Atypical strains of enteropathogenic E. coli are a leading cause of gastroenteritis in Melbourne.
Keywords: Diarrhea, enteropathogenic E. coli, Escherichia coli infections, gastroenteritis, research
Abstract
As part of a study to determine the effects of water filtration on the incidence of community-acquired gastroenteritis in Melbourne, Australia, we examined fecal samples from patients with gastroenteritis and asymptomatic persons for diarrheagenic strains of Escherichia coli. Atypical strains of enteropathogenic E. coli (EPEC) were the most frequently identified pathogens of all bacterial, viral, and parasitic agents in patients with gastroenteritis. Moreover, atypical EPEC were more common in patients with gastroenteritis (89 [12.8%] of 696) than in asymptomatic persons (11 [2.3%] of 489, p < 0.0001). Twenty-two random isolates of atypical EPEC that were characterized further showed marked heterogeneity in terms of serotype, genetic subtype, and carriage of virulence-associated determinants. Apart from the surface protein, intimin, no virulence determinant or phenotype was uniformly present in atypical EPEC strains. This study shows that atypical EPEC are an important cause of gastroenteritis in Melbourne.
Strains of Escherichia coli that cause diarrhea are classified into pathotypes (or virotypes) according to their specific virulence determinants (1). These virulence determinants give each pathotype the capacity to cause a clinical syndrome with distinctive epidemiologic and pathologic characteristics. For example, enterohemorrhagic E. coli (EHEC) may cause hemorrhagic colitis and the hemolytic uremic syndrome because of their production of Shiga toxins, whereas enteroaggregative E. coli (EAEC) are associated with persistent diarrhea in children in less-developed countries (1). Enteropathogenic E. coli (EPEC) share several key virulence determinants with the most common varieties of EHEC, but lack Shiga toxins, and cause nonspecific diarrhea in infants in less-developed countries (2,3). EPEC also differ from EHEC in that they typically carry an EPEC adherence factor plasmid (EAF). This plasmid encodes both bundle-forming pili (Bfp) that promote bacterial adherence to mammalian cells and are required for virulence (4) and a transcriptional activator, known as Per, that upregulates genes, such as eae, within a pathogenicity island termed the locus for enterocyte effacement (LEE) (5). LEE is required to produce attaching-effacing lesions, which are characteristic of EPEC-induced pathology. A subset of EPEC, known as atypical EPEC, does not carry EAF and hence does not produce Bfp (3). The role of EPEC in disease is uncertain.
The principal reservoir of EHEC is food animals, in particular, cattle, which harbor these bacteria in the distal intestinal tract and from which bacteria can spread to humans through fecally contaminated food or water (1). Although the other pathotypes of diarrheagenic E. coli generally do not originate in animals, they may also spread to humans through food or water contaminated with excrement. Recently, we conducted a study to determine if the water supply of Melbourne, Australia's second largest city with >3 million inhabitants, is a source of intestinal pathogens that are responsible for community-acquired gastroenteritis. Among the pathogens that were sought were diarrheagenic E. coli, including atypical EPEC, which emerged as the predominant cause of gastroenteritis in this community.
Materials and Methods
The design of the Water Quality Study (WQS), which was conducted from September 1997 to February 1999, has been reported previously (6). Briefly, 600 Melbourne families, with at least two children 1–15 years of age, were enrolled in the study. Each family was allocated at random to receive a real or sham water treatment unit, which was installed in the kitchen of their home and supplied water through a separate faucet. Family members, comprising 2,811 persons, were followed for 15 months (68 weeks). Each participating household had a nominated member who completed a weekly questionnaire regarding the presence, duration, and severity of gastrointestinal symptoms. The primary endpoint of the study was highly credible gastroenteritis, which was defined as exhibiting any of the following symptoms in a 24-hour period: two or more loose stools, two or more episodes of vomiting, one loose stool together with abdominal pain or nausea or vomiting, or one episode of vomiting with abdominal pain or nausea. Cases of highly credible gastroenteritis were deemed to be distinct if the participant was symptom-free for at least 6 days.
Sample Collection and Processing
Participants in the study were asked to collect fecal specimens during episodes of gastroenteritis. A total of 795 specimens collected during 2,669 reported episodes of gastroenteritis were examined for rotavirus, adenovirus, Norwalk-like viruses, Giardia spp., and Cryptosporidium spp. and were cultured for Salmonella spp., Shigella spp., Campylobacter spp., Vibrio spp., Yersinia spp., Aeromonas spp., Plesiomonas spp., and Clostridium difficile, as described previously (6,7). Baseline frequencies of these pathogens in the study population were determined during the 4-month period, May through August 1997, immediately preceding the WQS. Frequencies were examined by investigating 1,091 fecal specimens from a convenience sample of participants. Participants who provided a baseline specimen were similar to those who did not provide a specimen in age, sex, and family background.
Examination of Feces for E. coli
Sufficient funds were available to investigate 1,250 samples for diarrheagenic E. coli. Of these samples, 500 were randomly selected from 1,091 fecal samples obtained from healthy persons in the baseline study, and 750 samples were randomly selected from the 795 samples obtained from participants with highly credible gastroenteritis in the WQS.
Bacteria were isolated from fecal samples by direct plating on MacConkey agar (Oxoid Ltd., Basingstoke, UK). After overnight incubation at 37°C, a sterile cotton swab was used to transfer the entire growth from each plate into Luria broth containing 30% (vol/vol) glycerol, which was then frozen at –70°C until required. Diarrheagenic strains of E. coli were identified by polymerase chain reaction (PCR) and confirmed by Southern hybridization. Template DNA for use in PCR was prepared from bacteria grown in 2.5 mL of MacConkey broth that contained a loopful of stored frozen culture and was incubated with shaking at 37°C overnight. One milliliter of this culture was centrifuged to pellet the bacteria, the supernatant was removed, and then the pellet was washed in 1 mL of phosphate buffer, resuspended in 200 µL sterile distilled water, and heated for 10 min at 100°C. Samples were then placed on ice for 5 min and recentrifuged at 16,000 x g. Aliquots of the supernatant were pipetted into sterile tubes and stored for <1 week at –20°C before use.
PCR amplifications were performed in a Gene Amp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA) with AmpliTaq Gold polymerase (Applied Biosystems) and the primers listed in Table 1 in a reaction volume of 20 µL (for single reactions) or 50 µL (for multiplex PCR). The genes identified by these primers and their association with each pathotype of diarrheagenic E. coli are listed in Tables 1 and 2. PCR for the lacZ gene, which is found in almost all wild-type strains of E. coli, was included as a control to ensure that negative PCR assays were not a result of the absence of viable bacteria in the sample or the presence of inhibitors in the reaction mixture. Samples that were negative in the PCR for lacZ (3.6% of all those examined) were excluded from further analysis. At the conclusion of the PCR, 10 mL of the reaction mixture underwent electrophoresis on 2.5% 96-well format agarose gels (Electro-fast, ABgene, Epsom, UK). Gels were stained with ethidium bromide, visualized on a UV transilluminator, and photographed. A portion of the PCR product was retained for Southern blotting, which was performed by using capillary transfer of separated DNA fragments onto positively charged nylon membranes (Roche Diagnostics Ltd., Lewes, UK). Digoxigenin-labeled DNA probes were prepared by PCR (Roche Diagnostics) from the control strains (Table 2) by using the PCR primers listed in Table 1. The integrity of the probes was determined by nucleotide sequencing. Probes were hybridized overnight under conditions of high stringency at 65°C and detected by using chemiluminescence as recommended by the manufacturer. Probe-positive bacteria were assigned to a pathotype according the criteria in Table 2. Equivocal or ambiguous assays were repeated, and if results were still unclear, these specimens were excluded from further analysis.
Table 1. Characteristics of the polymerase chain reaction (PCR) primers used to detect pathogenic Escherichia coli in mixed culture.
| Target gene or virulence factor | Primera | Primer sequence (5´ to 3´) | PCR program (30 cycles)b | Product size (bp) | Reference |
|---|---|---|---|---|---|
| eae | 11 21 | GACCCGGCACAAGCATAAGC CCACCTGCAGCAACAAGAGG | 95°C, 30 s; 54°C, 90 s; 72°C, 90 s | 384 | (8) |
| ehxA | 11 21 | GCATCATCAAGCGTACGTTCC AATGAGCCAAGCTGGTTAAGCT | 534 | (8) | |
| stx1 | 12 22 | ATAAATCGCCATTCGTTGACTAC AGAACGCCCACTGAGATCATC | 95°C, 30 s; 52°C, 60 s; 72°C, 60 sc | 180 | (8) |
| stx2 | 12 22 | GGCACTGTCTGAAACTGCTCC TCGCCAGTTATCTGACATTCTG | 255 | (8) | |
| ltA | 13 23 | GGCGACAGATTATACCGTGC CCGAATTCTGTTATATATGTC | 94°C, 60 s; 50°C, 60 s; 72°C, 120 sc | 696 | (9) |
| st1A | 13 23 | TCTGTATTATCTTTCCCCTC ATAACATCCAGCACAGGC | 186 | (9) | |
| ipaC | 1 2 | CAGCAGATTGCAGCGCATAT CAAGAGCAGATGCATAACGC | 94°C, 30 s; 59°C, 90 s; 72°C, 90 s | 811 | This study |
| bfpA | 1 2 | ATTGAATCTGCAATGGTGC ATAGCAGTCGATTTAGCAGCC | 95°C, 40 s; 55°C, 40 s; 72°C, 40 s | 461 | This study |
| pCVD432 | 1 2 | CTGGCGAAAGACTGTATCAT CAATGTATAGAAATCCGCTGTT | 94°C, 40 s; 53°C, 60 s; 72°C, 60 s | 630 | (10) |
| aggA | 1 2 | ATGCATTACTTTGGGTTTAG TCAACCTTGACACTTGCC | 94°C, 60 s; 50°C, 60 s; 72°C, 120 sc | 414 | This study |
| lacZ | 1 2 | TGATTGAAGCAGAAGCCTGC CGCCAATCCACATCTGTGAA | 94°C, 30 s; 59°C, 90 s; 72°C, 90 s | 1,350 | This study |
aPrimers with the same superscript were used in duplex PCRs. bBefore the first cycle, the sample was denatured for 10 min at 95°C; after the last cycle, the sample was extended for 8 min at 72°C. c35 cycles.
Table 2. Classification of pathogenic Escherichia coli according to amplicon(s) generated by polymerase chain reaction for virulence-associated determinants.
| Interpretationb | Gene or virulence-associated determinanta |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pCVD432 | aggA | bfpA | eae | exhA | ipaC | stIA | ltA | stx1 | stx2 | Control strain | Reference | |
| EAECc | + | + | – | – | – | – | – | – | – | – | O42 | (11) |
| Typical EPEC | – | – | + | + | – | – | – | – | – | – | E2348/69 | (12) |
| Atypical EPEC | – | – | – | + | – | – | – | – | – | – | E128012 | (12) |
| EIEC | – | – | – | – | – | + | – | – | – | – | 223/83 | (13) |
| ETECc | – | – | – | – | – | – | + | + | – | – | H10407 | (14) |
| EHECd | – | – | – | + | + | – | – | – | + | + | EDL933 | (15) |
| STEC, not EHECe | – | – | – | – | – | – | – | – | + | + | ||
aFactors specified by virulence genes: aggA, aggregative fimbria, AAF/I; bfpA, bundle-forming pilus; eae, intimin; ipaC, invasion plasmid antigen; stIA, heat-stable enterotoxin; ltA, heat-labile enterotoxin; stx, Shiga toxin. bEAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; EIEC, enteroinvasive E. coli; ETEC, enterotoxigenic E. coli; STEC, Shiga toxin–producing E. coli; EHEC, enterohemorrhagic E. coli. cEither stlA or ltA positive. dEither eae or ehxA and stx1 or stx2 positive. eEither stx1 or stx2 positive.
Characterization of Isolates
Twenty-two EPEC isolates (11 from healthy persons and 11 from persons with diarrhea) were isolated in pure culture. Representative colonies of each were then serotyped with hyperimmune rabbit antisera (16). These strains were also subjected to PCR with the primers and conditions listed in Table 3 to determine intimin subtype and to investigate the presence of selected virulence-associated genes. The same 22 strains were also examined for their ability to adhere to and invade HEp-2 epithelial cells.
Table 3. Characteristics of the polymerase chain reaction (PCR) primers used in this study for strain characterization.
| Primer | Primer sequence (5´ to 3´) | Target | PCR program (30 cycles)a | Product size (bp) | Reference |
|---|---|---|---|---|---|
| P1b | CTGAACGGCGATTACGCGAA | ||||
| P2 | CCAGACGATACGATCCAG | eae c | 94°C, 30 s; 53°C, 30 s; 72°C, 60 s | 917 | (17) |
| P3 | CTGGAGTTGTCGATGTT | eae-α | 94°C, 30 s; 53°C, 30 s; 72°C, 120 s | 1,648 | (17) |
| P4 | GTAATTGTGGCACTCC | eae-β | 94°C, 30 s; 53°C, 30 s; 72°C, 120 s | 1,926 | (17) |
| P5 | GCCTCTGACATTGTTAC | eae-γ | 94°C, 30 s; 53°C, 30 s; 72°C, 120 s | 1,770 | (17) |
| ecsD-lower | TATTTTCAAAAAGAATGATGTC | eae | 94°C, 30 s; 56°C, 60 s; 72°C, 150 s | »2,990d | (18) |
| SK1e | CCCGAATTCGGCACAAGCATAAGC | ||||
| LP5 | AGCTCACTCGTAGATGACGGCAAGCG | eae-ε | 94°C, 30 s; 55°C, 60 s; 72°C, 120 s | 2,608 | (18) |
| LP6B | TAGTTGTACTCCCCTTATCC | eae-ζ | 94°C, 30 s; 53°C, 60 s; 72°C, 150 s | 2,430 | (18) |
| LP7 | TTTATCCTGCTCCGTTTGCT | eae-ι | 94°C, 30 s; 52°C, 60 s; 72°C, 150 s | 2,685 | (18) |
| LP8 | TAGATGACGGTAAGCGAC | eae-η | 94°C, 30 s; 52°C, 60 s; 72°C, 150 s | 2,590 | (18) |
| LP10 | GGCATTGTTATCTGTTGTCT | eae-κ | 94°C, 30 s; 52°C, 60 s; 72°C, 150 s | 2,769 | (18) |
| LP11B | GTTGATAACTCCTGATATTTTA | eae-θ | 94°C, 30 s; 50°C, 60 s; 72°C, 150 s | 2,686 | (18) |
| LPFDF LPFDR | GAACTGTAGATGGGTAC AGCAGGCATAACGCAAG | lpfD | 94°C, 60 s; 48°C, 50 s; 72°C, 60 s | 798 | (19) |
| Donne-280 Donne-281 | CGGAACAGTAGGTTCACCTTC AGTGCCCGTGTTCTTGAACTG | efa1 | 94°C, 30 s; 50°C, 30 s; 72°C, 120 s | 2,226 | (20) |
| EASTOS1 EASTOS2 | GCCATCAACACAGTATATCCG CGCGAGTGACGGCTTTGTAG | astA | 94°C, 30 s; 50°C, 60 s; 72°C, 90 s | 109 | (10) |
| AggRks1 AggRkas2 | GTATACACAAAAGAAGGAAGC ACAGAATCGTCAGCATCAGC | aggR | 94°C, 30 s; 50°C, 60 s; 72°C, 45 sf | 254 | (21) |
aBefore the first cycle, the sample was denatured for 10 min at 95°C; after the last cycle, the sample was extended for 7 min at 72°C. bPrimer P1 was used as forward primer in all PCR reactions in combination with P2, P3, P4, and escD-lower. cConserved region of eae. dAmplicon sequenced. ePrimer SK1 was used as forward primer in all PCR reactions in combination with LP5, LP6B, LP7, LP8, LP10, and LP11B. f35 cycles.
Bacterial Adhesion and Invasion of HEp-2 Cells
The Center for Vaccine Development (University of Maryland, Baltimore, MD) method was used to determine the pattern of bacterial adherence to HEp-2 epithelial cells (12). Bacterial strains were designated nonadherent if <10 of 200 HEp-2 cells had five or more bacteria attached. The fluorescent actin staining (FAS) assay, which correlates with the ability of E. coli to produce attaching-effacing lesions in the intestine, was performed by using a 6-h incubation period (22). At the completion of the assay, cells were examined by fluorescence and phase-contrast microscopy to confirm that fluorescent areas corresponded to attached bacteria. Bacterial strains that gave equivocal or negative results in the FAS assay were investigated for DNA corresponding to regions of the LEE, other than eae, by colony blotting using the LEE-A, LEE-B, and LEE-D DNA probes and hybridization conditions described previously (23). Quantitative assessment of the ability of E. coli to invade HEp-2 cells was performed with the gentamicin-protection assay (24). Results were expressed as the number of bacteria recovered from HEp-2 cells after treatment with gentamicin as a percentage of the total number of cell-associated bacteria (24).
Statistical analysis was performed with InStat version 3.0 (GraphPad Software Inc., San Diego, CA). A p value < 0.05 was considered significant.
Results
Association of E. coli Pathotypes with Gastroenteritis
After excluding samples for which patient data were incomplete (12 samples), which were negative by PCR for lacZ (45 samples), or which gave equivocal results in the PCR or DNA hybridization assays (8 samples), 1,185 (94.8%) samples of the original 1,250 were available for analysis: 696 from patients with gastroenteritis and 489 from healthy persons. The results of the assays are summarized in Table 4. Enterotoxigenic (ETEC) and enteroinvasive (EIEC) strains of E. coli and Bfp-positive EPEC were identified in <0.5% of healthy persons or patients with gastroenteritis. EHEC were present in 4 (0.6%) of 696 of samples from persons with gastroenteritis and in no healthy persons, but the difference between the two groups was not significant (p = 0.15, Fisher exact test, 2-tailed). In contrast, both EAEC and atypical (Bfp-negative) EPEC were identified in >5% of patients, and the difference between the symptomatic and baseline groups was significant for each of these pathotypes (Table 4). Analysis of the data pertaining to patients in whom EAEC and atypical EPEC were identified showed that atypical EPEC were more frequent in younger persons; patients with atypical EPEC were a median age of 3.4 years, compared with 7.4 years for the symptomatic group overall (p < 0.0001; Mann-Whitney test, 2-tailed). Of all atypical EPEC in study participants with gastroenteritis, 75 (84%) were identified in children <10 years old, compared with 14 (16%) in patients >10 years (relative rate [RR] 3.4, 95% confidence interval [CI] 2.0–5.9). In contrast, the median age of patients infected with EAEC was 7.3 years.
Table 4. Frequency of Escherichia coli pathotypes in study participants with and without gastroenteritis.
| E. coli pathotypea | Source |
||
|---|---|---|---|
| Symptomatic (n = 696) (%) | Symptom-free (n = 489) (%) | pb | |
| EAEC | 45 (6.5) | 15 (3.1) | 0.02 |
| Typical EPEC | 2 (0.3) | 1 (0.2) | NS |
| Atypical EPEC | 89 (12.8) | 11 (2.3) | < 0.0001 |
| EIEC | 0 | 0 | NS |
| ETEC | 2 (0.3) | 0 | NS |
| EHEC | 4 (0.6) | 0 | NS |
| STEC, not EHEC | 0 | 1 (0.2) | NS |
aEAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; EIEC, enteroinvasive E. coli; ETEC, enterotoxigenic E. coli; STEC, Shiga toxin–producing E. coli; EHEC, enterohemorrhagic E. coli; NS, not significant (p > 0.1). bFisher exact test, 2-tailed.
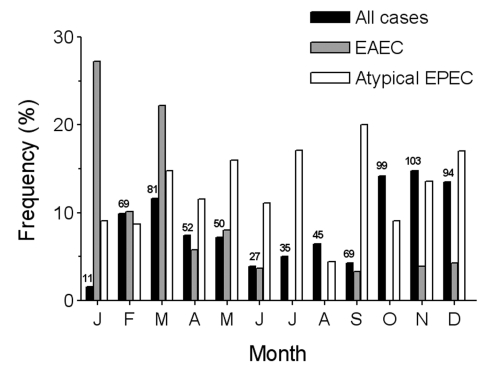
Examining the seasonal occurrence of gastroenteritis from all causes showed that gastroenteritis was more common in the warmer months; 65.7% of cases occurred in the 6 months from October through March (Figure 1). Infections with EAEC and atypical EPEC reflected this distribution with 80% and 65.5% of infections with these bacteria, respectively, occurring during the same period. The high incidence of EAEC in February and March may have been caused by small family outbreaks; 9 of 25 cases during this period originated in three households.
Figure 1.
Seasonal incidence in gastroenteritis in the Melbourne Water Quality Study, 1998. Solid black bars indicate all cases of gastroenteritis as a percentage of the total, with the number of cases indicated above each bar. The frequencies of enteroaggregative Escherichia coli (EAEC) and enteropathogenic E. coli (EPEC) are expressed as a percentage of all cases examined each month.
In view of the seasonal variation in the occurrence of EAEC and atypical EPEC, we reanalyzed the data and compared the symptomatic and baseline groups, because the EPEC were examined only during the 4-month period from May through August, 1997. This analysis showed that the occurrence of EAEC in persons with gastroenteritis (6 [3.8%] of 157) and in asymptomatic persons (15 [3.1%] of 489) was essentially the same during May through August of 1998 and 1997, respectively (RR 1.2, 95% CI 0.5–3.0). In contrast, atypical EPEC were more frequent in the gastroenteritis group during the same period; 19 (12.1%) of 157 of symptomatic patients were positive for these bacteria, compared with 11 (2.3%) of 489 for the asymptomatic group (RR 5.7, 95% CI 3.1–10.5, p < 0.0001; Fisher exact test, 2-tailed). The frequency of atypical EPEC in symptomatic patients from households with real and sham water treatment units was similar (42 [12.3%] of 341 and 47 [13.2%] of 355, respectively).
Characterization of EPEC Isolates
Pure cultures of PCR- and probe-positive atypical EPEC were obtained from 22 randomly selected samples that were positive in the original PCR. These isolates were characterized further to establish their identity as EPEC and to determine if they belonged to a limited number of clones. All strains were identified as atypical EPEC in that they were negative for bfpA, stx1, stx2, aggA, and aggR. Determination of O:H serotype and intimin subtype revealed that although strains from each sample were the same, those obtained from different persons were highly heterogeneous, and no two isolates belonged to the same serotype and intimin subtype (Table 5). Only three isolates were of serotypes (O55:H7 and O126:H6 [two isolates]) that commonly include atypical EPEC. In addition, one isolate, W145, was serotype O55:H6, which includes typical EPEC strains (2). Eight isolates were O-serogroups that were classified as nontypeable because they did not react with any of the available O-typing sera (O1–O181), and one isolate had an H antigen that did not react with any of the available H-typing sera (H1–H56) and was classified as nontypeable.
Table 5. Characteristics of 22 isolates of atypical enteropathogenic Escherichia coli identified during this study.
| Strain | Sou-rcea | Serotype | Intimin type | Result of PCRb |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| bfpA | aggR | efa1 | astA | lpfD | Adhesion patterna | FAS assay | HEp-2 cell invasionc | ||||
| W7 | N | O161:H40 | β | – | – | – | – | – | NA | – | 0.01 |
| W85-2 | N | OR:H- | γ | – | – | – | – | – | AA | + | 0.02 |
| W114 | N | O107:H8 | ι | – | – | – | – | – | IA | + | 0.02 |
| W143 | N | Ont:H5 | ε | – | – | – | – | – | IA | + | < 0.01 |
| W145 | N | O55:H6 | γ | – | – | + | + | – | AA | + | 0.03 |
| W154 | N | O139:H14 | β | – | – | – | – | – | AA | + | < 0.01 |
| W185 | N | Ont:H21 | θ | – | – | – | – | – | IA | + | 0.02 |
| W208 | N | Ont:H4 | β | – | – | – | – | – | IA | + | 0.08 |
| W761 | N | O124:H40 | λ | – | – | – | – | – | NA | – | 0.07 |
| W902 | N | O125:H6 | α2 | – | – | – | – | – | IA | + | 0.06 |
| W914 | N | O125:H6 | α | – | – | – | – | – | AA | + | 0.07 |
| W1040 | G | O15:Hnt | β | – | – | + | – | + | LAL | + | 0.10 |
| W1056 | G | O55:H7 | γ | – | – | + | + | – | IA | + | 0.77 |
| W1068 | G | O51:H49 | α | – | – | – | – | – | AA | + | < 0.01 |
| W1082 | G | Ont:H6 | β | – | – | – | – | – | AA | + | 0.01 |
| W1092 | G | OR:H- | η/ε | – | – | – | – | + | IA | + | < 0.01 |
| W1108 | G | O172:H4 | θ | – | – | – | – | – | IA | + | 0.03 |
| W1118 | G | O126:H6 | α | – | – | – | – | – | IA | + | 0.01 |
| W1120 | G | Ont:H34 | α | – | – | – | – | – | IA | + | 0.01 |
| W1134 | G | Ont:H6 | θ | – | – | – | – | – | NA | – | 0.01 |
| W1585 | G | Ont:H40 | θ | – | – | – | – | – | NA | – | 0.02 |
| W1706 | G | Ont:H6 | α | – | – | – | – | – | AA | + | 0.01 |
| E128012 | C | O114:H2 | β | – | – | + | – | + | IA | + | 0.32 |
aN, no symptoms; G, gastroenteritis; C, control strain; Ont, O nontypable (O1–O181); Hnt, H nontypable (H1–H56); NA, nonadherent; AA, aggregative adherence pattern; IA, indeterminate adherence pattern; LAL, localized-like adherence pattern; FAS, fluorescent actin staining. bPCR, polymerase chain reaction; +, positive; –, negative. cData are the number of bacteria recovered from HEp-2 cells after treatment with gentamicin as a percentage of the total number of cell-associated bacteria and are the mean of two separate assays.
Atypical EPEC strains were also heterogeneous in their carriage of putative accessory virulence determinants (Table 5). Although all bacteria were, by definition, negative for bfpA, nearly all were also negative for efa1 and lpfD, the genes for factors that have been implicated as adhesins of some attaching-effacing strains of E. coli (19,25). Only two strains were positive for astA, which has also been suggested to contribute to the virulence of atypical EPEC (26).
Adhesion to HEp-2 Cells
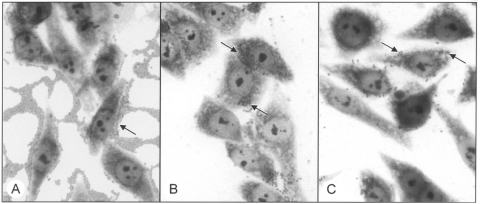
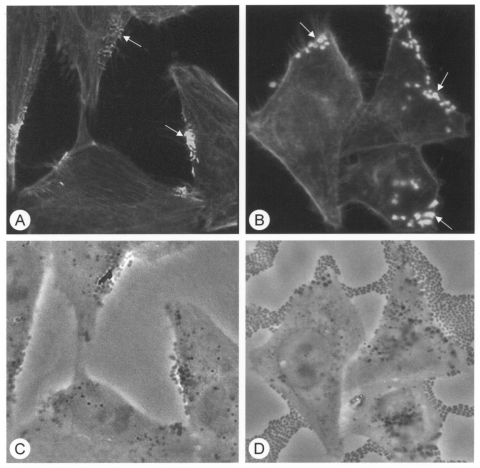
Typical EPEC adhere to HEp-2 cells in a distinctive pattern termed localized adherence, which requires the presence of Bfp (5). The 22 atypical EPEC strains isolated in this study showed variable patterns of adherence to HEp-2 cells, including aggregative adherence (7 strains) and a pattern previously termed localized-like adherence (1 strain) (27). Ten strains showed an indeterminate pattern of adherence, with small numbers of bacteria distributed apparently at random on the cell surface (Figure 2), and 4 strains were classified as nonadherent. The frequency of strains displaying each pattern of adherence was similar among isolates from patients with gastroenteritis and healthy persons. All atypical EPEC strains that adhered to HEp-2 cells, regardless of their pattern of adhesion, were positive in the FAS assay (Figure 3), which indicates that the LEE in these bacteria is functional. All nonadherent strains hybridized with DNA probes were derived from different regions of LEE, which suggests that they carry the entire pathogenicity island. We did not attempt to determine whether LEE was functional in these bacteria.
Figure 2.
Patterns of adherence of atypical enteropathogenic Escherichia coli strains (arrows) to HEp-2 epithelial cells. A) aggregative adherence, B) localized-like adherence, and C) indeterminate adherence. Magnification x1,000.
Figure 3.
Fluorescent actin staining (FAS) assay for attaching-effacing capacity of atypical enteropathogenic Escherichia coli (EPEC) strains with different patterns of adherence to HEp-2 cells. Fluorescent micrographs of HEp-2 cells (A and B) incubated with strains of atypical EPEC showing localized-like and aggregative adherence, respectively, and then reacted with fluorescein-labelled phalloidin. Note the foci of intense fluorescence (arrows) associated with adherent bacteria, which were also visualized by phase contrast microscopy of the same microscope fields (C and D). Magnification x1,000.
Previous reports of atypical EPEC have suggested that invasion of epithelial cells may contribute to the virulence of these bacteria (28). In this study, however, only one strain, W1056 (O55:H7), was able to invade HEp-2 cells to a noticeable extent (Table 5).
Discussion
The role of EPEC as a cause of diarrhea in children, particularly in developing countries, is now well established (2,3). The proven virulence determinants of EPEC include genes within the LEE, notably intimin (the outer membrane protein product of the eae gene), and Bfp, which is encoded by EAF (5). The key role of EAF in promoting the virulence of EPEC was established by Levine et al. (12), who showed that an EAF-negative derivative strain of EPEC, E2348/69, is markedly less virulent for adult volunteers than the wild-type strain. The same study showed that an atypical EPEC strain, E128012, which intrinsically lacks EAF, is also virulent in volunteers. This observation established that certain EPEC strains do not require EAF to cause disease. These intrinsically EAF-negative strains were originally called Class II EPEC (2,3) but are now more generally referred to as atypical EPEC. They are characterized by the presence of LEE and the absence of factors encoded by EAF, in particular, Bfp. In this way, atypical EPEC resemble EHEC, which are able to cause diarrhea despite their lack of Bfp. Indeed, persuasive evidence indicates that the most prevalent EHEC strain, serotype O157:H7, evolved from an atypical EPEC strain of serotype O55:H7 (29).
The aims of the WQS were to investigate the effect of household water treatment units on the incidence of gastroenteritis in Melbourne and to identify causative agents of gastroenteritis in the study population. The microbiologic investigations showed that the detection rate of EAEC (6.5%) and atypical EPEC (12.3%) in patients with diarrhea was greater than that of Campylobacter spp. (3%), Salmonella spp. (1.1%), adenovirus (1.1%), rotavirus (1.4%), Cryptosporidium spp. (1.6%), and Giardia spp. (2.5%) and was matched only by that of noroviruses (11.4%) (6,7,30).
Together, atypical EPEC and EAEC accounted for 19.3% of all cases; 21% of cases were attributable to all other bacterial, viral, and parasitic causes combined. However, the frequency of EAEC in patients with gastroenteritis and that in the baseline group without diarrhea was the same when matched for the time of year when the sample of feces was collected. In contrast, atypical EPEC was isolated significantly more often from patients with gastroenteritis than from those without symptoms, regardless of when the sample was collected. A subset of 22 randomly selected atypical EPEC strains was examined and found to be highly heterogeneous, which indicates that the high frequency of atypical EPEC in the study population was not the result of one or more outbreaks attributable to a small number of strains.
Despite the persuasive evidence of a volunteer study and reports of outbreaks of diarrhea with atypical EPEC (12,26,31), the role of atypical EPEC in disease is controversial. Originally, atypical EPEC were grouped with EPEC but were then segregated because they lack EAF. Justification for this division stemmed from the observation that EAF-bearing EPEC far outnumber atypical EPEC as the cause of infantile diarrhea in less-developed countries and of diarrhea outbreaks in general (3). In recent reports, however, from countries as diverse as Iran, Poland, South Africa, and the United Kingdom, atypical EPEC strains have outnumbered typical strains as a cause of gastroenteritis (32–35). Atypical EPEC were also more frequent than typical strains in aboriginal children hospitalized for diarrhea in the Northern Territory of Australia (36). These findings were reflected in the present study: 89 (94%) of eae-bearing strains identified in patients with gastroenteritis were atypical EPEC.
As for EPEC in general, atypical EPEC were originally incriminated as intestinal pathogens by virtue of their epidemiologic association with cases of diarrhea (2). Subsequently, these strains, which had been identified by serotype alone, were shown to be EPEC sensu stricto (37). Although atypical EPEC generally are serotypes which differ from EAF-positive EPEC (and other pathotypes of diarrheagenic E. coli), the 12 O-serogroups recognized by the World Health Organization as EPEC (i.e., serogroups O26, O55, O86, O111, O114, O119, O125, O126, O127, O128, O142, and O158) include both typical and atypical varieties (3). Some of the atypical EPEC strains within these serogroups carry accessory virulence-associated determinants such as the EHEC hemolysin (commonly found in serotypes O26:H11 and O111ac:H8). Some strains also carry astA, the gene for enteroaggregative heat-stable enterotoxin, EAST1, which is frequently found in serotypes O55:H7, O119:H2, and O128:H2 (3). Atypical EPEC strains of non-EPEC serogroups generally do not express these factors. In the present study, none of the 89 atypical strains was positive for ehxA, which is required for the production of EHEC hemolysin, and although two strains (both serogroup O55) tested positive for astA, both carried the previously described mutations in this gene, which would preclude the synthesis of biologically active enterotoxin (38).
Unlike EAF-bearing strains of EPEC, which show localized adherence to HEp-2 cells, atypical EPEC show different patterns of adherence. Although some investigators have reported localized-like adherence as a predominant adherence pattern of atypical EPEC (28,39), only 1 of 22 strains investigated here displayed this phenotype. The low frequency of localized-like adherence in this study may reflect differences in serotype distribution, as localized-like adherence seems to be most prevalent in atypical EPEC strains in EPEC serogroups, such as O26, O111, and O119 (28), which were infrequently found in this study. Ten of the 22 strains examined exhibited an adherence pattern that was distinct from localized, aggregative, or diffuse adherence (3) and was termed indeterminate adherence (Figure 1). This pattern may have been termed diffuse adherence by other investigators, which would account for the relatively high frequency of diffusely adherent atypical EPEC in some reports and their absence from this study. Seven of 22 strains displayed aggregative adherence despite their lack of known sequences associated with the production of fimbriae of EAEC.
The virulence of atypical EPEC despite their lack of Bfp, which typical EPEC require to cause severe disease, suggests that these bacteria carry an adhesin analogous to Bfp that augments their ability to colonize the intestine. Previous studies, however, have shown only a low frequency of known E. coli adhesins, including aggregative adherence fimbriae, P fimbriae, S fimbriae, PAP pili, and afimbrial adhesins in atypical EPEC strains (40). We extended these findings to show that atypical EPEC are also mostly negative for Efa1 and long polar fimbriae (encoded by lpfD), which contribute to the adhesive capacities of some attaching-effacing strains of E. coli (19,25). Although atypical EPEC may carry an adhesin equivalent to Bfp, which remains to be discovered, these bacteria may use any known E. coli adhesins to bind to the intestine. This suggestion is supported by the marked heterogeneity of atypical EPEC in terms of adhesion pattern and the observation that adhesins other than Bfp can restore cell-binding capacity to EAF-cured strains of typical EPEC (41).
In conclusion, atypical EPEC were the most commonly identified pathogens in a study of community-acquired diarrhea in Melbourne. Infections with atypical EPEC occurred throughout the year and were significantly more common in children. Characterization of a sample of atypical EPEC isolates revealed that these bacteria were antigenically heterogeneous and generally did not belong to O-serogroups associated with EPEC. Further studies are needed to determine the frequency of these bacteria in other communities, their reservoir, mode of spread, and mechanisms of virulence.
Acknowledgments
We are grateful to M.M. Levine and J.B. Kaper for providing the control strains and LEE probes used in the study and Alexander Kuzevski for skillful technical assistance.
This study was supported by grants from the Australian National Health and Medical Research Council, the Murdoch Children's Research Institute, the Cooperative Research Centre for Water Quality and Treatment, the Water Services Association of Australia, the Victorian Department of Human Services, Melbourne Water Corporation, South East Water Limited, Yarra Valley Water Ltd, and City West Water Ltd.
Biography
Dr. Robins-Browne is professor and head of the Department of Microbiology and Immunology at the University of Melbourne and head of microbiological research at the Royal Children's Hospital and the Murdoch Children's Research Institute in Melbourne, Australia. His major research interests are the etiologic agents of bacterial diarrhea and the pathogenesis of bacterial infections, in particular, those caused by Escherichia coli and related bacteria.
Suggested citation for this article: Robins-Browne RM, Bordun A-M, Tauschek M, Bennett-Wood VR, Russell J, Oppedisano F, et al. Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg Infect Dis [serial on the Internet]. 2004 Oct [date cited]. http://dx.doi.org/10.3201/eid1010.031086
Current affiliation: Burnet Institute, Melbourne, Victoria, Australia.
References
- 1.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robins-Browne RM. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev Infect Dis. 1987;9:28–53. 10.1093/clinids/9.1.28 [DOI] [PubMed] [Google Scholar]
- 3.Trabulsi LR, Keller R, Gomes TAT. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–8. 10.1126/science.280.5372.2114 [DOI] [PubMed] [Google Scholar]
- 5.Nougayrède JP, Fernandes PJ, Donnenberg MS. Adhesion of enteropathogenic Escherichia coli to host cells. Cell Microbiol. 2003;5:359–72. 10.1046/j.1462-5822.2003.00281.x [DOI] [PubMed] [Google Scholar]
- 6.Hellard ME, Sinclair MI, Forbes AB, Fairley CK. A randomized, blinded, controlled trial investigating the gastrointestinal health effects of drinking water quality. Environ Health Perspect. 2001;109:773–8. 10.1289/ehp.01109773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellard ME, Sinclair MI, Hogg GG, Fairley CK. Prevalence of enteric pathogens among community based asymptomatic individuals. J Gastroenterol Hepatol. 2000;15:290–3. 10.1046/j.1440-1746.2000.02089.x [DOI] [PubMed] [Google Scholar]
- 8.Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultsz C, Pool GJ, van Ketel R, de Wever B, Speelman P, Dankert J. Detection of enterotoxigenic Escherichia coli in stool samples by using nonradioactively labeled oligonucleotide DNA probes and PCR. J Clin Microbiol. 1994;32:2393–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vial PA, Mathewson JJ, Guers L, Levine MM, DuPont HL. Comparison of two assay methods for patterns of adherence to HEp-2 cells of Escherichia coli from patients with diarrhea. J Clin Microbiol. 1990;28:882–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine MM, Nataro JP, Karch H, Baldini MM, Kaper JB, Black RE, et al. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152:550–9. 10.1093/infdis/152.3.550 [DOI] [PubMed] [Google Scholar]
- 13.Wood PK, Morris JG Jr, Small PLC, Sethabutr O, Toledo MRF, Trabulsi L, et al. Comparison of DNA probes and the Sereny test for identification of invasive Shigella and Escherichia coli strains. J Clin Microbiol. 1986;24:498–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satterwhite TK, Evans DG, DuPont HL, Evans DJ Jr. Role of Escherichia coli colonisation factor antigen in acute diarrhoea. Lancet. 1978;2:181–4. 10.1016/S0140-6736(78)91921-9 [DOI] [PubMed] [Google Scholar]
- 15.Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–33. 10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- 16.Bettelheim KA, Thompson CJ. New method of serotyping Escherichia coli: implementation and verification. J Clin Microbiol. 1987;25:781–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid SD, Betting DJ, Whittam TS. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J Clin Microbiol. 1999;37:2719–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang WL, Kohler B, Oswald E, Beutin L, Karch H, Morabito S, et al. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J Clin Microbiol. 2002;40:4486–92. 10.1128/JCM.40.12.4486-4492.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton HJ, Sloan J, Bennett-Wood V, Adams LM, Robins-Browne RM, Hartland EL. Contribution of long polar fimbriae to the virulence of rabbit-specific enteropathogenic Escherichia coli. Infect Immun. 2004;72:1230–7. 10.1128/IAI.72.3.1230-1239.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klapproth JM, Scaletsky IC, McNamara BP, Lai LC, Malstrom C, James SP, et al. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect Immun. 2000;68:2148–55. 10.1128/IAI.68.4.2148-2155.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yatsuyanagi J, Saito S, Sato H, Miyajima Y, Amano K, Enomoto K. Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J Clin Microbiol. 2002;40:294–7. 10.1128/JCM.40.1.294-297.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutton S, Phillips AD, Smith HR, Gross RJ, Shaw R, Watson P, et al. Screening for enteropathogenic Escherichia coli in infants with diarrhea by the fluorescent-actin staining test. Infect Immun. 1991;59:365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–8. 10.1073/pnas.92.5.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robins-Browne RM, Bennett-Wood V. Quantitative assessment of the ability of Escherichia coli to invade cultured animal cells. Microb Pathog. 1992;12:159–64. 10.1016/0882-4010(92)90119-9 [DOI] [PubMed] [Google Scholar]
- 25.Nicholls L, Grant TH, Robins-Browne RM. Identification of a novel locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol Microbiol. 2000;35:275–88. 10.1046/j.1365-2958.2000.01690.x [DOI] [PubMed] [Google Scholar]
- 26.Yatsuyanagi J, Saito S, Miyajima Y, Amano K, Enomoto K. Characterization of atypical enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan. J Clin Microbiol. 2003;41:2033–9. 10.1128/JCM.41.5.2033-2039.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scaletsky IC, Pedroso MZ, Oliva CA, Carvalho RL, Morais MB, Fagundes-Neto U. A localized adherence-like pattern as a second pattern of adherence of classic enteropathogenic Escherichia coli to HEp-2 cells that is associated with infantile diarrhea. Infect Immun. 1999;67:3410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelayo JS, Scaletsky IC, Pedroso MZ, Sperandio V, Girón JA, Frankel G, et al. Virulence properties of atypical EPEC strains. J Med Microbiol. 1999;48:41–9. 10.1099/00222615-48-1-41 [DOI] [PubMed] [Google Scholar]
- 29.Feng P, Lampel KA, Karch H, Whittam TS. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–3. 10.1086/517438 [DOI] [PubMed] [Google Scholar]
- 30.Marshall JA, Hellard ME, Sinclair MI, Fairley CK, Cox BJ, Catton MG, et al. Incidence and characteristics of endemic Norwalk-like virus-associated gastroenteritis. J Med Virol. 2003;69:568–78. 10.1002/jmv.10346 [DOI] [PubMed] [Google Scholar]
- 31.Viljanen MK, Peltola T, Junnila SY, Olkkonen L, Jarvinen H, Kuistila M, et al. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet. 1990;336:831–4. 10.1016/0140-6736(90)92337-H [DOI] [PubMed] [Google Scholar]
- 32.Bouzari S, Jafari MN, Shokouhi F, Parsi M, Jafari A. Virulence-related DNA sequences and adherence patterns in strains of enteropathogenic Escherichia coli. FEMS Microbiol Lett. 2000;185:89–93. 10.1111/j.1574-6968.2000.tb09045.x [DOI] [PubMed] [Google Scholar]
- 33.Galane PM, Le Roux M. Molecular epidemiology of Escherichia coli isolated from young South African children with diarrhoeal diseases. J Health Popul Nutr. 2001;19:31–8. [PubMed] [Google Scholar]
- 34.Knutton S, Shaw R, Phillips AD, Smith HR, Willshaw GA, Watson P, et al. Phenotypic and genetic analysis of diarrhea-associated Escherichia coli isolated from children in the United Kingdom. J Pediatr Gastroenterol Nutr. 2001;33:32–40. 10.1097/00005176-200107000-00006 [DOI] [PubMed] [Google Scholar]
- 35.Paciorek J. Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol. 2002;51:548–56. [DOI] [PubMed] [Google Scholar]
- 36.Kukuruzovic R, Robins-Browne RM, Anstey NM, Brewster DR. Enteric pathogens, intestinal permeability and nitric oxide production in acute gastroenteritis. Pediatr Infect Dis J. 2002;21:730–9. 10.1097/00006454-200208000-00007 [DOI] [PubMed] [Google Scholar]
- 37.Robins-Browne RM, Yam WC, O'Gorman LE, Bettelheim KA. Examination of archetypal strains of enteropathogenic Escherichia coli for properties associated with bacterial virulence. J Med Microbiol. 1993;38:222–6. 10.1099/00222615-38-3-222 [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto T, Taneike I. The sequences of enterohemorrhagic Escherichia coli and Yersinia pestis that are homologous to the enteroaggregative E. coli heat-stable enterotoxin gene: cross-species transfer in evolution. FEBS Lett. 2000;472:22–6. 10.1016/S0014-5793(00)01414-9 [DOI] [PubMed] [Google Scholar]
- 39.Beutin L, Marches O, Bettelheim KA, Gleier K, Zimmermann S, Schmidt H, et al. HEp-2 cell adherence, actin aggregation, and intimin types of attaching and effacing Escherichia coli strains isolated from healthy infants in Germany and Australia. Infect Immun. 2003;71:3995–4002. 10.1128/IAI.71.7.3995-4002.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vieira MA, Andrade JR, Trabulsi LR, Rosa AC, Dias AM, Ramos SR, et al. Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry EAE and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J Infect Dis. 2001;183:762–72. 10.1086/318821 [DOI] [PubMed] [Google Scholar]
- 41.Francis CL, Jerse AE, Kaper JB, Falkow S. Characterization on interactions of enteropathogenic Escherichia coli O127:H6 with mammalian cells in vitro. J Infect Dis. 1991;164:693–703. 10.1093/infdis/164.4.693 [DOI] [PubMed] [Google Scholar]