Abstract
Cell cycle transitions depend on protein phosphorylation and dephosphorylation. The discovery of cyclin-dependent kinases (CDKs) and their mode of activation by their cyclin partners explained many important aspects of cell cycle control. As the cell cycle is basically a series of recurrences of a defined set of events, protein phosphatases must obviously be as important as kinases. However, our knowledge about phosphatases lags well behind that of kinases. We still do not know which phosphatase(s) is/are truly responsible for dephosphorylating CDK substrates, and we know very little about whether and how protein phosphatases are regulated. Here, we summarize our present understanding of the phosphatases that are important in the control of the cell cycle and pose the questions that need to be answered as regards the regulation of protein phosphatases.
Keywords: α-endosulfine, CDK, Greatwall, mitosis, phosphatase
See Glossary for abbreviations used in this article.
Glossary.
AGC kinases a family including PKA, PKG and PKC members
APC3 anaphase-promoting complex subunit 3
ARPP-16/-19 cyclic-AMP-regulated phosphoprotein of 16/19 kDa
CDK cyclin-dependent kinase
CSF cytostatic factor
DARPP-32 dopamine and cAMP-regulated phosphoproteins of 32 kDa
Ensa α-endosulfine
FEAR CDC fourteen early anaphase release
IC50 concentration that inhibits 50% of activity
INH inhibitor of post-translational activation of pre-MPF
MEN mitotic exit network
MPF maturation-promoting factor
PKA cyclic-AMP-activated protein kinase
PP1/2A type 1/2A protein phosphatase
RNAi RNA interference
TOR target of rapamycin
Background
After the discovery of CDK–cyclin complexes as the main regulators of the cell cycle [1, 2], various kinases were shown to have specialized functions during mitosis (Table 1). Analyses of their dynamic changes in activity and localization during the cell cycle, as well as the identification of their functional substrates significantly enhanced our understanding of how visible mitotic events—such as nuclear envelope breakdown, chromosome condensation and cohesion, and spindle assembly—are controlled by protein phosphorylation. As is easily imagined, once a phospho-dependent event is complete, dephosphorylation is required for cells to return to the basal state for the next cell cycle. Protein phosphatases hydrolyse phosphoesters on serine, threonine and/or tyrosine residues, thereby erasing the marks left by the kinases. In this regard, phosphatases are the main effectors to end mitosis. But in fact, as we shall see below, protein phosphatases also have important roles before and during mitosis. Finally, it is crucial in all cases to achieve good coordination of phosphatases with opposing kinases. This is achieved through the control of phosphatases, a topic that has thus far received little attention (Sidebar A).
Table 1. Kinases and phosphatases important for mitosis and their substrates.
| Kinase | Substrate (phosphosite) | Phosphatase | References |
|---|---|---|---|
| Cdk1–cyclin B | Broad range of substrates | PP2A-B55 | 15, 49, 64, 65, 66 |
| PP1 | 11, 67 | ||
| Cdc14 | 4 | ||
| NDEL1 (Thr 219*) | PP4 | 9 | |
| Fizzy/Cdc20 | PP2B/calcineurin | 12, 13 | |
| p90Rsk | Emi2 | PP2B/calcineurin | 12, 13 |
| Cdk2–cyclin E | Cdc25 (Thr 138#) | PP2A-B56 | 68 |
| Casein kinase 1 | Rec8 | PP2A-B56/Shugoshin | 5, 6, 7, 8 |
| Polo-like kinase 1 | Scc3/SA2 | PP2A-B56/Shugoshin | 5, 6, 7, 8 |
| Chk1 | Cdc25 (Ser 287#) | PP1 | 69, 70 |
| Aurora A | Aurora A (Thr 288 in T-loop*) | PP6-SAPs/ANKRDs | 71 |
| Aurora A and Aurora B | CENP-E (Thr 422*) | PP1 | 72 |
| Haspin | Histone H3 (Thr 3) | PP1γ-Repoman | 73 |
| Greatwall | α-Endosulfine/ARPP-19 (Ser 67#) | Unknown | 45, 46 |
| Wee1/Myt1 | Cdk1 (Thr 14/Tyr15) | Cdc25 | 74, 75 |
The number of each phosphosite shows its position either in
*human or in
#Xenopus protein.
Sidebar A | In need of answers.
It will be crucial to identify the phosphatases that dephosphorylate Gwl and Ensa/ARPP19 to explain how the Gwl–Ensa/ARPP-19 pathway is switched off, or reset, for the next round of the cell cycle.
It will be also important to explore the role of the Gwl–Ensa/ARPP-19 system in various biological contexts—such as the mammalian nervous system—and in various organisms (yeast and nematodes compared with insects and humans).
More generally, as it is becoming clear that protein phosphatases can be highly and specifically regulated, we need to elucidate the details of their control mechanisms, especially in terms of the balance with their partner kinases.
How many other of the PPP family of phosphatases can be switched on and off? Only biochemistry will tell!
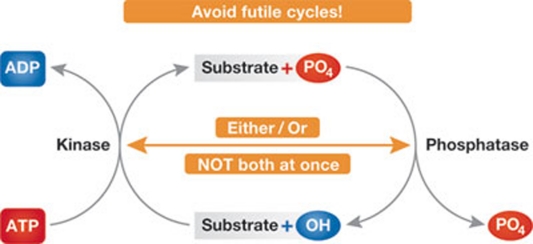
Avoid futile cycles!
We would expect to find mechanisms to avoid the futile cycles that would occur if kinases and their counteracting phosphatases were simultaneously active (Fig 1; Sidebar A). This applies especially to proteins that undergo almost complete conversion from an unphosphorylated state to a heavily phosphorylated state, as occurs to APC3 (Cdc27) as cells enter mitosis (Fig 2a). Phosphatases are clearly active at the end of mitosis to restore the phosphorylation state of such proteins to their interphase state of hypophosphorylation, and one or more kinases are activated at the onset of mitosis to bring about the mitotic hyperphosphorylated state. However, one cannot tell from simply looking at the fractional phosphorylation whether this interconversion necessarily entails reciprocal inhibition of phosphatases as the kinases are activated and activation of phosphatases when kinase activity is diminished.
Figure 1. Ensuring productive cycles.
A protein kinase adds, whereas a phosphatase removes, phosphate residues on substrates. If these mutually antagonistic enzymes work simultaneously, it not only results in a waste of ATP, but also renders impossible a full switch-like interconversion of the phosphorylation state of the substrate. To avoid this, the two enzymes should work alternatively, ideally while communicating with each other.
Figure 2. α-Endosulfine and ARPP-19 are Greatwall-dependent inhibitors of PP2A-B55.
(A) Schematic diagram of CDK1 and PP2A-B55 activity during the cell cycle. The patterns of CDK1 and PP2A-B55 activity are complementary to each other; CDK1 activity is shown in red and PP2A-B55 activity in green. The phosphorylation status of Apc3/Cdc27 reflects the ratio of kinase to phosphatase activity (upper bands indicate mitotic hyperphosphorylation). (B) Sequence alignment of the Ensa subfamily from yeast to human. Three possible phosphorylation sites are indicated with arrows. The CDK consensus site is found only in Xenopus Ensa, but is well conserved in the ARPP-19 subfamily. (C) Protein phosphatase (PPase) assay using a model CDK substrate and a catalytic C monomer, A+C dimer or heterotrimer holocomplex containing B55δ. Ensa phosphorylated by Gwl (red bars) inhibits PP2A trimeric holocomplexes that contain B55δ, but not dimeric or monomeric PP2A complexes. Fig 2c is a modified version of Figure 2A from Mochida et al [46]. APC3, anaphase-promoting complex subunit 3; ARPP-19, cyclic-AMP-regulated phosphoprotein of 19 kDa; CDK, cyclin-dependent kinase; Ensa, α-endosulfine; Gwl, Greatwall; PKA, cyclic-AMP-activated protein kinase A.
Spatial regulation of phosphatases
Some phosphatases have recently been found to be regulated by their intracellular localization (Sidebar A). For example, in budding yeast, Cdc14 is sequestered in the nucleolus until metaphase, then released into whole nucleus and cytoplasm by the FEAR and MEN systems (Table 1; [3]), mainly to dephosphorylate CDK substrates [4]. Another good example is the PP2A-B56–Shugoshin complex, which localizes to the pericentromeric region, where it keeps cohesin complexes dephosphorylated. Cohesin complexes on chromosome arms are phosphorylated by several kinases and thus removed from DNA well before the metaphase–anaphase transition (Table 1). The dephosphorylated population of cohesin at the pericentromeric region is sufficient to maintain sister chromatid attachment and allow chromosome separation—coordinated with CDK inactivation—in anaphase [5, 6, 7, 8]. PP4 is also regulated by its localization. During interphase, a population of PP4 localizes at the centrosome and suppresses unscheduled CDK1 activation. On entering mitosis, PP4 is dispersed into cytoplasm, thereby allowing its substrate NDEL1—a protein that is important for microtubule organization—to be phosphorylated by CDK1 [9].
Activity-level regulation of phosphatases
Another avenue in phosphatase research is the regulation of their enzymatic activity (Sidebar A). PP1 has a highly conserved CDK target motif at its carboxyl terminus, and phosphorylation of this site decreases its phosphatase activity in vitro [10]. In addition, inhibitor 1 of PP1 is phosphorylated and activated during mitosis [11]. These data support the idea that PP1 activity is reduced in mitosis, although the change in PP1 activity in vivo is still unknown. Another example is the activation of calcium/calmodulin-activated phosphatase—PP2B/calcineurin—upon exit from meiotic M phase II. This activation—owing to the fertilization-induced calcium ion flux—is important for proper cyclin degradation and timely entry into the embryonic cell cycle [12, 13]. The mechanism of calcineurin activation has been well studied [14]. We recently identified another phosphatase activity, which is high during interphase and very low in mitosis, and turned out to be a particular form of PP2A—namely, PP2A-B55δ (Fig 2a; [15]). Before discussing the regulatory mechanism of this phosphatase in detail, we note the use of phosphatase inhibitors in the study of mitosis control.
Avoid an abusive interpretation of useful inhibitors
When analysing and identifying phosphatases, it is important to understand and define the variety of protein phosphatase holocomplexes. For example, the active form of PP2A is a heterotrimer, composed of catalytic (C), scaffolding (A) and regulatory (B) subunits. In humans, there are two Cs (α & β), two As (α & β) and nearly 20 different Bs belonging to four families—B55/B, B56/B', B'' and B''' subfamilies [16]—suggesting that nearly 80 different PP2A holocomplexes could exist in a cell. All too often, researchers simply refer to ”PP2A“ without specifying the ‘flavour'. This variety applies equally to other PPP family members, including PP1, PP4 and PP6 [17, 18, 19, 20, 21, 22]. As each of the holocomplexes are likely to have specific functions in vivo, it is essential to analyse them individually. These considerations mean that often-used phosphatase inhibitors, such as okadaic acid and microcystin, are apt to give misleading results. First, these inhibitors do not distinguish among holocomplexes that contain identical catalytic subunits. Second, different phosphatases are expressed in vivo at various concentrations (sub-nanomolar to low micromolar levels), indicating that the IC50 values of these inhibitors are an unreliable guide for identifying phosphatases inhibited in vivo at a certain concentration of inhibitor. Those IC50s were originally calculated in vitro, by using diluted solutions of protein phosphatases [23, 24]. Third, some phosphatases show similar sensitivities to these inhibitors. For example, the sensitivities of PP2A and PP4 (and probably PP6) to okadaic acid are around 0.1–0.3 nM—too close to distinguish one from the other. Finally, we do not know how much inhibitor passes through the cell membrane. For all these reasons, interpretation of data using these inhibitors is necessarily limited and any results should be confirmed by using other methods, such as gene knockout. To analyse further specific functions of a particular phosphatase complex, it is necessary to identify the appropriate regulatory subunit. Of course, these inhibitors can still provide useful clues if they are used carefully.
Is PP2A-B55δ the phosphatase for CDK substrates?
Our attention was drawn to the question of which phosphatase(s) were responsible for mitotic exit by the discovery that PP2B/calcineurin was activated when crude Xenopus egg extracts are released from CSF arrest (arrested at meiotic metaphase II with high CDK activity by a combination of Mos kinase and Erp1/Emi2) by the addition of CaCl2 [12, 13]. Inhibition of PP2B (by cyclosporin A) seriously delayed the return to interphase in this setting, and we designed a substrate that could monitor phosphatase activity in these crude extracts. To our surprise, we found that the main role of PP2B/calcineurin was to allow the activation of a second phosphatase, not calcium activated, that was normally highly active in interphase and inhibited in meiosis and mitosis (Fig 2a; [12]). This regulation of phosphatase activity was abolished after addition of buffer to the concentrated egg extracts, so we had to use an immunodepletion technique—instead of standard biochemical fractionation and/or purification methods—to identify the fluctuating phosphatase activity. It proved to be a particular form of PP2A that contained a B55δ regulatory subunit [15]. In extracts that had been depleted of PP2A-B55δ, mitotic phosphorylation was accelerated at a lower-than-usual concentration of cyclin B [15]. Furthermore, histone 1 kinase activity, which reflects the level of Cdc2 kinase, was enhanced when PP2A-B55δ was depleted in interphase egg extracts. This is reminiscent of the experiments leading to the characterization of INH [25]. INH was originally defined as an activity that inhibited the activation of MPF in Xenopus oocytes, and was later identified as a form of PP2A [26, 27]. We found that depletion of PP2A-B55δ led to a failure to dephosphorylate mitotic CDK substrates at the end of mitosis, although cyclin degradation and CDK inactivation took place more or less normally [15]. These observations initially suggested that PP2A-B55δ was the main phosphatase for CDK substrates. However, when B55δ was depleted after the egg extracts entered mitosis, it was no longer required to exit mitosis [15]. This puzzling result suggested that although PP2A-B55δ is required for mitotic exit, its crucial role for mitotic exit is already complete before entering mitosis. We have no idea how PP2A-B55δ affects mitotic exit during the preceding interphase, nor how many CDK substrates are dephosphorylated by PP2A-B55δ. In any case, PP2A-B55δ is clearly not the only phosphatase that dephosphorylates CDK substrates. There must be other phosphatases acting at mitotic exit. For example, a form of PP1 is a good candidate, as a considerable body of data has already implicated it in mitotic exit. Cdc14 could be another candidate, although the functions of Cdc14 homologues in higher eukaryotes remain unclear [4, 28, 29].
Greatwall kinase regulates PP2A-B55δ activity in mitosis
The Greatwall (Gwl) gene was originally identified as the Scant (Scott of the Antarctic) mutation in Drosophila [30]. Scant later turned out to encode a protein kinase that is important for mitosis [31]. Drosophila mutants deficient in Gwl showed defects in chromosome condensation and delayed cell cycle progression throughout late G2 phase to mitosis. The kinase activity of Gwl increases as cells enter mitosis, during which Gwl itself is highly phosphorylated, at least in part by CDK1. Further analysis using Xenopus egg extracts revealed that Gwl is not only important for entering mitosis, but also required for maintaining the CSF-arrested mitotic state [32]. If Gwl is depleted from mitotic egg extracts (CSF), active CDK1 is inactivated by inhibitory phosphorylation on its Tyr 15 residue, rather than by cyclin proteolysis [32]. These findings suggest that the role of Gwl in mitosis is to control the CDK1 regulators Cdc25 and/or Wee1, which are themselves substrates of CDK1 [33, 34, 35]. Strikingly, even in the presence of high CDK1 activity, loss of Gwl induces dephosphorylation of mitotic phosphoproteins, strongly suggesting that Gwl acts as an inhibitor of the protein phosphatase(s) that antagonize CDK1. Indeed, PP2A-B55δ is activated after depletion of Gwl from CSF-arrested mitotic extracts [36, 37]. Oddly, however, Gwl does not phosphorylate any subunit of this phosphatase complex, so its mode of action was unknown for some time.
Ensa/ARPP-19, the first Gwl substrates, inhibit PP2A-B55δ
Two small, heat-stable proteins called Ensa and ARPP-19 have almost 70% sequence identity and are members of a highly conserved protein family (Fig 2b). ARPP-19 and its short form ARPP-16 were first identified as major substrates for protein kinase A in brain tissue [38, 39]. Therefore, they seemed to be involved in dopamine signalling in the postsynaptic neuron, where signalling cascades using protein phosphorylation are important [40]. Ensa was initially thought to be an endogenous ligand for the sulphonylurea receptor and was supposed to be involved in the control of insulin secretion [41]. But this original idea could not be confirmed and now seems unlikely, owing to the absence of a secretion-signal sequence in Ensa. In addition, little Ensa was found in biological membrane fractions [42]. Thus, for nearly 20 years after the identification of these proteins, no molecular function had been found. The first evidence for the importance of Ensa in cell cycle control came from a study in Drosophila, which has only one gene in this protein family. An RNAi screening using somatic S2 cells identified Endos (the Ensa homologue in Drosophila) as a protein important for mitotic chromosome alignment and normal spindle length [43]. Drosophila oocytes deficient in Endos show high CDK activity with low phosphorylation of CDK substrates, indicating that a lack of Ensa somehow changes the balance between kinase and phosphatase [44]. We and others independently discovered that Ensa and ARPP-19 are phosphorylated by Gwl at a highly conserved serine residue—Ser 67 in Xenopus Ensa (Fig 2b)—becoming potent inhibitors of PP2A-B55δ (Fig 2c; [45, 46]). Importantly, this inhibition is highly specific for PP2A-B55δ; other forms of PP2A are unaffected [46]. Although the exact sequence that is phosphorylated by Gwl (KYFDSGDYNM) is found only in these two small proteins, Gwl could have other substrates, as the stringency of its substrate recognition sequence is unknown.
Reversing the balance of CDK1 and PP2A-B55δ
Phosphorylation of Ensa/ARPP-19 by Gwl is essential for CDK substrates to be highly phosphorylated in Xenopus embryonic mitosis. In cycling egg extracts that lack Ensa, although Tyr 15 dephosphorylation and full CDK activation occur to the same level as in control extracts (albeit somewhat delayed) [46], PP2A-B55δ activity is not suppressed and CDK substrates are never fully phosphorylated. A threefold increase in PP2A-B55δ concentration induces a similar phenotype, presumably because the increased PP2A titrates out endogenous Ensa [15]. Furthermore, the addition of active—thiophosphorylated—Ensa is enough to induce significant phosphorylation of CDK substrates even at low levels of cyclin, below those required for normal mitosis. These observations collectively suggest that even full CDK1 activation is unable to promote entry into mitosis—inactivation of phosphatase(s) is also required. The Gwl pathway evolved to achieve this seesaw-like relationship.
A word of caution is necessary, however, because not all cell divisions seem to depend on the Gwl–Ensa/ARPP-19 system. For example, although Drosophila that lack Endos are inviable, loss of one copy of the twins/aar gene that encodes the B55 subunit of PP2A rescues the lethality, although not the female sterility [47]. It is difficult to interpret these data with our current understanding of the pathway.
Specificity and regulation of the Ensa/ARPP-19 family
Unlike okadaic acid, Ensa is highly specific for the particular species of PP2A that contains the B55δ subunit (Fig 2c) and does not bind to other forms of PP2A containing the B56ε, B56γ or B''/PR48 regulatory subunits [46]. Thus, different PP2A holocomplexes are distinctly regulated. It is highly probable, however, that other isoforms of B55 (α and β) are also targeted by Ensa/ARPP-19 [48, 49].
In addition to the Gwl phosphorylation site, Ensa/ARPP-19 family proteins have another highly conserved phosphorylation site at their carboxyl terminus [50]. This site—Ser 109 in Xenopus Ensa—seems to be phosphorylated by protein kinases that prefer basic residues preceding the phosphorylation site, such as PKA and Chk1 (Fig 1b). Xenopus Ensa and ARPP-19 have one more possible phosphorylation site in their amino-terminal region, Thr 28 (Fig 1b), which matches the CDK consensus (S/T-P-X-K/R, where X can be any amino acid) and can be phosphorylated by CDK2 in vitro [46]. The functions of these additional phosphorylation sites are of great interest, and it is important to know when they are phosphorylated in vivo. Multiple phosphorylation sites in such small phosphatase inhibitors are similar to PP1 inhibitor proteins, such as DARPP-32 and inhibitor 1 [51, 52, 53]. Like them the Ensa/ARPP-19 family could be an integrator of multiple signals.
Alternative functions of Gwl/Ensa/ARPP-19
In Xenopus and Drosophila (and probably also human cells), Gwl and Ensa/ARPP-19 family proteins are clearly involved in mitotic regulation [45, 46]. In budding yeast, however, Rim15 kinase (the closest homologue of Gwl) and its substrates Igo1 and Igo2 (homologues of Ensa and ARPP-19), are important for the response to nutritional deprivation under the control of TOR [54]. The phosphatase targeted by Igo1 and Igo2 in yeast remains to be identified, but Rim15 phosphorylates Igo1 and Igo2 at the same sites as does Gwl in Xenopus (Fig 2b). It will be important to analyse whether Igo1 and Igo2 inhibit PP2A-Cdc55—the budding yeast homologue of the B55 family [55]—and to characterize the substrates of this phosphatase and identify which kinase phosphorylates these sites (Sidebar A). If this kinase is activated by a TOR signal (or by TOR itself), a picture analogous to cell cycle control would emerge in a different context of biological function. That is, the Rim15 pathway might act by changing the balance of a paired protein kinase and phosphatase in the context of the response to starvation.
It should be noted that ARPP-19 was first identified in the brain, where many signals are rapidly changing [39, 56]. The balance of protein kinase and phosphatase could be changed rapidly and coordinately by using the MAST-L kinase—the Gwl homologue in humans—and ARPP-19 [57, 58]. For example, CDK5 could be a candidate antagonizing kinase and tau protein a substrate in this context (Sidebar A; [59]).
Key factors for the suddenness of mitotic entry
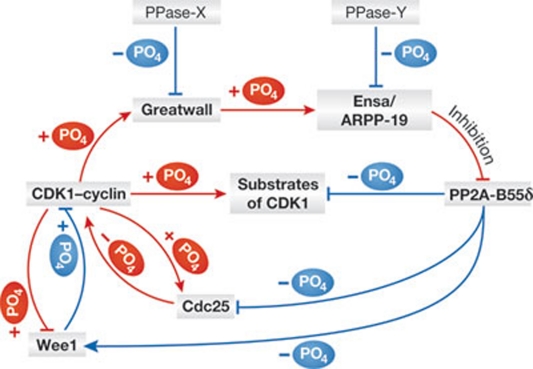
Wee1 is the main kinase that phosphorylates the Tyr 15 residue of CDK1, the dephosphorylation of which by the Cdc25 phosphatase is essential for the activation of CDK1. CDK1-mediated phosphorylation activates Cdc25, whereas it turns off Wee1 (Fig 3). Thus, Cdc25 and Wee1 form positive and negative feedback loops, respectively, with CDK1 [33, 34, 35]. As we originally identified PP2A-B55δ as a phosphatase able to act on CDK substrates, Cdc25 and Wee1 could be two physiological targets of PP2A-B55δ [15]. If this is the case, then PP2A-B55δ contributes to the suppression of premature CDK activation by maintaining these two major CDK regulators in their hypophosphorylated state (Fig 3). This model raises the question of how and what triggers the transition from interphase to mitosis. Given that the balance between CDK1 and its phosphatases is the target of this unknown triggering mechanism, protein phosphatases that dephosphorylate Gwl and Ensa/ARPP-19 during interphase must be important (labelled PPase-X and PPase-Y in Fig 3; Sidebar A). PP1 is probably involved in the reversal of either Gwl or Ensa/ARPP-19, or both, in addition to the multiple roles of the different PP1 complexes in mitosis [60]. When the balance between kinase and phosphatase for Gwl and/or Ensa/ARPP-19 is changed, the Gwl pathway would be fired to induce rapid mitotic phosphorylation. Thus, the activating and deactivating mechanisms of Gwl and Ensa/ARPP-19 are extremely important not only for the occurrence of, but also for the kinetics of mitotic entry. Evidence indicates that CDK is essential but not sufficient for Gwl activation. A report from the Montpellier group about AGC kinase activation [61] is probably not the last word on the subject. The observation that a small population of PP2A-B55 was found associated with Gwl in interphase, but not in mitosis, raises the possibility that PP2A-B55 itself is involved in keeping the Gwl pathway turned off in interphase [62]. The existence of all these positive and negative feedback loops is probably to be expected of a reversible flip-flop switch, but from a biological perspective we need to know whose finger is on the trigger, so to speak (Fig 3).
Figure 3. Factors that control the Greatwall pathway during the cell cycle.
Red arrows show factors that promote mitosis, whereas blue ones support interphase. +PO4 and –PO4 denote phosphorylation and dephosphorylation, respectively. PPase-X and PPase-Y are the as-yet-unidentified protein phosphatases that deactivate the Gwl pathway. ARPP-19, cyclic-AMP-regulated phosphoprotein of 19 kDa; CDK1, cyclin-dependent kinase 1; Ensa, α-endosulfine; Gwl, Greatwall.
Conclusion and perspectives
Considering that CDK1 has hundreds of substrates and that several other protein kinases, such as Aurora A and B, Polo, Wee1 and Myt1 are involved in entry into mitosis and mitotic progression, several different protein phosphatases are probably involved in the reversal or regulation of these processes (Table 1). A systematic survey in Drosophila using RNAi implicated no fewer than 22 protein phosphatases, although PP1 and PP2A were prominent among them [63]. Phosphatases, in addition to kinases, would contribute to the fine-tuning of cellular events. We obviously need to pay fresh attention to protein phosphatases and refine our view of them.

Satoru Mochida & Tim Hunt
Acknowledgments
S.M. is supported by a Grant-in-Aid for challenging Exploratory Research and in part by the Global COE Program (Cell Fate Regulation Research and Education Unit), MEXT (Ministry of Education, Culture, Sports, Science and Technology, Japan).
Footnotes
The authors declare that they have no conflict of interest.
References
- Meijer L, Arion D, Golsteyn R, Pines J, Brizuela L, Hunt T, Beach D (1989) Cyclin is a component of the sea urchin egg M-phase specific histone H1 kinase. EMBO J 8: 2275–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V, Nurse P (1986) The cell cycle control gene cdc2+ of fission yeast encodes a protein kinase potentially regulated by phosphorylation. Cell 45: 261–268 [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Charbonneau H, Deshaies RJ (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244 [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 2: 709–718 [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y (2010) Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol 12: 500–506 [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W (2010) Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell 18: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y (2006) Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52 [DOI] [PubMed] [Google Scholar]
- Toyo-oka K et al. (2008) Protein phosphatase 4 catalytic subunit regulates Cdk1 activity and microtubule organization via NDEL1 dephosphorylation. J Cell Biol 180: 1133–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano H, Ishii K, Yanagida M (1994) Phosphorylation of dis2 protein phosphatase at the C-terminal cdc2 consensus and its potential role in cell cycle regulation. EMBO J 13: 5310–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Guo JY, Tang W, Yang CS, Freel CD, Chen C, Nairn AC, Kornbluth S (2009) PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat Cell Biol 11: 644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Hunt T (2007) Calcineurin is required to release Xenopus egg extracts from meiotic M phase. Nature 449: 336–340 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Yoshizaki N, Kishimoto T, Ohsumi K (2007) Transient activation of calcineurin is essential to initiate embryonic development in Xenopus laevis. Nature 449: 341–345 [DOI] [PubMed] [Google Scholar]
- Klee CB, Ren H, Wang X (1998) Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J Biol Chem 273: 13367–13370 [DOI] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T (2009) Regulated activity of PP2A-B55δ is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J 28: 2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn PJ, Creyghton MP, Bernards R (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 1795: 1–15 [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84: 1–39 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R (2005) A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics 4: 1725–1740 [DOI] [PubMed] [Google Scholar]
- Hastie CJ, Carnegie GK, Morrice N, Cohen PT (2000) A novel 50 kDa protein forms complexes with protein phosphatase 4 and is located at centrosomal microtubule organizing centres. Biochem J 347: 845–855 [PMC free article] [PubMed] [Google Scholar]
- Kloeker S, Wadzinski BE (1999) Purification and identification of a novel subunit of protein serine/threonine phosphatase 4. J Biol Chem 274: 5339–5347 [DOI] [PubMed] [Google Scholar]
- Luke MM, Della Seta F, Di Como CJ, Sugimoto H, Kobayashi R, Arndt KT (1996) The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol 16: 2744–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson B, Ohama T, Daugherty AE, Brautigan DL (2008) Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 47: 1442–1451 [DOI] [PubMed] [Google Scholar]
- Takai A, Bialojan C, Troschka M, Ruegg JC (1987) Smooth muscle myosin phosphatase inhibition and force enhancement by black sponge toxin. FEBS Lett 217: 81–84 [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Matsushima R, Watanabe MF, Harada K, Ichihara A, Carmichael WW, Fujiki H (1990) Inhibition of protein phosphatases by microcystins and nodularin associated with hepatotoxicity. J Cancer Res Clin Oncol 116: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW (1990) Cyclin activation of p34cdc2. Cell 63: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Cyert MS, Kirschner MW (1988) Regulation of MPF activity in vitro. Cell 53: 185–195 [DOI] [PubMed] [Google Scholar]
- Lee TH, Solomon MJ, Mumby MC, Kirschner MW (1991) INH, a negative regulator of MPF, is a form of protein phosphatase 2A. Cell 64: 415–423 [DOI] [PubMed] [Google Scholar]
- Mocciaro A, Schiebel E (2010) Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci 123: 2867–2876 [DOI] [PubMed] [Google Scholar]
- Taylor GS, Liu Y, Baskerville C, Charbonneau H (1997) The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem 272: 24054–24063 [DOI] [PubMed] [Google Scholar]
- White-Cooper H, Carmena M, Gonzalez C, Glover DM (1996) Mutations in new cell cycle genes that fail to complement a multiply mutant third chromosome of Drosophila. Genetics 144: 1097–1111 [DOI] [PubMed] [Google Scholar]
- Yu J, Fleming SL, Williams B, Williams EV, Li Z, Somma P, Rieder CL, Goldberg ML (2004) Greatwall kinase: a nuclear protein required for proper chromosome condensation and mitotic progression in Drosophila. J Cell Biol 164: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhao Y, Li Z, Galas S, Goldberg ML (2006) Greatwall kinase participates in the Cdc2 autoregulatory loop in Xenopus egg extracts. Mol Cell 22: 83–91 [DOI] [PubMed] [Google Scholar]
- Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G (1993) Phosphorylation and activation of human cdc25-C by cdc2–cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J 12: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Maller JL (1993) Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell 4: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Dunphy WG (1995) Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell 6: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho PV, Williams BC, Mochida S, Zhao Y, Goldberg ML (2009) The M phase kinase Greatwall (Gwl) promotes inactivation of PP2A/B55δ, a phosphatase directed against CDK phosphosites. Mol Biol Cell 20: 4777–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron S, Brioudes E, Burgess A, Labbe JC, Lorca T, Castro A (2009) Greatwall maintains mitosis through regulation of PP2A. EMBO J 28: 2786–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault JA, Horiuchi A, Gustafson EL, Rosen NL, Greengard P (1990) Differential expression of ARPP-16 and ARPP-19, two highly related cAMP-regulated phosphoproteins, one of which is specifically associated with dopamine-innervated brain regions. J Neurosci 10: 1124–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi A, Williams KR, Kurihara T, Nairn AC, Greengard P (1990) Purification and cDNA cloning of ARPP-16, a cAMP-regulated phosphoprotein enriched in basal ganglia, and of a related phosphoprotein, ARPP-19. J Biol Chem 265: 9476–9484 [PubMed] [Google Scholar]
- Dulubova I, Horiuchi A, Snyder GL, Girault JA, Czernik AJ, Shao L, Ramabhadran R, Greengard P, Nairn AC (2001) ARPP-16/ARPP-19: a highly conserved family of cAMP-regulated phosphoproteins. J Neurochem 77: 229–238 [DOI] [PubMed] [Google Scholar]
- Virsolvy-Vergine A, Leray H, Kuroki S, Lupo B, Dufour M, Bataille D (1992) Endosulfine, an endogenous peptidic ligand for the sulfonylurea receptor: purification and partial characterization from ovine brain. Proc Natl Acad Sci USA 89: 6629–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros L, Breant B, Duchene B, Leroy C, Fauconnier G, Bataille D, Virsolvy A (2002) Localization of α-endosulphine in pancreatic somatostatin δ cells and expression during rat pancreas development. Diabetologia 45: 703–710 [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N (2007) Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina JR, Tranguch S, Dey SK, Lee LA, Cha B, Drummond-Barbosa D (2008) α-Endosulfine is a conserved protein required for oocyte meiotic maturation in Drosophila. Development 135: 3697–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi-Ayachi A, Labbe JC, Burgess A, Vigneron S, Strub JM, Brioudes E, Van-Dorsselaer A, Castro A, Lorca T (2010) The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330: 1673–1677 [DOI] [PubMed] [Google Scholar]
- Mochida S, Maslen SL, Skehel M, Hunt T (2010) Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330: 1670–1673 [DOI] [PubMed] [Google Scholar]
- Rangone H, Wegel E, Gatt MK, Yeung E, Flowers A, Debski J, Dadlez M, Janssens V, Carpenter AT, Glover DM (2011) Suppression of Scant identifies Endos as a substrate of Greatwall kinase and a negative regulator of protein phosphatase 2A in mitosis. PLoS Genet 7: e1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Guillamot M, de Carcer G, Eguren M, Trickey M, Garcia-Higuera I, Moreno S, Yamano H, Canamero M, Malumbres M (2010) Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55α, δ phosphatase. Cancer Cell 18: 641–654 [DOI] [PubMed] [Google Scholar]
- Schmitz MH et al. (2010) Live-cell imaging RNAi screen identifies PP2A-B55α and importin-β1 as key mitotic exit regulators in human cells. Nat Cell Biol 12: 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault JA, Shalaby IA, Rosen NL, Greengard P (1988) Regulation by cAMP and vasoactive intestinal peptide of phosphorylation of specific proteins in striatal cells in culture. Proc Natl Acad Sci USA 85: 7790–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings HC Jr, Greengard P, Tung HY, Cohen P (1984) DARPP-32, a dopamine-regulated neuronal phosphoprotein, is a potent inhibitor of protein phosphatase-1. Nature 310: 503–505 [DOI] [PubMed] [Google Scholar]
- Huang FL, Glinsmann WH (1976) Separation and characterization of two phosphorylase phosphatase inhibitors from rabbit skeletal muscle. Eur J Biochem 70: 419–426 [DOI] [PubMed] [Google Scholar]
- Oliver CJ, Shenolikar S (1998) Physiologic importance of protein phosphatase inhibitors. Front Biosci 3: D961–D972 [DOI] [PubMed] [Google Scholar]
- Talarek N, Cameroni E, Jaquenoud M, Luo X, Bontron S, Lippman S, Devgan G, Snyder M, Broach JR, De Virgilio C (2010) Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5'-3' mRNA decay pathway. Mol Cell 38: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy AM, Zolnierowicz S, Stapleton AE, Goebl M, DePaoli-Roach AA, Pringle JR (1991) CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol Cell Biol 11: 5767–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walaas SI, Hemmings HC Jr, Greengard P, Nairn AC (2011) Beyond the dopamine receptor: regulation and roles of serine/threonine protein phosphatases. Front Neuroanat 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A (2010) Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci USA 107: 12564–12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voets E, Wolthuis RM (2010) MASTL is the human orthologue of Greatwall kinase that facilitates mitotic entry, anaphase and cytokinesis. Cell Cycle 9: 3591–3601 [DOI] [PubMed] [Google Scholar]
- Hellmich MR, Pant HC, Wada E, Battey JF (1992) Neuronal cdc2-like kinase: a cdc2-related protein kinase with predominantly neuronal expression. Proc Natl Acad Sci USA 89: 10867–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW (2011) Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol 12: 469–482 [DOI] [PubMed] [Google Scholar]
- Vigneron S, Gharbi-Ayachi A, Raymond AA, Burgess A, Labbe JC, Labesse G, Monsarrat B, Lorca T, Castro A (2011) Characterization of the mechanisms controlling Greatwall activity. Mol Cell Biol 31: 2262–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto TM, Blake-Hodek K, Williams BC, Lewellyn AL, Goldberg ML, Maller JL (2011) Regulation of Greatwall kinase during Xenopus oocyte maturation. Mol Biol Cell 22: 2157–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F et al. (2007) Multiple protein phosphatases are required for mitosis in Drosophila. Curr Biol 17: 293–303 [DOI] [PubMed] [Google Scholar]
- Agostinis P, Derua R, Sarno S, Goris J, Merlevede W (1992) Specificity of the polycation-stimulated (type-2A) and ATP, Mg-dependent (type-1) protein phosphatases toward substrates phosphorylated by P34cdc2 kinase. Eur J Biochem 205: 241–248 [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel RE, Ohkura H, Gomes R, Sunkel CE, Baumgartner S, Hemmings BA, Glover DM (1993) The 55 kd regulatory subunit of Drosophila protein phosphatase 2A is required for anaphase. Cell 72: 621–633 [DOI] [PubMed] [Google Scholar]
- Sola MM, Langan T, Cohen P (1991) p34cdc2 phosphorylation sites in histone H1 are dephosphorylated by protein phosphatase 2A1. Biochim Biophys Acta 1094: 211–216 [DOI] [PubMed] [Google Scholar]
- Stone EM, Yamano H, Kinoshita N, Yanagida M (1993) Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr Biol 3: 13–26 [DOI] [PubMed] [Google Scholar]
- Margolis SS et al. (2006) Role for the PP2A/B56δ phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell 127: 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SS, Perry JA, Weitzel DH, Freel CD, Yoshida M, Haystead TA, Kornbluth S (2006) A role for PP1 in the Cdc2/Cyclin B-mediated positive feedback activation of Cdc25. Mol Biol Cell 17: 1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277: 1501–1505 [DOI] [PubMed] [Google Scholar]
- Zeng K, Bastos RN, Barr FA, Gruneberg U (2010) Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J Cell Biol 191: 1315–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW (2010) Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 142: 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M (2011) PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr Biol 21: 766–773 [DOI] [PubMed] [Google Scholar]
- Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D (1991) mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Strausfeld U, Labbe JC, Fesquet D, Cavadore JC, Picard A, Sadhu K, Russell P, Doree M (1991) Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human CDC25 protein. Nature 351: 242–245 [DOI] [PubMed] [Google Scholar]