Abstract
Apolipoprotein E (APOE)-ɛ4 is associated with a deleterious outcome after ischemic brain injury, which may involve abnormal regulation of mitochondrial function. We have assessed the mitochondrial proteomic response of APOE-ɛ3 and APOE-ɛ4 transgenic mice to transient global ischemic injury in the hippocampus. A genotype-dependent increase in ApoE levels in mitochondria was observed after ischemia, with APOE-ɛ4 mice showing significantly greater increases than APOE-ɛ3 mice. Quantitative analysis of the mitochondria-enriched fractions was performed using liquid-chromatography mass spectrometry coupled to label-free analysis. Of the 1,067 identified proteins, 274 were mitochondria associated. Mitochondrial protein expression was significantly different between genotypes under basal conditions as well as in response to global ischemia. A total of 12 mitochondrial proteins (including respiratory chain proteins NDUFA11, NDUFS3, NDUF5B, ATP5J, as well as ETFA, CYB5B, ATP6V1A, HSPA1B, OXR1, GLUL, IARS2, and PHYHIPL) were significantly altered with respect to genotype, global ischemia, or their interaction (P<0.01). A compelling interactome, created using proteins found to be significantly modulated by global ischemia (P<0.05), involved proteins that regulate energy production and oxidative stress. Thus, APOE genotype has a differential effect on the mitochondrial protein expression in the absence and presence of an injury, which may underlie the differing genotype susceptibility.
Keywords: apolipoprotein E, ischemia, mitochondria, proteomics
Introduction
Of the different forms of apolipoprotein E (ApoE 2, 3, and 4), ApoE4 has a deleterious role in ischemic brain injury and is associated with a worse response and poorer long-term outcome in response to acute focal ischemic stroke, transient global ischemia, and hemorrhage (Horsburgh et al, 2000a, 2000b; Sheng et al, 1998). Possession of an APOE-ɛ4 allele is also associated with a deleterious influence on cognitive aging and is the major genetic risk factor for Alzheimer's disease (Saunders et al, 1993). Although the underlying mechanism for the deleterious effect of ApoE4 is not yet known, there are strong links between ApoE4 and mitochondrial dysfunction that might explain a general influence of ApoE4 on injury and disease. Mitochondria are critical in regulating normal neuronal functions and are disrupted in injury and disease (Mattson et al, 2008), often influenced by the presence of APOE-ɛ4. Notably, one of the earliest features detectable in the brains of young and elderly APOE-ɛ4 individuals (de Leon et al, 2001) is regional cerebral hypometabolism, a reflection of reduced mitochondrial function. This regional hypometabolism is further exacerbated in disease in APOE-ɛ4 carriers (Mielke et al, 1998). Transgenic APOE-ɛ4 mice also exhibit structural (Shenk et al, 2009) and functional (Strum et al, 2007; Choi et al, 2004) mitochondrial abnormalities. It has also been shown that there are structural and subsequent biophysical differences between ApoE protein isoforms, which may explain differential influences on mitochondrial function (Huang et al, 2004). The structural conformation of ApoE4 ensures more efficient neuronal proteolytic cleavage compared with ApoE2 or ApoE3 with generated ApoE4 fragments causing neurodegeneration and cognitive deficits in transgenic mice (Harris et al, 2003). There is direct evidence from in vitro cell-culture studies that overexpression of a truncated ApoE4 fragment, localizes to mitochondria and impairs mitochondrial function (Chang et al, 2005). Similarly, a number of mitochondrial proteins have been identified as ApoE4-binding partners from cytosolic and membrane fractions of mouse brain (Nakamura et al, 2009). Thus, ApoE isoforms may have differential effects on mitochondrial function that, in the presence of ApoE4, could render mitochondria susceptible to the effects of brain injury.
At present, the protein signature of mitochondria in different APOE genotypes is not known and is important as it could provide a basis on which to further explore biological differences between the different ApoE isoforms. To address this, we used liquid-chromatography mass spectrometry (LC-MS), which permits the quantitative assessment of large numbers of proteins simultaneously in brain tissue samples, and obtained a measure of mitochondrial proteins at baseline and in response to an ischemic challenge in APOE-ɛ3 and APOE-ɛ4 transgenic mice. The data demonstrate that not only is the protein signature different between ApoE isoforms at basal conditions but that there is a differential response of these proteins in response to injury.
Materials and methods
APOE Transgenic Mice
APOE-ɛ3 and APOE-ɛ4 transgenic mice were produced as described previously (Xu et al, 1996). Briefly, APOE knockout mice were generated by inactivating the endogenous APOE gene. Cosmid libraries were constructed from lymphoblasts of humans who were homozygous for APOE-ɛ3 and APOE-ɛ4. Fragments containing human regulatory sequences and the coding sequences for human APOE (ɛ3 and ɛ4) were isolated and injected into single-cell embryos from homozygous APOE-deficient mice and transferred to female recipients (a cross of C57BL/6J × DBA/2J) to produce founder transgenic mice. Founder mice were backbred to APOE knockout mice to produce mice hemizygous for human APOE transgenes and homozygous for inactivated mouse APOE gene. Animals were bred to homozygosity for human APOE transgenes and backbred into APOE knockout strains to remove the genetic influence of DBA/2J background strains. The presence of human APOE was confirmed by PCR. The mice were backcrossed at least 10 times to C57BL/6J mice in the USA before transport to Edinburgh, UK and thereafter several backcrosses to C57BL/6J were made. For the present study, the APOE-ɛ3-437 and APOE-ɛ4-81 lines were used, which carry two copies of the respective transgenes and are best matched for ApoE levels from the original lines bred (Xu et al, 1996). All procedures were authorized under the Home Office approved project license number 60/3722. This license was approved by the University of Edinburgh's Ethical Review Committee and the Home Office and adhered to regulations specified in the Animals (Scientific Procedures) Act (1986). Mouse genotype was confirmed by PCR (Horsburgh et al, 2000b).
Transient Global Ischemia
Adult, male mice were initially anesthetized with 5% isofluorane with oxygen, and maintained in 1.5% isofluorane. Temperature was monitored using a rectal probe and controlled to 37±0.5°C. A midline cervical incision was made and both common carotid arteries exposed and isolated using 4/0 silk thread. The common carotid arteries were occluded using microaneurysm clips applied bilaterally for 20 minutes. The duration of ischemia was based on previous observations, indicating that this duration was sufficient to induce ischemic neuronal damage in the caudate nucleus (Horsburgh et al, 2000b; Kelly et al, 2001). After the period of occlusion, the clips were removed and blood flow through the arteries was confirmed before the incision was sutured closed (n=9, APOE-ɛ3 mice; n=9, APOE-ɛ4 mice). Sham-operated animals underwent an identical procedure except clips were not applied to the common carotid arteries (n=5, APOE-ɛ3 mice; n=6, APOE-ɛ4). The animals were placed in an incubator for 1 hour after which they were returned to the animal housing and allowed to recover. At 72 hours after treatment, the animals were deeply anesthetized with 5% isofluorane with oxygen, and perfused transcardially with buffered saline and the brain removed and halved. One hemisphere was fixed using 4% paraformaldehyde and then paraffin embedded for histological analyses. From the other hemisphere, the hippocampus was dissected out and used to prepare mitochondrial fractions. At the outset of the study, mice were coded and randomized for surgery with the operator blind to the genotype of the mice. The study was conducted with a priori hypothesis.
Hematoxylin and Eosin Staining to Determine Ischemic Damage
Paraffin-embedded sections (6 μmol/L) were cut and stained with hematoxylin and eosin. The numbers of morphologically normal neurons and neurons showing features of ischemic cell change, defined by intense hematoxylin-stained pyknotic nuclei surrounded by eosinophilic cytoplasm, were counted in five randomly selected fields within the CA1 region of the hippocampus, using a 1-cm2 grid at × 400 magnification. The percentage of ischemic neurons was calculated for the area of interest.
Preparation of Mitochondrial Fractions
From each animal (n=29), mitochondrial fractions were prepared from a single hippocampus. Mitochondria were prepared from fresh tissue and all steps were performed on ice. Mitochondrial fractions were prepared using the Benchtop Mitochondria Isolation Kit for Rodent Tissue (Mitosciences, Eugene, OR, USA), generating two fractions: a mitochondrial fraction (M) and a postmitochondrial supernatant fraction (PM). The mitochondrial fraction is enriched in mitochondrial proteins, while the supernatant fraction is predominantly cytosolic, containing soluble and membrane-associated proteins. Protein concentrations of the fractions were determined using Pierce BCA assay.
Immunoblotting and Antibody Detection
Proteins were separated by SDS–PAGE and transferred onto PVDF membrane (GE Healthcare, Buckinghamshire, UK). Immunoblotting was performed using the Odyssey Infrared Imaging System (LiCor Biosciences, Lincoln, NE, USA). Membranes were blocked in Odyssey blocking buffer (diluted 1:1 with phosphate-buffered saline) and washed in phosphate-buffered saline–Tween (phosphate-buffered saline with 0.1% Tween). Primary antibodies were detected using fluorescently labelled secondary antibodies (LiCor Biosciences). Proteins were visualized by scanning antibody-labelled blots in the Odyssey Imager under the appropriate channel. Antibodies for western blotting were obtained from relevant manufacturer's: ApoE (Calbiochem, Darmstadt, Germany), COXI (Mitosciences, Eugene, OR, USA), COXIV (Cellular Signalling, Herfordshire, UK) and VDAC1 (AbCam, Cambridge, UK).
Analysis of Proteins from Immunoblots
Quantification was performed using the Odyssey image analysis program (LiCor Biosciences). In brief, the appropriate band was selected, background subtraction applied, and a resulting optical density (OD) value computed. To determine enrichment of mitochondrial markers (VDAC1, COXI, and COXIV), OD intensities were obtained from both mitochondrial and postmitochondrial fractions. These were used to determine the percentage of each protein in the mitochondrial fraction and allowed the assessment of mitochondrial integrity. For ApoE quantification, all raw OD values were normalized to recombinant ApoE3 (Chemicon, Watford, UK), loaded to enable intergel comparisons. This resulted in a normalized OD value (norm OD) that was analyzed statistically.
Liquid-Chromatography Mass Spectrometry Analysis of Mitochondria-Enriched Fractions
High-throughput proteomics using liquid chromatography in tandem with mass spectrometry has emerged as a valuable tool in understanding global protein expression without prior knowledge of the studied system (for recent review, see Nilsson et al, 2010). To examine the proteomic response in APOE transgenic mice after ischemic injury, we conducted a quantitative analysis of the mitochondria-enriched fractions using a shotgun proteomic LC-MS approach coupled to a label-free analysis using MaxQuant software.
Four groups were analyzed with the number of animals within each group corresponding to that already examined for ApoE levels (ApoE3 sham n=5, ApoE3 global ischemia n=9; ApoE4 sham n=6, ApoE4 global ischemia n=9). All chemicals were purchased from Sigma-Aldrich (Gillingham, UK) unless otherwise stated. Acetonitrile and water for LC-MS/MS were HPLC grade (Fisher, Loughborough, UK). Formic acid was Suprapure 98% to 100% (Merck, Darmstadt, Germany) and trifluoroacetic acid was 99% purity sequencing grade. All HPLC-MS connector fittings were from Upchurch Scientific or Valco (Hichrom, Theale, UK and RESTEK, Saunderton, UK).
In all, 100 μg of protein extract from mitochondrial fractions was resuspended in 12.5 μL of 6 mol/L urea, 2.5 μL of 200 mmol/L DTT, 2.5 μL of 1 mol/L ammonium bicarbonate, and 1 μL of 5% CHAPS. The samples were reduced at room temperature for 30 minutes then 2.5 μL of 500 mmol/L iodoacetamide was added. Samples were diluted to 50 μL, 2 μg of trypsin was added and digested overnight. A fraction of the tryptically digested samples (10 μg) was cleaned using an in-house SCX column (200 μm × 6 cm) packed with 10 μm PolySulfoethyl (Poly LC Inc., Columbia, MD, USA) as follows: SCX columns were conditioned with 100 μL of 10 mmol/L potassium phosphate buffer (KPB), pH 3. Tryptic digests (5 μL) were diluted with 45 μL of 50 mmol/L H3PO4, 10% acetonitrile and loaded on the SCX column. SCX columns were washed with 40 mmol/L KPB, 10% acetonitrile and eluted with 75 μL of 200 mmol/L ammonium bicarbonate.
Capillary-HPLC-MSMS data were collected on an online system consisting of a micropump (1,200 binary HPLC system, Agilent, Edinburgh, UK) coupled to a hybrid LTQ-Orbitrap XL instrument (Thermo-Fisher, Loughborough, UK). The LTQ was controlled through Xcalibur 2.0.7 and LTQ Orbitrap XL MS2.4SPI (Thermo-Fisher, Loughborough, UK). HPLC-MS methods have been described previously (Le Bihan et al, 2010). Peptides were reconstituted in 10 μL of loading buffer before injection and 8 μL was loaded. The peptide mixture was separated using a 2-hour gradient from 0% to 100% Buffer B (90% acetonitrile, 10% H2O, 0.025% trifluoroacetic acid, and 0.1% formic acid).
The mass spectrometer was operated in ‘data-dependent mode,' with a single MS scan (400 to 2,000 m/z) in FT mode 60K resolution followed by MS/MS scans in the linear ion trap on the three most abundant ions and dynamic excluded for 120 seconds.
Protein Identification and Quantification
Conversion from RAW to MGF files was performed as described previously (Le Bihan et al, 2010). MS/MS data were searched using MASCOT Versions 2.3 (Matrix Science Ltd., London, UK) against a mouse plus contaminant IPI database with 55,413 sequences downloaded from http://www.ebi.ac.uk (version v3.42) and amended with the human ApoE and ApoE4 sequences due to the transgenic nature of the mouse model. Variable methionine oxidation, STY phosphorylation, protein N-terminal acetylation, and fixed cysteine carbamidomethylation were used in all searches. Precursor mass tolerance was set to 7 p.p.m. and MS/MS tolerance to 0.4 a.m.u. The significance threshold (P) was set <0.05 (MudPIT scoring in Mascot). All LC-MS runs were combined using MaxQuant (version 1.0.13.8, Max Plank Institute of Biochemistry, Martinsried, Germany), assuming a false positive rate of 0.01. The MaxQuant output of protein group intensities, calculated as the sum of MS-peak intensities of identified peptides, was used as a relative abundance measure between samples. The data were converted using the Pride converter v2.4.2 (Barsnes et al, 2009) and are available on the public data repository PRIDE (http://www.ebi.ac.uk/pride/; project ‘Mouse APOE,' accessions 15885 to 15913).
MaxQuant Label-Free Analysis Validation
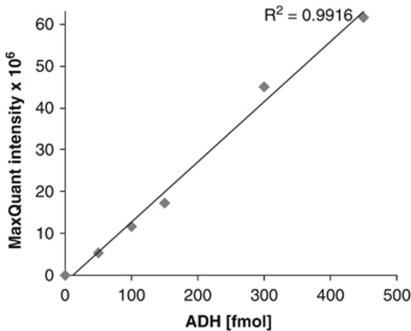
The quantitation of relative protein abundance using MaxQuant was validated by spiking alcohol dehydrogenase (ADH Saccharomyces cerevisiae, Sigma, Gillingham, UK) into 10 mL HeLa cell extract (Dundee Cell, Dundee, UK, see Le Bihan et al, 2010). Spiked samples were analyzed back-to-back in 2-hour LC gradients. Three unspiked samples were run at the start, middle, and end of the queue to check for any carryover. The intensity of ADH in the unspiked samples was zero, indicating no carryover issues for ADH peptides. Spiked ADH concentrations were correlated with protein group intensities. A linear fit showed a positive correlation of spiked concentration with the protein group intensity computed by MaxQuant, with an R2 value of 0.992 (see Figure 1). This control experiment provides validation for the use of MaxQuant as a label-free analysis tool for the results presented below.
Figure 1.
Validation for the use of MaxQuant as a label-free analysis tool. Quantitation of relative protein abundance using MaxQuant was validated by spiking alcohol dehydrogenase (ADH Saccharomyces cerevisiae, Sigma) into 10 mL HeLa cell extracts. A linear fit showed a correlation of spiked ADH concentrations with the protein group intensity computed by MaxQuant, with an R2 value of 0.992.
Bioinformatics Analysis
All proteins identified by LC-MS and Maxquant software (version 1.0.13.8) by at least two or more peptides were uploaded to the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov; Huang da et al, 2009) to determine subcellular localization based on gene ontology (GO). The experimental interest and focus for this study were changes in the levels of mitochondrial proteins as a result of global ischemia in the context of two different APOE genotypes. Cognizant of the fact that mitochondrial preparations are enriched in, but are not exclusively mitochondrial proteins, proteins included in the GO term mitochondrion (GO:0005739) were identified from the master list, filtered out and subsequently analyzed statistically to determine differences in expression levels for this select protein pool.
Statistical Analysis
Immunoblot data are presented as mean±s.e.m. Data were analyzed using GraphPad Prism version 5 (GraphPad Software Inc., La Jolla, CA, USA). Analysis of mitochondrial marker enrichment and quantification of ApoE levels from mitochondrial fractions were performed using two-way analyses of variance (ANOVAs) to allow simultaneous comparison between treatment groups (sham operated or global ischemia) and genotype (APOE-ɛ3 or APOE-ɛ4). Post hoc comparisons of two-way ANOVAs were made using t-test with Bonferroni correction unless otherwise noted. A value of P<0.05 was considered to be statistically significant.
The primary statistical analysis for proteomic data focused on mitochondrial proteins only, as extracted from DAVID based on GO:0005739. Signal intensities of the mitochondria-associated proteins involved calculations with two-way ANOVA using the MultiExperiment Viewer: http://www.tm4.org/mev/ (Saeed et al, 2003). Results of the two-way ANOVA are presented within functional categories (DAVID, Uniprot) along with the F-ratio for each factor of each protein found to have an F-ratio greater than the critical value of 7.77 (P<0.01) with respect to APOE genotype, global ischemia or their interaction. A conservative probability was selected to reduce the rate of identifying false positives, but was relaxed slightly for network analysis (see below). Protein functions were determined primarily from annotations in Uniprot (http://www.uniprot.org/) and DAVID protein databases. For indicative purposes, selected biologically relevant proteins including human ApoE found to differ significantly by two-way ANOVA are presented with accompanying histograms along with results of post hoc comparisons using t-test with Bonferroni correction.
Network Analysis
Three separate data sets containing only proteins considered to be significantly affected by a given factor (APOE genotype, global ischemia, or their interaction) with F-ratio values >4.24 were uploaded with their corresponding P values to Ingenuity Pathway Analysis (IPA) software. Protein identifiers were mapped to corresponding objects in Ingenuity's Knowledge Base. Networks were algorithmically generated based on direct relationships (physical interaction) between eligible proteins. The shading of network objects is negatively correlated with their P value. IPA generates a score which is putatively a measure of probability (but see Deighton et al, 2010 for critical analysis) for the network.
Results
Integrity of Mitochondrial Fractions
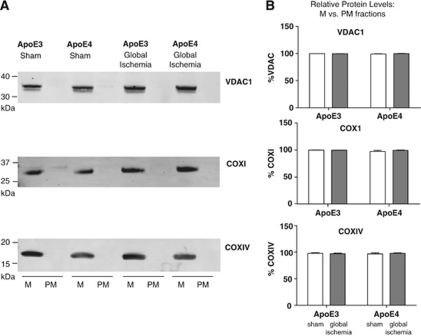
Mitochondrial markers (VDAC1, COXI, and COXIV) were used to determine the enrichment of mitochondrial proteins in mitochondria fractions isolated from hippocampal tissue of APOE transgenic mice as well as their absence in postmitochondrial fraction supernatants (Figure 2A). VDAC1 is an outer mitochondrial membrane protein involved in metabolite diffusion, while both COXI and COXIV are components of the electron transport chain, located within the inner mitochondrial membrane. All three antibodies show presence of mitochondrial proteins in the enriched fraction. There was no significant effect of either ischemia or genotype on the percentage of VDAC1, COXI, or COXIV in the mitochondrial fraction (Figure 2B). None of these proteins were found in the postmitochondrial fractions. These data indicate there has been minimal breakdown of mitochondrial membranes during fractionation, and neither ischemia nor APOE genotype influence mitochondrial fractionation or integrity.
Figure 2.
Mitochondrial integrity of fractions. (A) Representative images of VDAC1, COX1, and COX1V immunoblots of sham and global ischemia apolipoprotein E (APOE)-ɛ3 and APOE-ɛ4 hippocampal tissue. The hippocampus of each was fractionated into mitochondrial (M) and postmitochondrial supernatant (PM) fractions. Mitochondrial proteins are detected in the mitochondrial fraction only as evident by detection of outer membrane marker VDAC1 or inner membrane markers COXI and COXIV. (B) The percentage of VDAC1, COXI, and COXIV measured in immunoblots in the mitochondrial fraction as compared with postmitochondrial fraction was calculated. Two-way analysis of variance (ANOVA) determined no significant effect of either treatment or APOE genotype.
Genotype-Dependent Increase in Hippocampal Apolipoprotein E Levels of APOE-ɛ4 Transgenic Mice After Transient Global Ischemia
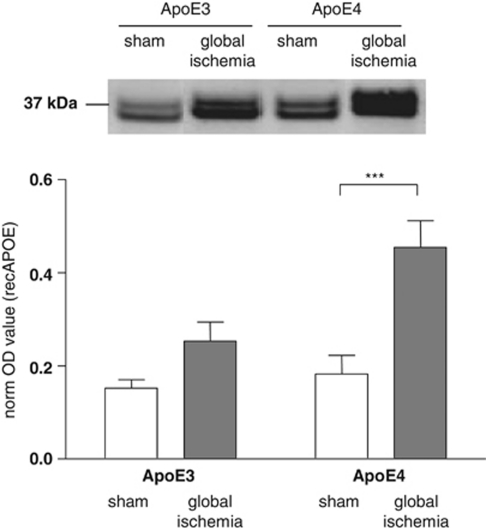
Levels of full-length ApoE were measured by immunoblot in mitochondria-enriched fractions prepared from the hippocampus by immunoblot in shams and after ischemia. There were similar levels of ApoE in sham APOE-ɛ3 and APOE-ɛ4 mice. However in response to ischemia, there was a marked increase in ApoE levels in APOE-ɛ3 and APOE-ɛ4 transgenic mice (Figure 3). The levels of ApoE were significantly greater in APOE-ɛ4 as compared with APOE-ɛ3 mice after ischemia. Two-way ANOVA yielded a main effect for treatment (F(1, 25)=14.30, P<0.001) and genotype (F(1, 25)=5.49, P<0.05). No interaction effect was observed (F(1, 25)=3.02, P>0.05). Post hoc comparisons identified that ApoE levels were significantly higher in APOE-ɛ4 mice with ischemic injury (2.5-fold higher) than in sham-operated APOE-ɛ4 mice (P<0.001).
Figure 3.
Full-length ApoE protein is increased in mitochondrial fractions after global ischemia. Full-length apolipoprotein E (ApoE) was detected in immunoblots of mitochondria prepared from the hippocampus of APOE transgenic mice after global ischemia (APOE-ɛ3 n=9; APOE-ɛ4 n=9) or in sham-operated controls (APOE-ɛ3 n=5; APOE-ɛ4 n=6). ApoE is present as a doublet at 34 kDa. ApoE protein expression levels were quantified. Post hoc comparisons showing statistical significance are shown. ***P<0.001.
Ischemic Neuronal Damage in the Hippocampus
The percentage of ischemic neuronal damage was assessed within the CA1 region of the hippocampus. In all sham-operated APOE-ɛ3 and APOE-ɛ4 transgenic mice, there was no ischemic neuronal damage. Transient global ischemia for 20 minutes led to minimal neuronal damage in the hippocampus of APOE-ɛ3 and APOE-ɛ4 transgenic mice (APOE-ɛ3 1.5±1.0%, APOE-ɛ4 1.6±1.2% ischemic neuronal damage), with no significant difference between genotypes (P=0.50). The low level of histological damage in the study limits definitive exploration of genotype influences (see Horsburgh et al, 2000b).
Quantitative Proteomic Analysis of Mitochondria-Enriched Fractions
Using Maxquant software as a protein clustering and validation tool, and assuming a false positive rate of 0.01 based on a decoy database search, a total of 1,283 proteins were identified across all four groups, and of this, 1,067 were identified with a minimum of 2 peptides, the criterion for analytical inclusion (see Supplementary Table 1 for full list of proteins).
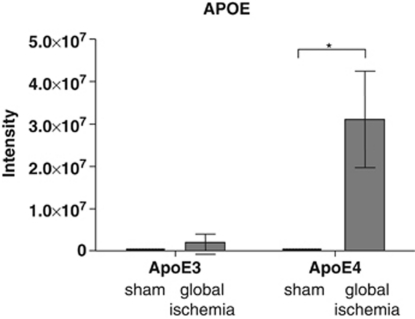
LC-MS confirmed that human ApoE levels were significantly higher in mitochondrial fractions from APOE-ɛ4 transgenic mice after global ischemia (Figure 4). There was a significant effect of global ischemia on ApoE levels (F(1, 25)=4.798, P<0.05) and a significant difference between APOE-ɛ4 mice with ischemic injury and sham-operated APOE-ɛ4 mice (P<0.05), whereas in both sham groups ApoE was undetectable.
Figure 4.
Liquid-chromatography mass spectrometry (LC-MS) detected increased apolipoprotein E (ApoE) protein intensity levels in mitochondrial fractions after global ischemia. ApoE was detected using LC-MS in mitochondria prepared from the hippocampus of APOE transgenic mice after global ischemia (APOE-ɛ3 n=9; APOE-ɛ4 n=9) or in sham-operated controls (APOE-ɛ3 n=5; APOE-ɛ4 n=6) using LC-MS. There was a significant effect of global ischemia. Post hoc comparisons showing statistical significance are shown. *P<0.05.
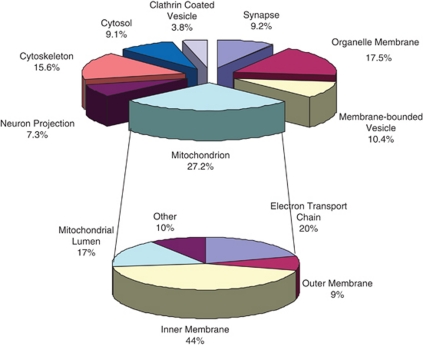
In all, 274 proteins were identified as mitochondria associated based on the GO designation mitochondrion, from the 1,067 analyzed using DAVID software (see Supplementary Table 2 for full list of mitochondrial proteins). This distribution of mitochondrial versus nonmitochondrial proteins is in agreement with other published studies using similar mitochondrial preparations (Forner et al, 2006). The categorization based on function and subcellular localization as determined by DAVID analysis for all the proteins identified is shown in Figure 5 along with the functional annotation of the mitochondrial proteins.
Figure 5.
Functional annotation of all identified proteins as well as mitochondrial proteins across all four cohorts. Functional groupings of identified proteins based on Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis are shown in the top pie chart. Of the 1,067 proteins uploaded to DAVID, the largest functional category, including 274 proteins, was identified as mitochondrion. Further DAVID analysis of these 274 proteins is shown in the lower pie chart with the majority of identified proteins localized to the inner membrane.
In all, 12 of the 274 mitochondrial proteins were significantly altered (two-way ANOVA; P<0.01) in relation to APOE genotype (two proteins: ETFA and ATP6V1A), global ischemia (five proteins: NDUFB5, CYB5B, HSPA1B, GLUL, and PHYHIPL), and interaction (five proteins: NDUFA11, NDUFS3, ATP5J, OXR1, and IARS2). Functional categorization of these proteins show a bias toward identifying changes in proteins involved in oxidative phosphorylation and stress (see Table 1).
Table 1. Altered mitochondrial proteins.
| IPI | Gene name | F-ratio (genotype) | F-ratio (global ischemia) | F-ratio (interaction) | Protein function |
|---|---|---|---|---|---|
| Oxidative phosphorylation/tricarboxylic acid cycle | |||||
| IPI00318645 | Ndufa11 | 2.06 | 3.13 | 19.75 | Complex I subunit |
| IPI00132531 | Ndufb5 | 5.69 | 8.07 | 0.12 | Complex I subunit |
| IPI00121288 | Ndufb10 | 4.33 | 0.00 | 1.91 | Complex I subunit |
| IPI00121309 | Ndufs3 | 0.55 | 3.02 | 8.90 | Complex I subunit |
| IPI00129516 | Uqcrh | 0.16 | 0.42 | 4.55 | Complex III subunit |
| IPI00111885 | Uqcrc1 | 0.73 | 4.70 | 0.89 | Complex III subunit |
| IPI00114377 | Cox7a2 | 0.87 | 0.38 | 5.60 | Complex IV subunit |
| IPI00230507 | Atp5h | 1.26 | 0.59 | 7.34 | Complex V subunit |
| IPI00125460 | Atp5j | 0.00 | 0.12 | 7.88 | Complex V subunit |
| IPI00453777 | Atp5d | 2.11 | 0.12 | 4.69 | Complex V subunit |
| IPI00315794 | Cyb5b | 1.10 | 8.98 | 2.53 | Electron transport |
| IPI00116753 | Etfa | 10.33 | 0.55 | 1.84 | Electron transport |
| IPI00261627 | Sucla2 | 5.01 | 1.28 | 0.33 | Tricarboxylic acid cycle enzyme |
| IPI00323592 | Mdh | 0.55 | 4.69 | 3.48 | Tricarboxylic acid cycle enzyme |
| IPI00230754 | Slc25a11 | 5.07 | 0.56 | 0.14 | Carrier protein |
| Reactive oxygen species control/oxidative stress | |||||
| IPI00660262 | Gpx4 | 0.10 | 0.98 | 5.94 | Antioxidant |
| IPI00121788 | Prdx1 | 0.03 | 6.85 | 0.57 | Antioxidant |
| IPI00130589 | Sod1 | 0.08 | 4.39 | 4.70 | Antioxidant |
| IPI00117929 | Oxr1 | 5.80 | 7.64 | 14.79 | Protects from oxidative damage |
| IPI00319518 | Lonp1 | 6.44 | 4.36 | 0.16 | Proteolysis of oxidized proteins |
| IPI00346073 | Hspa1b | 0.74 | 24.26 | 0.74 | Chaperone protein |
| IPI00229080 | Hsp90ab1 | 2.04 | 5.94 | 0.41 | Chaperone protein |
| Other metabolic processes | |||||
| IPI00626790 | Glul | 1.68 | 13.02 | 1.65 | Glutamate metabolism |
| IPI00132593 | Gls2 | 1.04 | 0.02 | 4.40 | Glutamine catabolism |
| IPI00154047 | Hibch | 5.15 | 2.22 | 6.92 | Amino-acid metabolism |
| IPI00116222 | Hibadh | 5.63 | 0.59 | 0.47 | Amino-acid metabolism |
| IPI00227773 | Ppp1cc | 0.01 | 3.16 | 5.30 | Glycogen metabolism |
| IPI00750256 | Ak1 | 0.36 | 5.33 | 2.53 | ATP/ADP interconversion |
| IPI00221769 | Ak3 | 4.10 | 3.02 | 6.14 | AMP phosphorylation |
| IPI00453499 | Iars2 | 1.41 | 1.68 | 7.86 | Protein synthesis |
| Lipid metabolism | |||||
| IPI00119114 | Acadl | 0.33 | 0.00 | 4.25 | Lipid metabolism |
| IPI00154054 | Acat | 5.11 | 0.44 | 2.31 | Lipid metabolism |
| IPI00468653 | Pccb | 5.36 | 5.36 | 5.85 | Lipid metabolism |
| Apoptosis | |||||
| IPI00404551 | Ctsd | 1.85 | 6.46 | 1.80 | Protease/Apoptosis |
| Protein import | |||||
| IPI00134484 | Timm13 | 2.07 | 6.38 | 0.00 | Small transporter inner membrane complex |
| IPI00165902 | Timm10 | 2.96 | 0.39 | 5.73 | Small transporter inner membrane complex |
| Mitochondrial transport | |||||
| IPI00515349 | Rhot1 | 5.09 | 0.69 | 2.22 | Attachment to microtubules |
| IPI00473646 | Rhot2 | 2.16 | 3.20 | 4.31 | Attachment to microtubules |
| Unknown mitochondrial function | |||||
| IPI00115827 | Gbas | 0.09 | 1.37 | 5.50 | NipSnap family/vesicular transport |
| IPI00407692 | Atp6v1a | 8.14 | 0.14 | 0.01 | Subunit of V-ATPase/proton transport |
| IPI00648312 | Nt5dc3 | 2.37 | 1.79 | 6.77 | ? |
| IPI00224093 | Phyhipl | 2.07 | 14.02 | 2.07 | ? Central nervous system development |
| IPI00655156 | Ccdc109a | 4.75 | 1.15 | 1.40 | Coiled-coil domain family |
| IPI00331549 | Dhrs1 | 1.33 | 4.82 | 0.66 | Oxidoreductase |
| IPI00135106 | Mobp | 1.60 | 0.08 | 7.30 | Myelin sheath stability |
| IPI00131614 | Sncb | 0.85 | 0.03 | 5.59 | ? Neuronal plasticity |
| IPI00131896 | Brp44 | 0.04 | 0.09 | 4.41 | ? |
| IPI00153792 | Qil1 | 0.16 | 0.04 | 4.94 | ? |
The critical values F (1, 25) for P<0.01 and P<0.05 are 7.77 and 4.24, respectively. Of the 275 mitochondrial proteins, 12 presented here have F values >7.77 in respect to genotype (2), global ischemia (5), or their interaction (5). All mitochondrial proteins with an F value >4.24 for at least one comparison are included in the table and highlighted in bold. Proteins are presented in functional categories as they relate to mitochondrial function.
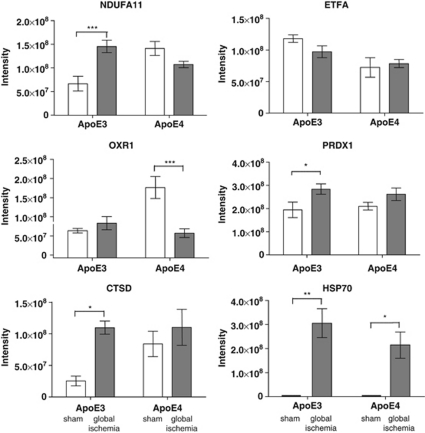
Illustrative changes for some proteins found to have relevance to ischemia are shown in Figure 6 (Chan et al, 1998; Kelly et al, 2001). For example, marked increases in HSPA1B (HSP70) were observed in APOE-ɛ3 and APOE-ɛ4 mice after global ischemia. OXR1 was reduced in APOE-ɛ4 with global ischemia, but was not significantly different in APOE-ɛ3 mice. Certain patterns of protein expression are indicative of differential regulation in response to ischemic injury as a function of genotype.
Figure 6.
Protein intensity levels of selected mitochondrial proteins significantly modulated as a result of global ischemia, apolipoprotein E (APOE) genotype, or both. Normalized peptide protein intensity levels of selected proteins across all four experimental cohorts are indicative of differential protein regulation. Post hoc comparisons are shown. *P<0.05, **P<0.01, ***P<0.001.
Ingenuity Pathway Analysis Network Analysis
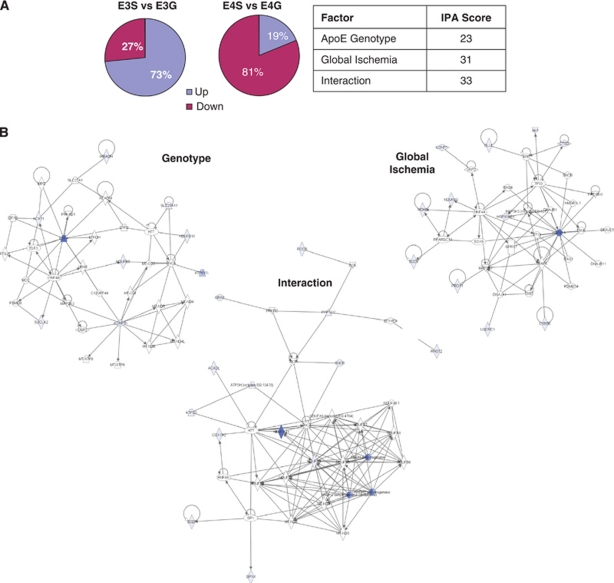
IPA, applied judiciously, allows the synthesis of protein interactions from expansive protein lists and facilitates a better understanding of complex biological systems based on the creation of interactome networks. IPA network analysis was performed on three protein data sets referred to as APOE genotype, global ischemia, and interaction (Table 1) and comprising proteins with F-ratio values greater than the critical value of 4.17 (P<0.05) and included 14, 18, and 27 proteins, respectively. Each network was scored and attributed functions (see Figure 7B).
Figure 7.
(A, B) Apolipoprotein E (APOE) genotype differential regulation of mitochondrial protein expression after global ischemia. (A) The percentage of upregulated and downregulated proteins in ApoE3 and ApoE4 transgenic mice after global ischemia (G) versus sham (S) indicates a differential regulation of protein levels. IPA network analysis was completed with proteins whose differential expression was a result of APOE genotype, global ischemia, or their interaction as determined by two-way analysis of variance (ANOVA). All proteins with an F-ratio value >4.24 within a given category were included. Network scores are indicative of a more robust fit to a given network. (B) The top networks from each data category are presented. Blue shaded nodes are those proteins uploaded to IPA, whereas unshaded nodes have been inserted by IPA to create a network with a maximum number of 35 molecules. Only those molecules with direct relationships (physical interaction) indicated by solid edges were included in the analysis. IPA, Ingenuity Pathway Analysis.
The APOE genotype network (IPA score=23), generated from 14 proteins, was attributed the functions of ‘cellular compromise, genetic disorder, and neurologic disease.' The APOE genotype network included ETFA as a major hub protein as well as inner membrane components of complex 1 (NDUFB5 and NDUFB10), but required the insertion of multiple MT-ND family members as well as the hub proteins SNCA and HTT to allow for cohesion. The IPA score was substantially lower than the other 2 networks.
The global ischemia network (IPA score=31), generated from 17 proteins, was attributed the functions of ‘cellular growth and proliferation, embryonic development and cell death.' The global ischemia network included HSP1AB as a major hub protein and included multiple proteins involved in oxidative stress (e.g., PRDX1, SOD1, and HSPA1B).
The interaction network (IPA score=33), generated from 25 proteins, was attributed the functions of ‘cardiovascular disease, genetic disorder, neurologic disease.' The interaction network had the highest IPA score and like the genotype network includes components of complex 1, albeit different NDUF family members (NDUFA11 and NDUFS3), that appear to make up the core of the functional interactions. Distinct to this network are the ‘peripheral' proteins that are involved in various metabolic roles that are lacking in the other networks (e.g., DLAT, IVD, and ACADL).
Overall, in terms of significantly modulated mitochondrial proteins (Table 1), the effect of global ischemia had a slightly larger effect on mitochondria than APOE genotype. Moreover, after global ischemia, of those mitochondria-associated proteins found to be significantly altered within a given cohort, 73% are found to increase in APOE-ɛ3 transgenic mice compared with basal levels while only 19% are increased in APOE-ɛ4 mice (Figure 7A), indicating that mitochondria in APOE transgenic mice are responding differently to the stress of ischemic injury.
Discussion
The present study demonstrates that not only are the levels of ApoE increased to a greater extent in mitochondria of APOE-ɛ4 mice as compared with APOE-ɛ3 mice, after an ischemic challenge, but that there are clear APOE genotype differences in the proteomic response of mitochondria in response to this challenge. The study additionally highlights that, even in the absence of injury, the proteomic signature is different between APOE genotypes.
ApoE isoform effects were investigated in response to a transient ischemic insult, which results in selective and delayed neuronal degeneration. In the mouse model used in this study and others (Horsburgh et al, 2000b; Kelly et al, 2001), there is a regional hierarchy of damage such that the striatum exhibits a greater extent of damage as compared with the hippocampus. The extent of damage can be influenced by a number of variables notably, the duration of ischemia, temperature, and anesthetic. In the current study, in the hippocampus we detected minimal neuronal damage in response to transient global ischemia. Thus, our results reflect the hippocampal mitochondrial response in APOE transgenic mice to ischemic injury, and not overt neuronal cell death, with mitochondrial dysfunction a likely consequence of the ischemic insult. This work builds on our previous studies in which we demonstrated that ApoE levels are markedly increased in neurons in response to a severe ischemic injury with ApoE levels being greatest in APOE-ɛ4 compared with APOE-ɛ3 transgenic mice. This increase in ApoE parallels the more extensive hippocampal damage in APOE-ɛ4 as compared with APOE-ɛ3 mice in response to injury, suggesting that the increase in ApoE4 may be contributing to the cell death (Horsburgh et al, 2000b). In the present study, the marked elevation of ApoE levels in mitochondria may be important to the pathogenic effect of ApoE4.
Previous studies have indicated structural and subsequent biophysical differences between ApoE protein isoforms which may explain differential influences on mitochondrial function (Huang et al, 2004). The present study sought to identify whether the protein signature is different between APOE genotypes in the presence and absence of injury. We used mass spectrometry-based proteomic techniques that offer a powerful approach to identify and quantify proteins in a complex mixture. Label-free quantification requires neither prior knowledge nor labelling strategy, and protein expression is determined from peptide intensities (Ong and Mann, 2005; Stewart et al, 2004; Wang et al, 2003). The ADH spiking experiments (see Figure 1) validate the use of MaxQuant to quantitate relative protein abundance in our hippocampal fractions without the use of isotope labelling. Using this technique, we were able to identify 274 mitochondria-associated proteins from a total of 1,067 proteins identified with at least two peptides. Based on recent work suggesting there are 1,075 mitochondrial proteins in brain (Kislinger et al, 2006), we have identified ∼26% of the ‘complete' mitochondrial proteome based on current GO annotation. Given our experimental design, with each sample subjected to a single round of LC-MS, in contrast to more comprehensive interrogations of the mitochondrial proteome (Kislinger et al, 2006; Mootha et al, 2003), which typically use replicate analysis of the same sample or more complex protein fractionation methods before LC-MS analysis, we have detected a reasonable percentage of the mitochondrial proteome. Furthermore, the detection of human ApoE at levels that are reminiscent of earlier observed differences in ApoE levels within the immunoblot data lend credence to the use of label-free quantification techniques not only to identify proteins, but do so in different biological contexts, i.e., an ischemic challenge. In the present study, the largest functional category was identified as mitochondrion but other categories were identified including the cytoskeleton and synapse. ApoE is reported to have diverse roles within the central nervous system including regulation of synaptic plasticity and these other functions could be studied further using the present approach by isolation of different cellular compartments (e.g., synapses).
Our proteomic results show that under basal and injury conditions, APOE genotype has a differential effect on mitochondrial protein levels indicative of significant alterations in the regulation of energy production, metabolism, redox control/oxidative stress, and organelle dynamics (Table 1). One of the primary functions of mitochondria is energy production, with glucose the primary fuel for the brain. Glucose is converted into ATP via a number of metabolic reactions encompassing glycolysis, the pyruvate dehydrogenase complex, and tricarboxylic acid cycle, before oxidative phosphorylation and the subsequent generation of ATP. Although glycolysis generates ATP under basal conditions, and in particular in times of cellular stress, oxidative phosphorylation is the most efficient means of ATP production (Magistretti and Pellerin, 1999); and therefore, neuronal function is critically dependent on oxidative phosphorylation, which itself is dependent on the efficient functioning of various metabolic pathways. In the present study, we were able to demonstrate that the isolated mitochondria remained relatively intact after cerebral ischemia as compared with those from control tissue allowing the identification of their proteomic signature. Within the isolated mitochondria, we were able to detect even small molecules (including cytochrome C) although these were not found to be significantly altered across the cohorts. In response to ischemia, it is recognized that dynamic alterations occur within mitochondria resulting in generation of reactive oxygen species and release of proteins such as cytochrome C and Smac, alterations in which we would not be able to detect in the preparation used (for review, see Niizuma et al, 2010). Although we cannot directly correlate expression levels with enzyme activity, assumptions can be made of the functional capacity of the affected mitochondria.
Approximately the same number of proteins, albeit distinctly different proteins, categorized as being functionally involved in oxidative phosphorylation, are affected by genotype, global ischemia, or interaction as seen in the respective data sets in Table 1. The lack of congruency is suggestive of differential regulation of key inner mitochondrial proteins. For example, NDUFA11 is increased in APOE-ɛ3 mice after global ischemia (see Figure 6), but is decreased in APOE-ɛ4 after the same insult, leading to a relatively high F-ratio (19.75) for the interactive effect of both genotype and ischemia on this component of complex 1 of the electron transport chain (P<0.01). Various other proteins located within the inner mitochondrial membrane were also found to be significantly modulated across the groups. Many of these, like NDUFA11 are found within the electron transport chain (e.g., NDUFB5, NDUFB10, and COX7A2) and are major components of the genotype and interaction functional networks (Figure 7B). Others are involved in protein/ion transport across the inner membrane (e.g., TIMM10, TIMM13, and SLC25A11; Lutz et al, 2003; Iacobazzi et al, 1992) and are differentially expressed across the different cohorts in similar ways. These data may suggest that the inner mitochondrial dynamics of the two genotypes are fundamentally different. Increased expression of a truncated form of ApoE4 associated with mitochondrial dysfunction (Chang et al, 2005) has been shown to reduce the activities of complex III and IV of the inner mitochondrial membrane (Nakamura et al, 2009), and likewise, we see differential regulation of proteins that represent both complexes (e.g., UQCRH and Cox7a2, respectively).
Unsurprisingly, after the ischemic challenge, both APOE-ɛ3 and APOE-ɛ4 mice show modulation of proteins involved in oxidative stress, including HSPA1B (or HSP70), SOD1, OXR1, and PRDX1 (Kelly et al, 2001; Hwang et al, 2010; Chan et al, 1998) but alterations in these proteins were APOE genotype dependent. For example, OXR1 which protects against oxidative damage and has been shown to be induced after oxidative stress (Elliott and Volkert, 2004), is increased in APOE-ɛ3 mice after ischemia but is downregulated after ischemia in APOE-ɛ4 mice (see Figure 6). Similarly, PRDX1 was significantly increased in the APOE-ɛ3 mice after global ischemia, whereas no such change was seen in the APOE-ɛ4 mice. This may suggest a decreased capacity to respond to oxidative damage under basal conditions (increased expression in sham-operated APOE-ɛ4 compared with APOE-ɛ3 mice), with less effective protection afforded after ischemic injury (decreased expression in APOE-ɛ4 ischemic compared with sham mice). This is reflected in the global ischemia functional network as well, which comprising multiple proteins implicated in the cellular response to global ischemia and attributed the associated function of ‘cell death.'
Various metabolic proteins were also found to be altered, some of which can be found within the interaction functional network (Figure 7B) as ‘peripheral' nodes (e.g., PCCB, ACADL, and ACAT) and are involved in different facets of lipid metabolism vital for normal cell functioning. Other proteins (GLUL, HIBCH, AK1, and AK3) are involved in different aspects of energy metabolism. This suggests that APOE genotype has an effect on the metabolic capacity of the mitochondria after ischemia, which in turn may have downstream effects on ATP production.
IPA networks also indicated certain molecules that are potentially involved in the regulatory control of proteins that were found to be upregulated and downregulated as a result of either genotype or ischemia. This includes the transcription factor HNF4A, which appears in all three generated networks. This molecule as well as other inserted nodes allow for the generation of new hypothesis pertaining to mechanisms underlying pathology as well as how to potentially intervene.
Besides the efficient production of ATP by individual mitochondria, the proper distribution of mitochondria along the neuron is important to provide local ATP supplies as well as buffer calcium transients in both the synapse and dendrite (Cai and Sheng, 2009; Ly and Verstreken, 2006). Therefore, the efficient transport of mitochondria along the neuronal cytoskeleton is critical. Disturbances to mitochondrial motility can impact on neuronal energy demands and viability (Rintoul and Reynolds, 2010), which are shown to be disturbed in APOE transgenic mice (Tesseur et al, 2000). RHOT1 and RHOT2 are two modulated proteins (genotype and interaction, respectively) identified with involvement in mitochondrial transport (Koutsopoulos et al, 2010; Misko et al, 2010; Russo et al, 2009). These changes may also have relevance to understanding potential apoE isoform differences in regulating aspects of synaptic plasticity.
Proteomic analysis has allowed us to identify functional pathways that are likely more susceptible to dysfunction in the context of a particular APOE genotype. Future studies could also elucidate whether the association between ApoE and mitochondria is direct or mediated through a secondary interaction, and if ApoE can translocate across the mitochondrial membrane. Additionally, it remains to be determined whether increases in ApoE levels or isoform are the main contributor to the pathogenic effect of ApoE4 after injury. Irrespective of this, the present study provides a basis on which to allow for further exploration into the mechanisms behind the differential regulation of mitochondrial function by ApoE isoforms.
Acknowledgments
The provision of APOE transgenic mice via a collaboration with Dr Allen Roses is gratefully acknowledged.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
This work was supported by a Wellcome Trust University Award to KH, and Age UK which has funded ‘The Disconnected Mind' project to which the present study contributes. KH has received funding from GlaxoWellcome (USA).
Supplementary Material
References
- Barsnes H, Vizcaino JA, Eidhammer I, Martens L. PRIDE converter: making proteomics data-sharing easy. Nat Biotechnol. 2009;27:598–599. doi: 10.1038/nbt0709-598. [DOI] [PubMed] [Google Scholar]
- Cai Q, Sheng ZH. Mitochondrial transport and docking in axons. Exp Neurol. 2009;218:257–267. doi: 10.1016/j.expneurol.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PH, Kawase M, Murakami K, Chen SF, Li Y, Calagui B, Reola L, Carlson E, Epstein CJ. Overexpression of SOD1 in transgenic rats protects vulnerable neurons against ischemic damage after global cerebral ischemia and reperfusion. J Neurosci. 1998;18:8292–8299. doi: 10.1523/JNEUROSCI.18-20-08292.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci USA. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Forster MJ, McDonald SR, Weintraub ST, Carroll CA, Gracy RW. Proteomic identification of specific oxidized proteins in ApoE-knockout mice: relevance to Alzheimer's disease. Free Radic Biol Med. 2004;36:1155–1162. doi: 10.1016/j.freeradbiomed.2004.02.002. [DOI] [PubMed] [Google Scholar]
- de Leon MJ, Convit A, Wolf OT, Tarshish CY, DeSanti S, Rusinek H, Tsui W, Kandil E, Scherer AJ, Roche A, Imossi A, Thorn E, Bobinski M, Caraos C, Lesbre P, Schlyer D, Poirier J, Reisberg B, Fowler J. Prediction of cognitive decline in normal elderly subjects with 2-[(18)F]fluoro-2-deoxy--glucose/poitron-emission tomography (FDG/PET) Proc Natl Acad Sci USA. 2001;98:10966–10971. doi: 10.1073/pnas.191044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton RF, Kerr LE, Short DM, Allerhand M, Whittle IR, McCulloch J. Network generation enhances interpretation of proteomic data from induced apoptosis. Proteomics. 2010;10:1307–1315. doi: 10.1002/pmic.200900112. [DOI] [PubMed] [Google Scholar]
- Elliott NA, Volkert MR. Stress induction and mitochondrial localization of Oxr1 proteins in yeast and humans. Mol Cell Biol. 2004;24:3180–3187. doi: 10.1128/MCB.24.8.3180-3187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner F, Arriaga EA, Mann M. Mild protease treatment as a small-scale biochemical method for mitochondria purification and proteomic mapping of cytoplasm-exposed mitochondrial proteins. J Proteome Res. 2006;5:3277–3287. doi: 10.1021/pr060361z. [DOI] [PubMed] [Google Scholar]
- Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, Fish JD, Masliah E, Hopkins PC, Scearce-Levie K, Weisgraber KH, Mucke L, Mahley RW, Huang Y. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci USA. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh K, McCarron MO, White F, Nicoll JA. The role of apolipoprotein E in Alzheimer's disease, acute brain injury and cerebrovascular disease: evidence of common mechanisms and utility of animal models. Neurobiol Aging. 2000a;21:245–255. doi: 10.1016/s0197-4580(00)00097-x. [DOI] [PubMed] [Google Scholar]
- Horsburgh K, McCulloch J, Nilsen M, Roses AD, Nicoll JA. Increased neuronal damage and apoE immunoreactivity in human apolipoprotein E, E4 isoform-specific, transgenic mice after global cerebral ischaemia. Eur J Neurosci. 2000b;12:4309–4317. [PubMed] [Google Scholar]
- Huang Y, Weisgraber KH, Mucke L, Mahley RW. Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease. J Mol Neurosci. 2004;23:189–204. doi: 10.1385/JMN:23:3:189. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hwang IK, Yoo KY, Kim DW, Lee CH, Choi JH, Kwon YG, Kim YM, Choi SY, Won MH. Changes in the expression of mitochondrial peroxiredoxin and thioredoxin in neurons and glia and their protective effects in experimental cerebral ischemic damage. Free Radic Biol Med. 2010;48:1242–1251. doi: 10.1016/j.freeradbiomed.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Iacobazzi V, Palmieri F, Runswick MJ, Walker JE. Sequences of the human and bovine genes for the mitochondrial 2-oxoglutarate carrier. DNA Seq. 1992;3:79–88. doi: 10.3109/10425179209034000. [DOI] [PubMed] [Google Scholar]
- Kelly S, McCulloch J, Horsburgh K. Minimal ischaemic neuronal damage and HSP70 expression in MF1 strain mice following bilateral common carotid artery occlusion. Brain Res. 2001;914:185–195. doi: 10.1016/s0006-8993(01)02801-3. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Koutsopoulos OS, Laine D, Osellame L, Chudakov DM, Parton RG, Frazier AE, Ryan MT. Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim Biophys Acta. 2010;1803:564–574. doi: 10.1016/j.bbamcr.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Le Bihan T, Grima R, Martin S, Forster T, Le Bihan Y. Quantitative analysis of low-abundance peptides in HeLa cell cytoplasm by targeted liquid chromatography/mass spectrometry and stable isotope dilution: emphasising the distinction between peptide detection and peptide identification. Rapid Commun Mass Spectrom. 2010;24:1093–1104. doi: 10.1002/rcm.4487. [DOI] [PubMed] [Google Scholar]
- Lutz T, Neupert W, Herrmann JM. Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J. 2003;22:4400–4408. doi: 10.1093/emboj/cdg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly CV, Verstreken P. Mitochondria at the synapse. Neuroscientist. 2006;12:291–299. doi: 10.1177/1073858406287661. [DOI] [PubMed] [Google Scholar]
- Magistretti PJ, Pellerin L. Cellular mechanisms of brain energy metabolism and their relevance to functional brain imaging. Philos Trans R Soc Lond B Biol Sci. 1999;354:1155–1163. doi: 10.1098/rstb.1999.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke R, Zerres K, Uhlhaas S, Kessler J, Heiss WD. Apolipoprotein E polymorphism influences the cerebral metabolic pattern in Alzheimer's disease. Neurosci Lett. 1998;254:49–52. doi: 10.1016/s0304-3940(98)00673-9. [DOI] [PubMed] [Google Scholar]
- Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Watanabe A, Fujino T, Hosono T, Michikawa M. Apolipoprotein E4 (1–272) fragment is associated with mitochondrial proteins and affects mitochondrial function in neuronal cells. Mol Neurodegener. 2009;4:35. doi: 10.1186/1750-1326-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Mann M, Aebersold R, Yates JR, III, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- Niizuma K, Yoshioka H, Chen H, Kim GS, Jung JE, Katsu M, Okami N, Chan PH. Mitochondrial and apoptotic neuronal death signaling pathways in cerebral ischemia. Biochim Biophys Acta. 2010;1802:92–99. doi: 10.1016/j.bbadis.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Rintoul GL, Reynolds IJ. Mitochondrial trafficking and morphology in neuronal injury. Biochim Biophys Acta. 2010;1802:143–150. doi: 10.1016/j.bbadis.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Russo GJ, Louie K, Wellington A, Macleod GT, Hu F, Panchumarthi S, Zinsmaier KE. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J Neurosci. 2009;29:5443–5455. doi: 10.1523/JNEUROSCI.5417-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, Hulette C, Crain B, Goldgaber D, Roses AD. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Sheng H, Laskowitz DT, Bennett E, Schmechel DE, Bart RD, Saunders AM, Pearlstein RD, Roses AD, Warner DS. Apolipoprotein E isoform-specific differences in outcome from focal ischemia in transgenic mice. J Cereb Blood Flow Metab. 1998;18:361–366. doi: 10.1097/00004647-199804000-00003. [DOI] [PubMed] [Google Scholar]
- Shenk JC, Liu J, Fischbach K, Xu K, Puchowicz M, Obrenovich ME, Gasimov E, Alvarez LM, Ames BN, Lamanna JC, Aliev G. The effect of acetyl--carnitine and R-alpha-lipoic acid treatment in ApoE4 mouse as a model of human Alzheimer's disease. J Neurol Sci. 2009;283:199–206. doi: 10.1016/j.jns.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strum JC, Shehee R, Virley D, Richardson J, Mattie M, Selley P, Ghosh S, Nock C, Saunders A, Roses A. Rosiglitazone induces mitochondrial biogenesis in mouse brain. J Alzheimers Dis. 2007;11:45–51. doi: 10.3233/jad-2007-11108. [DOI] [PubMed] [Google Scholar]
- Stewart II, Zhao L, Le Bihan T, Larsen B, Scozzaro S, Figeys D, Mao GD, Ornatsky O, Dharsee M, Orsi C, Ewing R, Goh T. The reproducible acquisition of comparative liquid chromatography/tandem mass spectrometry data from complex biological samples. Rapid Commun Mass Spectrom. 2004;18:1697–1710. doi: 10.1002/rcm.1538. [DOI] [PubMed] [Google Scholar]
- Tesseur I, Van Dorpe J, Bruynseels K, Bronfman F, Sciot R, Van Lommel A, Van Leuven F. Prominent axonopathy and disruption of axonal transport in transgenic mice expressing human apolipoprotein E4 in neurons of brain and spinal cord. Am J Pathol. 2000;157:1495–1510. doi: 10.1016/S0002-9440(10)64788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- Xu PT, Schmechel D, Rothrock-Christian T, Burkhart DS, Qiu HL, Popko B, Sullivan P, Maeda N, Saunders AM, Roses AD, Gilbert JR. Human apolipoprotein E2, E3, and E4 isoform-specific transgenic mice: human-like pattern of glial and neuronal immunoreactivity in central nervous system not observed in wildtype mice. Neurobiol Dis. 1996;3:229–245. doi: 10.1006/nbdi.1996.0023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.