cancer Society in Stockholm, Children Cancer Society, Lundberg's Research Foundation in Gothenburg, Stockholm County Council and Karolinska Institutet.
Author contributions: D.W., S.S., S.A. and B.S. performed experiments. B.S. designed the experiments. D.W., S.S. and B.S. analysed the data. D.W., S.S., O.L. and B.S. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baserga R, Hongo A, Rubini M, Prisco M, Valentinis B (1997) The IGF-I receptor in cell growth, transformation and apoptosis. Biochim Biophys Acta 1332: F105–F126 [DOI] [PubMed] [Google Scholar]
- Werner H, Le Roith D (1997) The insulin-like growth factor-I receptor signaling pathways are important for tumorigenesis and inhibition of apoptosis. Crit Rev Oncog 8: 71–92 [DOI] [PubMed] [Google Scholar]
- Yan D et al. (2001) Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc Natl Acad Sci USA 98: 14973–14978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F (1999) Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422–426 [DOI] [PubMed] [Google Scholar]
- Giese K, Kingsley C, Kirshner JR, Grosschedl R (1995) Assembly and function of a TCR alpha enhancer complex is dependent on LEF1-induced DNA bending and multiple protein-protein interactions. Genes Dev 9: 995–1008 [DOI] [PubMed] [Google Scholar]
- Bruhn L, Munnerlyn A, Grosschedl R (1997) ALY, a context-dependent coactivator of LEF1 and AML-1, is required for TCRalpha enhancer function. Genes Dev 11: 640–653 [DOI] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y (1998) Transcriptional repression by AML1 and LEF1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA 95: 11590–11595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M (2008) Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer 8: 915–928 [DOI] [PubMed] [Google Scholar]
- Baserga R (2009) Customizing the targeting of IGF1 receptor. Future Oncol 5: 43–50 [DOI] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241 [DOI] [PubMed] [Google Scholar]
- Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Pawson T, Pelicci PG (1992) A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell 70: 93–104 [DOI] [PubMed] [Google Scholar]
- Sasaoka T, Rose DW, Jhun BH, Saltiel AR, Draznin B, Olefsky JM (1994) Evidence for a functional role of Shc proteins in mitogenic signaling induced by insulin, insulin-like growth factor-1, and epidermal growth factor. J Biol Chem 269: 13689–13694 [PubMed] [Google Scholar]
- Eastman Q, Grosschedl R (1999) Regulation of LEF1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol 11: 233–240 [DOI] [PubMed] [Google Scholar]
- Playford MP, Bicknell D, Bodmer WF, Macaulay VM (2000) IGF-1 regulates the location, stability, and transcriptional activity of beta-catenin. Proc Natl Acad Sci USA 97: 12103–12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morali OG, Delmas V, Moore R, Jeanney C, Thiery JP, Larue L (2001) IGF-II induces rapid beta-catenin relocation to the nucleus during epithelium to mesenchyme transition. Oncogene 20: 4942–4950 [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, Li G, Vaidya B, Patel D, Herlyn M (2001) IGF-I induces survival and growth of biologically early melanoma cells through both the mitogen-activated protein kinase and beta-catenin pathways. Cancer Res 61: 7318–7324 [PubMed] [Google Scholar]
- Prisco M, Santini F, Baffa R, Liu M, Drakas R, Wu A, Baserga R (2002) Nuclear translocation of insulin receptor substrate-1 by the simian virus 40 T antigen and the activated type 1 insulin-like growth factor receptor. J Biol Chem 277: 32078–32085 [DOI] [PubMed] [Google Scholar]
- Chen J, Wu A, Sun H, Drakas R, Garofalo C, Cascio S, Surmacz E, Baserga R (2005) Functional significance of type 1 insulin-like growth factor-mediated nuclear translocation of the insulin receptor substrate-1 and beta-catenin. J Biol Chem 280: 29912–29920 [DOI] [PubMed] [Google Scholar]
- Wu A, Chen J, Baserga R (2008) Nuclear insulin receptor substrate-1 activates promoters of cell cycle progression genes. Oncogene 27: 397–403 [DOI] [PubMed] [Google Scholar]
- Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J, Larsson O (2010) SUMOylation mediates the nuclear translocation and signaling of the IGF1 receptor. Sci Signal 3: ra10. [DOI] [PubMed] [Google Scholar]
- Aleksic T, Chitnis MM, Perestenko OV, Gao S, Thomas PH, Turner GD, Protheroe AS, Howarth M, Macaulay VM (2010) Type 1 insulin-like growth factor receptor translocates to the nucleus of human tumor cells. Cancer Res 70: 6412–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvakova MA, Surmacz E (1997) Overexpressed IGF-I receptors reduce estrogen growth requirements, enhance survival, and promote E-cadherin-mediated cell-cell adhesion in human breast cancer cells. Exp Cell Res 231: 149–162 [DOI] [PubMed] [Google Scholar]
- Tartare-Deckert S, Sawka-Verhelle D, Murdaca J, Van Obberghen E (1995) Evidence for a differential interaction of SHC and the insulin receptor substrate-1 (IRS-1) with the insulin-like growth factor-I (IGF-I) receptor in the yeast two-hybrid system. J Biol Chem 270: 23456–23460 [DOI] [PubMed] [Google Scholar]
- O'Connor R, Kauffmann-Zeh A, Liu Y, Lehar S, Evan GI, Baserga R, Blattler WA (1997) Identification of domains of the insulin-like growth factor I receptor that are required for protection from apoptosis. Mol Cell Biol 17: 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacca L, Mineo R, Pandini G, Murabito A, Vigneri R, Belfiore A (2002) In IGF-I receptor-deficient leiomyosarcoma cells autocrine IGF-II induces cell invasion and protection from apoptosis via the insulin receptor isoform A. Oncogene 21: 8240–8250 [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A (1999) The cyclin D1 gene is a target of the beta-catenin/LEF1 pathway. Proc Natl Acad Sci USA 96: 5522–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F (2002) Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22: 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.