Abstract
[11C]PBR28 binds the 18-kDa Translocator Protein (TSPO) and is used in positron emission tomography (PET) to detect microglial activation. However, quantitative interpretations of signal are confounded by large interindividual variability in binding affinity, which displays a trimodal distribution compatible with a codominant genetic trait. Here, we tested directly for an underlying genetic mechanism to explain this. Binding affinity of PBR28 was measured in platelets isolated from 41 human subjects and tested for association with polymorphisms in TSPO and genes encoding other proteins in the TSPO complex. Complete agreement was observed between the TSPO Ala147Thr genotype and PBR28 binding affinity phenotype (P value=3.1 × 10−13). The TSPO Ala147Thr polymorphism predicts PBR28 binding affinity in human platelets. As all second-generation TSPO PET radioligands tested hitherto display a trimodal distribution in binding affinity analogous to PBR28, testing for this polymorphism may allow quantitative interpretation of TSPO PET studies with these radioligands.
Keywords: Ala147Thr, PBR28, polymorphism, radioligand binding, TSPO
Introduction
The 18-kDa Translocator Protein TSPO (also called the Peripheral Benzodiazepine Receptor) is expressed within monocyte-derived cells and has been proposed as a marker of brain microglial activation (Venneti et al, 2006). Quantitative imaging of TSPO with positron emission tomography (PET) has been technically challenging. The poor signal-to-noise ratio and high nonspecific binding of the first-generation ligand [11C]PK11195 limit accurate quantification (Banati et al, 2000). Several second-generation TSPO ligands with improved signal-to-noise ratio, including [11C]PBR28, [18F]PBR06, [18F]FEPPA, [11C]DAA1106, [11C]DPA713, and [18F]PBR111, have been investigated in human (Chauveau et al, 2008).
However, PET (Kreisl et al, 2010) and recent in-vitro studies have revealed substantial heterogeneity in binding potential due to intersubject variability in the affinity of the second-generation PET ligands for the TSPO (Owen et al, 2010, 2011a). These tracers bind TSPO in brain tissue from different subjects in one of three ways: high-affinity binders and low-affinity binders (HABs and LABs) express a single binding site for TSPO with either high or low affinity, respectively, whereas mixed affinity binders (MABs) express approximately equal numbers of the HAB and LAB binding sites (Owen et al, 2011a). Subjects remain in the same binding class regardless of which second-generation radioligand is used for classification, although the exact Ki values of the binding classes vary between ligands (Owen et al, 2011a). For example, PBR28 Ki values for HABs and LABs are 4 and 200 nmol/L, respectively, whereas for PBR111 these values are 16 and 62 nmol/L. Prior knowledge of binding affinity, therefore, is required for quantitative comparisons of TSPO expression between subjects in PET studies with these radioligands. This may not be necessary for [11C]PK11195 as in-vitro studies suggest that this radioligand binds to a different site on the TSPO, with no apparent difference in affinity between HABs and LABs (Owen et al, 2010).
The mechanisms responsible for the different TSPO binding behaviors are not understood. Based on our data which suggests that MABs appear to express the HAB and LAB sites in equal proportion (Owen et al, 2011a), we hypothesized that codominant expression of an underlying genetic trait explains variation in binding phenotype and that this behavior could arise from polymorphisms in either TSPO or other genes encoding proteins in the TSPO complex.
Here, we present the results of a genetic association study between PBR28 binding affinity to human platelets and polymorphisms in genes encoding TSPO and associated proteins. Our work provides the basis for a simple genetic test for determination of TSPO binding class, which will contribute to quantitative interpretation of PET studies of TSPO expression.
Materials and methods
The study protocol, volunteer information, and informed consent forms were approved by the North West London Research Ethics Committee. Forty-one healthy volunteers (29 male, 12 female, mean age 36.3±1.4 years) were recruited. Ethnicity was self-reported as 37/41 Caucasian, 2/41 Asian, 1/41 mixed Caucasian/Asian, and 1/41 Hispanic. Venous blood (50 ml) was drawn into EDTA-containing tubes, and separated into a lymphocyte-rich bottom layer (for genetic analysis) and platelet-rich top layer by centrifugation (180 × g, 15 min, room temperature (RT)). The platelet-rich layer (for binding assays) was re-centrifuged (1800 × g, 15 min, RT) to produce a platelet-containing pellet. Platelet membranes were prepared as previously described (Owen et al, 2011a). To measure binding affinity, aliquots of membrane suspension were incubated with 5 nmol/L [3H]PK11195 (Perkin-Elmer, Beaconsfield, UK) and one of 12 concentrations (0.1 nmol/L to 100 μmol/L) of unlabelled PBR28 (Borochem, Caen, France) as previously described (Owen et al, 2011a).
DNA Extraction
Genomic DNA was extracted (by Gen-Probe, Manchester, UK) from ∼20 ml of lymphocyte-enriched blood product (see membrane preparation) using a chlorinated DNA extraction protocol and resuspended in 10 mmol/L Tris/0.1 mmol/L EDTA pH 8.0. Twenty nanograms of each genomic DNA sample was plated and lyophilized in a 96-well microtiter plate for each polymorphism tested. All samples were duplicated on each plate as a quality control measure.
Polymorphism Selection and Genotyping
A total of 58 polymorphisms (both single nucleotide changes and insertions/deletions) with a perceived effect on protein function were selected from known polymorphisms in the TSPO gene and in genes encoding proteins directly associated with TSPO in the TSPO complex (VDAC1, VDAC2, VDAC3, ANT (SLC25A4), PRAX1 (BZRAP1), and PAP7 (ACBD3)) (Supplementary Table 1). The polymorphisms were genotyped using TaqMan (Applied Biosystems, Foster City, CA, USA), Luminex-based Flow Assorted SNP Typing (Taylor et al, 2001), direct sequencing or PCR fragment analysis (see Supplementary Methods for more details on assay conditions and quality control measures). Of the 58 genotyped polymorphisms, 38 were found to be monomorphic in the study sample; therefore, only 20 polymorphisms were analyzed for association with the ligand binding phenotype (Supplementary Table 1).
Data Analysis
Platelet binding affinity
Binding data were analyzed using GraphPad Prism 5.0 (GraphPad Software Inc, La Jolla, CA, USA). Single site and two site competition models were fitted using the least squares algorithm and model selection was compared using an F test. The null hypothesis, that the data fitted a single site model, was rejected if the P value was <0.05. A Kd for [3H]PK11195 of 29.25 nmol/L (Owen et al, 2010) was used to generate the Ki for PBR28. HABs and LABs were defined as subjects with a single binding site with Ki <15 or >100 nmol/L, respectively. MABs were defined as subjects with two binding sites. Data are expressed as the mean±standard error of the mean.
Genetic Polymorphisms
The PBR28 ligand binding classification end point was analyzed using three groups: LABs, MABs, and HABs. Single marker analysis was performed by Fisher's exact test using SAS 9.2 (Cary, NC, USA). The two-tailed P value and estimated odds ratio with 95% confidence interval were calculated. A 5% significance threshold was used by applying a Bonferroni correction for the number of markers analyzed (n=20) (significance threshold P=0.0025). Linkage disequilibrium analysis (VanLiere and Rosenberg, 2008) and departure from Hardy–Weinberg Equilibrium (Wigginton et al, 2005) were tested on data from Caucasian subjects. See Supplementary Methods for more detail.
Results
Binding Class
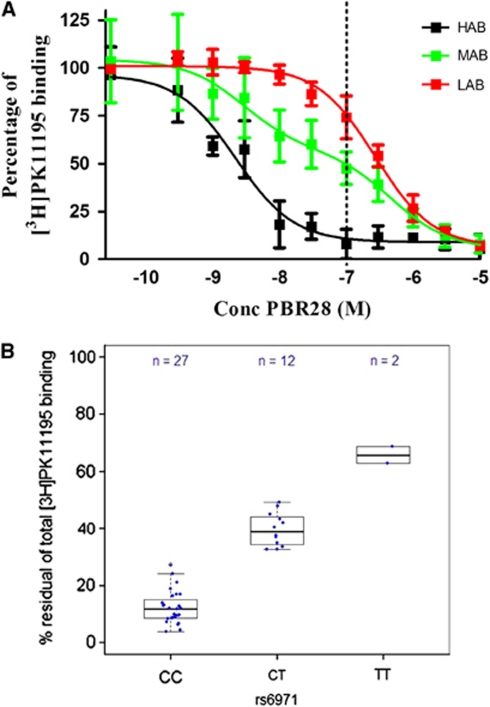
Protein in platelets from 27/41 subjects (66%) showed ligand binding to a single class of high affinity sites (Ki=2.17±0.17 nmol/L). These subjects were classified as HABs. In 12/41 subjects (29%), the data fitted best to a two site model with affinities of 2.23±0.31 and 297±43 nmol/L. These subjects were classified as MABs. In the remaining 2/41 subjects (5%), the ligand bound to a single class of low affinity sites (Ki=187±20 nmol/L). These subjects were classified as LABs (Figure 1A).
Figure 1.
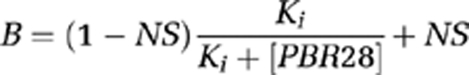 |
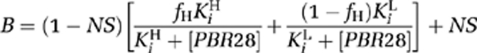 |
Genetic Polymorphisms
Single marker analysis revealed that only one polymorphism (rs6971) surpassed the multiple testing P value threshold of 0.0025 (Supplementary Figure 1a; Supplementary Table 1). This polymorphism is located in exon 4 of the TSPO gene and is predicted to result in a nonconservative amino-acid substitution at position 147 from alanine to threonine (Ala147Thr) in the fifth transmembrane domain of the TSPO protein. Complete agreement was observed between the PBR28 binding phenotype for individual subjects and their rs6971 genotype (P value 3.1 × 10−13; Table 1; Figure 1B). A linkage disequilibrium plot for the five TSPO markers analyzed indicated that the low level of linkage disequilibrium (Supplementary Figure 1b) could account for the weak, statistically nonsignificant associations observed for flanking TSPO polymorphisms, rs3937387 and rs6972 (Supplementary Figure 1a). This pattern of association is what would be expected for a codominant monogenic trait.
Table 1. Distribution of rs6971 genotypes against ligand binding classification.
|
TSPO genotype |
Binding phenotype (subject, n) |
|||
|---|---|---|---|---|
| DNA (polymorphism rs6971) | Protein (position 147) | HAB | MAB | LAB |
| C/C | Ala/Ala | 27 | ||
| C/T | Ala/Thr | 12 | ||
| T/T | Thr/Thr | 2 | ||
Ala=alanine, Thr=threonine, HAB=high affinity binder, MAB=mixed affinity binder, LAB=low affinity binder.
Genotypes correspond to carriage of the 147 amino acid as follows: CC=Ala147/Ala147; CT=Ala147/Thr147; TT=Thr147/Thr147.
Discussion
Here, we demonstrate complete agreement between TSPO binding affinity class measured in human platelets with PBR28, and variation at a common polymorphism (rs6971) in the TSPO gene which leads to an amino-acid substitution (Ala147Thr). These data indicate that variation in binding affinity of PBR28 for human platelets is a codominant monogenic trait.
This finding is highly significant for the interpretation of PET studies using [11C]PBR28. We have not formally demonstrated concordance in binding class between platelets and brain or other organs, but agreement seems highly likely as PET data with [11C]PBR28 strongly suggests that the LAB phenotype is consistent across all tissues within the same subject (Kreisl et al, 2010). We, therefore, believe that PBR28 binding affinity class in the brain (or other tissues) can be predicted simply by genotyping the TSPO rs6971 polymorphism. In the absence of an available TSPO radioligand which binds with equivalent affinity in all subjects and has a high signal-to-noise ratio, genotyping the TSPO rs6971 polymorphism will enable confident, quantitative comparisons of [11C]PBR28 PET data between groups of patients. This can be achieved either by screening out certain subjects to ensure all study participants are from the same binding class, or by including all subjects but correcting PET data based on binding class.
Our results have the same implications for PET studies using [18F]PBR06, [11C]DAA1106, [11C]DPA713, [18F]PBR111, and [11C]AC-5216. Although there is no data confirming that these ligands bind at the same site as [11C]PBR28, we have previously demonstrated that binding class shows complete consistency between radioligands; in other words, all tissue samples classified as HABs with PBR28 are also classified as HABs with the other radioligands (Owen et al, 2011a).
The results also could help to better understand pharmacokinetic–pharmacodynamic relationships for drugs targeting TSPO, as we have suggested previously based on data from direct binding affinity assays with XBD173 (AC-5216) (Owen et al, 2011b).
This binding affinity variation has greatest impact for studies of Caucasians, for whom the rs6971 polymorphism has a reported minor allele (Thr147) frequency of 30% and a major allele (Ala147) frequency of 70% (11). The minor allele is less prevalent in other populations, such as African American (25%), Han Chinese (2%), and Japanese (4%) (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/snp_details_phase3?name=rs6971&source=hapmap28_B36&tmpl=snp_details_phase3). In our small predominantly Caucasian sample, the observed percentage of MABs and LABs (29% and 5%, respectively) was lower than expected (42% and 9%, respectively) based on published frequencies, although they are not outside the 95% confidence bounds for sampling variation. This discrepancy could be explained by some subjects inaccurately reporting their own ancestral background, or the result of an unknown bias in ascertainment.
Structural modelling using a general platform in wide use (PolyPhen software; Ramensky et al, 2002) suggests that substitution of threonine (neutral and polar) for alanine (neutral and hydrophobic) at position 147 of TSPO could alter the protein tertiary structure (PolyPhen score 0.999, data not shown). Alanine 147 is highly conserved across most species (Murail et al, 2008), and likely contributes to maintaining the helical structure of the fifth transmembrane domain of the protein. Protein structure data based on mouse and bacterial TSPO suggest that this helical conformation could have a key role in TSPO function (Korkhov et al, 2010; Murail et al, 2008). We, therefore, hypothesize that the Ala147Thr amino-acid substitution results in a conformational change affecting the interaction between TSPO and the variety of molecules for which affinity differences have been demonstrated (Owen et al, 2010, 2011a, 2011b).
There is some evidence suggesting that the Ala147Thr substitution has an impact on biological functions of TSPO. An association between the polymorphism and variation in pregnenolone production and plasma levels of LDL cholesterol has been reported in healthy individuals (Costa et al, 2009a). A small pilot study in patients with a diagnosis of depression also found an association between the polymorphism and separation anxiety (Costa et al, 2009b). However, neither of these findings has been replicated yet.
While the relatively small size of our sample is a potential limitation of this study, the perfect concordance between binding affinity class and the rs6791 polymorphism is striking. Direct testing of the relationships between genetic variation, platelet binding and [11C]PBR28 PET signal in vivo is needed now.
The relative affinity of the PET radioligand [11C]PBR28 for TSPO in human platelets is determined by a single polymorphism (rs6971) in the TSPO gene. Our results, therefore, suggest that a simple test of genotype will enable determination of TSPO ligand binding class to allow quantitative assessments of TSPO density using PET.
Acknowledgments
The authors are grateful to the healthy volunteers for their participation in this study. DRO was a clinical fellow in The Wellcome Trust-GSK Translational Medicine Training Programme at Imperial College. The authors thank our colleagues at the GSK Human BioSample Repository and the GSK Genetics Data Generation, Data Quality and Data Managements groups for their contributions to the sample preparation and generation of the genotyping data. Additionally, The authors thank Samiul Hasan from the GSK Computational Biology group for providing the rs6971 PolyPhen data.
AJY, RNG, KS, GW, DJP, IB, LRC, CAP, PLS, VEM, PMM, EAR, and JR are GSK employees and hold GSK stock.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
The work described in this paper has been supported by GlaxoSmithKline and Imperial College London.
Supplementary Material
References
- Banati RB, Newcombe J, Gunn RN, Cagnin A, Turkheimer F, Heppner F, Price G, Wegner F, Giovannoni G, Miller DH, Perkin GD, Smith T, Hewson AK, Bydder G, Kreutzberg GW, Jones T, Cuzner ML, Myers R. The peripheral benzodiazepine binding site in the brain in multiple sclerosis: quantitative in vivo imaging of microglia as a measure of disease activity. Brain. 2000;123 (Pt 11:2321–2337. doi: 10.1093/brain/123.11.2321. [DOI] [PubMed] [Google Scholar]
- Chauveau F, Boutin H, Van CN, Dolle F, Tavitian B. Nuclear imaging of neuroinflammation: a comprehensive review of [11C]PK11195 challengers. Eur J Nucl Med Mol Imaging. 2008;35:2304–2319. doi: 10.1007/s00259-008-0908-9. [DOI] [PubMed] [Google Scholar]
- Costa B, Pini S, Gabelloni P, Da PE, Abelli M, Lari L, Preve M, Lucacchini A, Cassano GB, Martini C. The spontaneous Ala147Thr amino acid substitution within the translocator protein influences pregnenolone production in lymphomonocytes of healthy individuals. Endocrinology. 2009a;150:5438–5445. doi: 10.1210/en.2009-0752. [DOI] [PubMed] [Google Scholar]
- Costa B, Pini S, Martini C, Abelli M, Gabelloni P, Landi S, Muti M, Gesi C, Lari L, Cardini A, Galderisi S, Mucci A, Lucacchini A, Cassano GB. Ala147Thr substitution in translocator protein is associated with adult separation anxiety in patients with depression. Psychiatr Genet. 2009b;19:110–111. doi: 10.1097/YPG.0b013e32832080f6. [DOI] [PubMed] [Google Scholar]
- Korkhov VM, Sachse C, Short JM, Tate CG. Three-dimensional structure of TspO by electron cryomicroscopy of helical crystals. Structure. 2010;18:677–687. doi: 10.1016/j.str.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl WC, Fujita M, Fujimura Y, Kimura N, Jenko KJ, Kannan P, Hong J, Morse CL, Zoghbi SS, Gladding RL, Jacobson S, Oh U, Pike VW, Innis RB. Comparison of [(11)C]-(R)-PK 11195 and [(11)C]PBR28, two radioligands for translocator protein (18 kDa) in human and monkey: Implications for positron emission tomographic imaging of this inflammation biomarker. Neuroimage. 2010;49:2924–2932. doi: 10.1016/j.neuroimage.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murail S, Robert JC, Coic YM, Neumann JM, Ostuni MA, Yao ZX, Papadopoulos V, Jamin N, Lacapere JJ. Secondary and tertiary structures of the transmembrane domains of the translocator protein TSPO determined by NMR. Stabilization of the TSPO tertiary fold upon ligand binding. Biochim Biophys Acta. 2008;1778:1375–1381. doi: 10.1016/j.bbamem.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Owen DR, Gunn RN, Rabiner EA, Bennacef I, Fujita M, Kreisl WC, Innis RB, Pike VW, Reynolds R, Matthews PM, Parker CA. Mixed-affinity binding in humans with 18-kDa translocator protein ligands. J Nucl Med. 2011a;52:24–32. doi: 10.2967/jnumed.110.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DR, Howell OW, Tang SP, Wells LA, Bennacef I, Bergstrom M, Gunn RN, Rabiner EA, Wilkins MR, Reynolds R, Matthews PM, Parker CA. Two binding sites for [(3)H]PBR28 in human brain: implications for TSPO PET imaging of neuroinflammation. J Cereb Blood Flow Metab. 2010;30:1608–1618. doi: 10.1038/jcbfm.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DR, Lewis AJ, Reynolds R, Rupprecht R, Eser D, Wilkins MR, Bennacef I, Nutt DJ, Parker CA. Variation in binding affinity of the novel anxiolytic XBD173 for the 18 kDa translocator protein in human brain. Synapse. 2011b;65:257–259. doi: 10.1002/syn.20884. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JD, Briley D, Nguyen Q, Long K, Iannone MA, Li MS, Ye F, Afshari A, Lai E, Wagner M, Chen J, Weiner MP. Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. Biotechniques. 2001;30:661–669. doi: 10.2144/01303dd04. [DOI] [PubMed] [Google Scholar]
- VanLiere JM, Rosenberg NA. Mathematical properties of the r2 measure of linkage disequilibrium. Theor Popul Biol. 2008;74:130–137. doi: 10.1016/j.tpb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Lopresti BJ, Wiley CA. The peripheral benzodiazepine receptor (Translocator protein 18 kDa) in microglia: from pathology to imaging. Prog Neurobiol. 2006;80:308–322. doi: 10.1016/j.pneurobio.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.