Cats can shed antimicrobial drug−resistant Salmonella serotypes in the environment.
Keywords : zoonoses, Salmonella Infections, cats, disease reservoirs, research
Abstract
To determine whether cats were a risk for transmission of Salmonella to humans, we evaluated the excretion of Salmonella by pet cats. Rectal-swab specimens were taken from 278 healthy house cats, from 58 cats that died of disease, and from 35 group-housed cats. Group-housed cats were kept in one room with three cat trays and a common water and feed tray. Eighteen (51.4%) of 35 group-housed cats, 5 (8.6%) of 58 diseased cats (5/58), and 1 (0.36%) of 278 healthy house cats excreted Salmonella. Salmonella isolates were of serotypes Typhimurium, Enteritidis, Bovismorbificans and 4:i:-. Acquired antimicrobial resistance was found in serotype Typhimurium (resistance to ampicillin, chloramphenicol, and tetracycline; to ampicillin; and to chloramphenicol) and 4:i:- strains (resistance to ampicillin, chloramphenicol, sulfonamides, trimethoprim, and sulfamethoxazole/trimethoprim). Cats that excrete Salmonella can pose a public health hazard to people who are highly susceptible to Salmonella, such as children, the elderly, and immunocompromised persons.
Salmonella infections are still a leading cause of human foodborne infections in the world (1,2). These infections primarily originate from eating contaminated food, especially chicken eggs and egg products, and also meat products from pigs and chickens (3,4). Considering the high frequency of food contamination and the emergence of multidrug-resistant Salmonella strains, control of Salmonella in food-producing animals has become a worldwide challenge. Other environmental sources can lead to accidental human infections with Salmonella as well. The role of pet animals as a source of Salmonella has not been fully investigated, but severe human infections originating from reptiles, especially pet turtles, have been reported (5).
Cats and dogs are the most widely kept pet animals, yet the incidence of Salmonella in these animals is largely unknown, and the risk that these animals pose for transmission of Salmonella to humans is unclear. In particular, cats that can freely roam outside, and are therefore able to scavenge or hunt food of unknown quality, are potential candidates for Salmonella carriage. Most reports concerning Salmonella and cats are case studies of clinical salmonellosis, which resulted in septicemia and death (6,7). Subclinical infections and carrier animals, however, are much more important with respect to transmission to humans. In this study, rectal swabs from cats of different origin (house cats, group-housed cats, diseased cats) were cultured for Salmonella. The serotype and phage type of the Salmonella isolates were determined, and the isolates were characterized with respect to their antimicrobial drug–resistance pattern and interaction with human intestinal epithelial cells.
Methods
Collection of Fecal Samples
A total of 278 rectal swab samples from house cats of different age, sex, and breed were taken between July and November 2003. All house cats came from different owners. The animals came from all over the Dutch-speaking part of Belgium, i.e., north of Brussels. Rectal swab specimens were also taken from 58 cats that were submitted for autopsy to the Faculty of Veterinary Medicine, Ghent University. The latter died or were euthanized because of incurable disease. All cats came from different owners, except three cats that had feline immunodeficiency virus (FIV), which came from one owner. Finally, rectal samples of 35 kittens (all <4 months of age) were taken at a facility where the animals were group-housed, waiting to be adopted. These animals came from 16 different owners.
Bacteriologic Analysis
Bacteriologic analysis was performed by enrichment of the rectal swabs. The samples were first pre-enriched in buffered peptone water (BPW) (Oxoid, Basingstoke, Hampshire, UK) overnight at 37°C, after which 1 mL of this suspension was added to 9 mL of tetrathionate brilliant green broth (Oxoid) (enrichment). After incubation overnight at 37°C, a drop of this suspension was spread on brilliant green agar (BGA) (Oxoid). Both the serotype and phage type of positive isolates were determined.
Antimicrobial Susceptibility Testing
Resistance to antimicrobial agents was tested by using the disk diffusion assay on Mueller-Hinton agar with commercial antimicrobial susceptibility disks (Oxoid) according to the international standards of the National Council for Clinical Laboratory Standards (NCCLS) (8). The following antimicrobial agents were tested: ampicillin (A, 10 μg), chloramphenicol (C, 30 μg), streptomycin (S, 10 μg), sulfonamide (Su, 300 μg), tetracycline (T, 30 μg), ciprofloxacin (Cip, 5 μg), kanamycin (K, 30 μg), gentamicin (Gn, 10 μg), sulfamethoxazole-trimethoprim (Sxt, 25 μg), cefotaxime (Cxt, 30 μg), nalidixic acid (Na, 30 μg), and amoxicillin-clavulanic acid (Amc, 30 μg). Salmonella enterica serovar Typhimurium 8420 (resistance type ACSSuT), 6237 (sensitive), 3520 (resistance type T), 2200 (resistance type ASSuT), and 5833 (sensitive) isolates from human patients in Belgium were used as control strains in antimicrobial susceptibility testing.
Polymerase Chain Reaction (PCR)
For PCR, a loop of bacterial culture was resuspended in 50 μL of water, and DNA was released from bacterial cells by boiling for 20 min. After the mixture was spun for 1 min in a microfuge at 14,000 x g, 2 μL of the supernatant was taken as a template DNA for PCR. PCR was carried out in 20-μL volumes by using PCR Master Mix from Qiagen (Hilden, Germany), according to the manufacturer's instructions. All the resistant strains were tested for the presence of the genes typical for particular resistance. The genes determined and primers used are listed in Table 1. Cycling consisted of 50-s incubations at 92°C, 55°C, and 72°C, which were repeated 25 times. After PCR, amplification products were detected by electrophoresis in 2% agarose gel, stained with ethidium bromide, and visualized under UV light. Antimicrobial drug-sensitive strain S. Typhimurium F98 was used as a negative control in all the amplifications. S. Typhimurium strains 8420, 6237, 3520, 2200, and 5833 were used as positive controls.
Table 1. List of primers used in the PCR reactions for detection of resistance genes.
| Resistance | Gene | Primer | Sequence (5´– 3´) | Size (bp) | Reference |
|---|---|---|---|---|---|
| Ampicillin | blaPSE1 | PSEF | TAG CCA TAT TAT GGA GCC TC | 321 | AF261825 |
| PSER | TTA ACT TTT CCT TGC TCA GC | ||||
| blaTEM | TEMF | GCA CGA GTG GGT TAC ATC GA | 310 | 9 | |
| TEMR | GGT CCT CCG ATC GTT GTC AG | ||||
| blaoxa1 | oxa1F | AGC AGC GCC AGT GCA TCA | 708 | 10 | |
|
|
|
oxa1R |
ATT CGA CCC CAA GTT TCC |
|
|
| Chloramphenicol | floR | floRF | GCG ATA TTC ATT ACT TTG GC | 425 | 11 |
| floRR | TAG GAT GAA GGT GAG GAA TG | ||||
| Cat | catF | CCT GCC ACT CAT CGC AGT | 623 | 10 | |
|
|
|
catR |
CCA CCG TTG ATA TAT CCC |
|
|
| Streptomycin | aadA1 | aad1For | CGA CTC AAC TAT CAG AGG TA | 384 | AY534545 |
| aad1Rev | CTT TTG TCA GCA AGA TAG CC | ||||
| aadA2 | aadA2F | CGG TGA CCA TCG AAA TTT CG | 249 | 12 | |
| aadA2R | CTA TAG CGC GGA GCG TCT CGC | ||||
| strA | strAF | CCT ATC GGT TGA TCA ATG TC | 250 | 11 | |
|
|
|
strAR |
GAA GAG TTT TAG GGT CCA CC |
|
|
| Tetracycline | tetA | tetAF | GCT ACA TCC TGC TTG CCT TC | 210 | 13 |
| tetAR | CAT AGA TCG CCG TGA AGA GG | ||||
| tetB | tetBF | TTG GTT AGG GGC AAG TTT TG | 659 | 13 | |
| tetBR | GTA ATG GGC CAA TAA CAC CG | ||||
| tetC | tetCF | GCG GGA TAT CGT CCA TTC CG | 207 | 14 | |
| tetCR | GCG TAG AGG ATC CAC AGG ACG | ||||
| tetG | tetGF | GCT CGG TGG TAT CTC TGC TC | 468 | 13 | |
|
|
|
tetGR |
AGC AAC AGA ATC GGG AAC AC |
|
|
| Sulfonamide | sul1 | sul1F | ATG GTG ACG GTG TTC GGC ATT CTG | 841 | 15 |
| sul1R | GCT AGG CAT GAT CTA ACC CTC GG | ||||
| sul2 | sul2F | AGG GGG CAG ATG TGA TCG AC | 249 | 11 | |
|
|
|
sul2R |
GCA GAT GAT TTC GCC AAT TG |
|
|
| Trimethoprim | dfrA1 | dfrA1F | GTG AAA CTA TCA CTA ATG G | 470 | 10 |
| dfrA1R | CCC TTT TGC CAG ATT TGG | ||||
| dfrA10 | dfrA10F | TTA ATT ACC AGA GCA TTC GG | 374 | AY049746 | |
| dfrA10R | TAC ACA TCA GCA TGA ACA GG | ||||
| dfrA12 | dfrA12F | ACT CGG AAT CAG TAC GCA | 463 | 10 | |
|
|
|
dfrA12R |
GTG TAC GGA ATT ACA GCT |
|
|
| Kanamycin | aadD | aadD¬F | ATA TTG GAT AAA TAT GGG GAT | 161 | 12 |
| aadD¬R | TCC ACC TTC CAC TCA CCG GTT | ||||
| aphA/aph(3´)¬Id | aphAaphIdF | ATG GGC GCC TAT CAC AAT TGG | 257 | 12 | |
| aphAaphIdR | TCG CCT CCA GCT CTT CGT AGA | ||||
| aphAI¬IAB | aphAI¬IABF | AAA CGT CTT GCT CGA GGC | 461 | 12 | |
| aphAI¬IABR | CAA ACC GTT ATT CAT TCG TGA | ||||
| aph(3´)¬IIa | KanAphF | GAG AAA GTA TCC ATC ATG GC | 465 | L19385 | |
| KanAphR | GCT CAG AAG AAC TCG TCA AG |
All Salmonella strains were tested for the presence of the SopB gene. The primers were GATAGGAAAGATTGAGCACCTCTG and TACAGAGCTTCTATCACTCAGCTTC, and the PCR cycle consisted of 30 cycles of (30 s 95°C, 1 min 58°C, 1 min 72°C).
Pulsed-Field Gel Electrophoresis (PFGE)
The bacteria were grown while being shaken overnight at 37°C in Luria-Bertani broth (LB). The XbaI PFGE patterns were determined for all 21 S. Typhimurium strains by using previously described PFGE methods (16,17) with some slight modifications. The patterns were grouped in a dendrogram with GelCompar II software (Applied Maths, St.-Martens-Latem, Belgium) by using the Dice coefficient and the unweighted pair group method with an arithmetic averages clustering algorithm.
Invasion of the Human intestinal Epithelial Cell Line T84
The capacity of all cat Salmonella isolates and the human S. Typhimurium isolates 8420, 6237, 3520, 2200, and 5833 to invade human intestinal epithelial cells was determined. Cells of the human colon carcinoma cell line T84 were seeded in 96-well cell culture plates (Greiner, Frickenhausen, Germany) at a density of 5.105 cells/mL culture medium (DMEM + 10% fetal calf serum + 2% L-glutamine, without antimicrobial drugs) and grown for 24 h. Bacteria were grown for 20 h in LB-medium, after which the suspension was diluted 1:50 in fresh LB-medium. After 4 h of incubation at 37°C, suspensions were centrifuged and resuspended in DMEM with 10% fetal calf serum (FCS). The number of colony-forming units (CFU)/mL was determined by plating 10-fold dilutions on BGA. The suspensions were stored overnight at 4°C. The next day, 106 CFU in 200 μL were added to the T84 cell cultures, which were then centrifuged for 10 min at 1,500 rpm to make close contact between the bacteria and the colon cells. The plates were incubated for 1 h at 37°C and 5% CO2. The cells were then rinsed three times with Hanks' Balanced Salt Solution (HBSS, Life Technologies, Paisley, Scotland). Cell culture medium with gentamicin (50 μg/mL) was added, and plates were incubated for 1 h at 37°C and 5% CO2. Hereafter, the cells were rinsed three times with PBS and analyzed with 1% Triton X-100 (Sigma, St. Louis, MO) in distilled water. From this lysate, 10-fold dilution series were made. From each dilution, 6 x 20 μL was added to BGA, to determine the number of CFU Salmonella per mL. The assays were performed in triplicate. The percentage of intracellular bacteria, relative to the number of Salmonella bacteria, initially incubated with the cells, was calculated. The previously mentioned human isolates of S. Typhimurium were used for comparison between the cat isolates and human isolates. Statistical analysis was performed by analysis of variance methods using the SPSS 11.0 software.
Results
Characterization of Salmonella Isolates from Cats
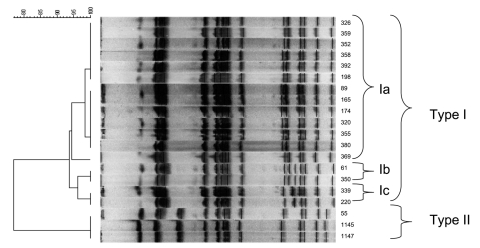
Of 278 healthy house cats, 1 Salmonella strain was isolated, an S. Enteritidis phage type 21 strain, sensitive to all tested antimicrobial drugs. Five strains were isolated from cats that died from or were euthanized because of incurable disease. Feline AIDS (caused by feline immunodeficiency virus [FIV]) was diagnosed in three cats, one died due to feline panleukopenia parvovirus infection, and one was poisoned. Three isolates were identified as being ampicillin-resistant S. Typhimurium phage type 193, harboring the blaTEM gene. They had the same pulsed-field gel electrophoresis (XbaI) pattern, indicating that the isolates were of clonal origin (Figure 1). The three cats came from the same owner. One isolate was an antimicrobial drug–sensitive Salmonella Bovismorbificans strain. One isolate was Salmonella 4:i:-, which was resistant to ampicillin, chloramphenicol, sulfonamides, tetracycline and sulfamethoxazole-trimethoprim (ACSuTSxt), harboring the blaTEM, cat, sul2, tet(A), and dfrA1 antimicrobial drug–resistance genes. Eighteen strains were isolated from the group-housed cats. All of these were S.Typhimurium phage type 120/ad. Fourteen of these strains showed acquired resistance to ampicillin, chloramphenicol and tetracycline and harbored the blaTEM, cat, and tet(A) antimicrobial drug-resistance genes, while four isolates were resistant to chloramphenicol only and only harbored the cat gene (Table 2). Pulsed-field gel electrophoresis showed that the isolates from the group-housed cats were of the same XbaI PFGE type, and that three subtypes within this type were present, indicating a clonal origin (Figure 1). One subtype contained the 14 strains that were resistant to the three mentioned antimicrobial drugs. All Salmonella strains harbored the SopB gene.
Figure 1.
Dendrogram obtained by unweighted pair group method using arithmetic averages clustering of the pulsed-field gel electrophoresis–XbaI patterns of serovar Typhimurium strains isolated from cats on the basis of the Dice coefficient.
Table 2. Characteristics of Salmonella isolates from catsa.
| Isolate no. | Source | Serotype | Phage type | PFGE patternb | Resistance phenotypeb | Resistance genotype |
|---|---|---|---|---|---|---|
| 11 | Diseased house cat | 4:i:- | – | ND | ACSuTSxt | blaTEM, cat, sul2, tet(A), dfrA1 |
| 40 | Diseased house cat | Bovismorbificans | – | ND | – | - |
| 109 | House cat | Enteritidis | 21 | ND | – | - |
| 1145, 1147, 55 | Diseased house cats | Typhimurium | 193 | II | A | blaTEM |
| 89, 165, 174, 198, 320, 326, 352, 355, 358, 359, 369, 380, 390, 392 | Group-housed cats | Typhimurium | 120/ad | Ia | ACT | blaTEM, cat, tet(A) |
| 161, 350 | Group-housed cats | Typhimurium | 120/ad | Ib | C | cat |
| 220, 339 | Group-housed cats | Typhimurium | 120/ad | Ic | C | cat |
aA, ampicillin; C, chloramphenicol; Su, sulfonamides, T, tetracycline; Sxt, sulfamethoxazole-trimethoprim; ND, not determined; PFGE, pulsed-field gel electrophoresis. bRoman numerals indicate the major types of fragment patterns; lowercase letters indicate minor variations in the respective fragment pattern.
Invasion of the Human Intestinal Epithelial Cell Line T84
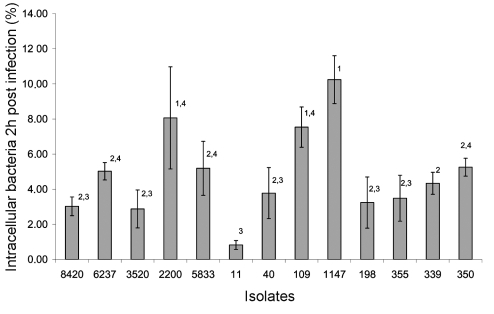
All isolates invaded T84 cells, with the cat isolates of S. Typhimurium PT193 (strains 1147, 1145, and 55, which belong to the same clone) and the human isolate S. Typhimurium strain 2200, the most invasive, yielding a percentage of invasion of 8% to 10%. The multidrug-resistant cat isolate Salmonella 4:i:- (strain 11) was the least invasive strain, having an invasion percentage of about 0.5%. Invasion percentages of the different isolates are shown in Figure 2. Of the strains of the same PFGE type, only one was shown in Figure 2, since no significant differences were detected between the invasion percentages of these strains. Statistically significant differences are shown in the figure.
Figure 2.
Invasion of Salmonella strains in the human intestinal epithelial cell line T84. The y-axis shows the percentages of intracellular bacteria 2 hours postinfection, relative to the initial number of bacteria, incubated with the cells. The x-axis shows isolate numbers. All isolates derived from the group-housed cats had the same invasion percentage as strains 198 and 355 (data not shown). Isolates 55 and 1145 had the same invasion percentage as strain 1147 (data not shown). Data not sharing superscript numbers indicate statistically significant differences (p < 0.05).
Discussion
This study concluded that, although cats can transmit Salmonella strains, healthy house cats are generally safe. Earlier reports regarding isolation of pathogens from healthy cats showed low percentages (mostly around 1%) of Salmonella-positive rectal swabs (18,19). In our study, 1 of 278 healthy cats was found to be positive. Immunodeficiency and nonhygienic housing can be predisposing factors for cats to shed Salmonella in the feces, resulting in contamination of the environment. Rectal swabs from 18 of 35 group-housed kittens were Salmonella-positive in our study. The fact that the 35 kittens were derived from more than 10 different owners before being group-housed and that one PFGE type (three subtypes) of S. Typhimurium 120/ad was isolated, indicates spread of the Salmonella strain between the cats or a common source. The age of these animals may also play a role, since all animals in this group were <4 months. Young animals are more susceptible to Salmonella infection. Also immunodeficiency can result in Salmonella excretion. One outbreak of fatal salmonellosis in cats has been reported after mild immunosuppression induced by live panleukopenia virus vaccination (7). In our study, animals infected with FIV and one animal that had panleukopenia shed Salmonella. Three animals that were infected with FIV were derived from the same owner, which indicates that the animals were infected with Salmonella from the same source or that one animal contaminated the others.
In our study, serotypes Typhimurium, Enteritidis, Bovismorbificans, and 4:i:- were isolated from cats. The isolated serotypes indicate that the cats were infected from the same sources compared with other animals and man. Indeed, serotypes Typhimurium and Enteritidis are the most widespread serotypes and the serotype Bovismorbificans is not uncommon in other animals, including humans (2,20).
Generally, invasion in the human intestinal epithelial cell line T84 was comparable between the cat isolates and isolates from humans. Invasion in intestinal epithelial cells is the primary step in the pathogenesis of Salmonella that causes gastrointestinal problems (21). This finding implies that the cat isolates are potentially pathogenic for humans. Moreover, all cat isolates harbored the SopB gene, which is involved in blocking the closure of chloride channels in gut epithelium and thus in inducing diarrhea. As in most other animal species, the cat isolates of the serotype Typhimurium harbored antimicrobial drug–resistant genes, raising concerns about spreading antimicrobial drug–resistant strains to humans.
Since the 1990s, concerns have arisen about the emergence and spread of multidrug-resistant Typhimurium strains, especially the multidrug-resistant ACSSuT type, which is resistant to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (2). In our study, some S. Typhimurium isolates from cats were resistant to a single drug such as ampicillin or chloramphenicol, while most isolates from the group-housed cats (same clone) were resistant to ampicillin, chloramphenicol, and tetracycline. Resistance genes were found to be blaTEM (ampicillin), cat (chloramphenicol), and tet(A) (tetracycline). The genes in the class 1 integron of the multidrug-resistant genomic island in ACSSuT type S. Typhimurium, required for the resistances to the above three mentioned antimicrobial drugs, are blaPSE1, floR, and tet(G) (22). This illustrates that these isolates did not acquire their resistance genes from horizontal transfer from pentadrug-resistant ACSSuT type strains. The isolate Salmonella 4:i:- was resistant to ampicillin, chloramphenicol, sulfonamides, tetracycline, and sulfamethoxazole/trimethoprim (ACSuTSxt-type), encoded by blaTEM (ampicillin), cat (chloramphenicol), sul2 (sulfonamides), tet(A) (tetracycline), and dfrA1 (trimethoprim). Also the resistance shown by this example had no relationship to the typical S. Typhimurium DT104 multidrug-resistant genomic island.
In conclusion, healthy house cats are generally safe with regard to excretion of Salmonella in the environment. Cats that are sick or are receiving medication resulting in immune deficiencies can potentially pose a threat to public health. Young children, the elderly, and immunocompromised persons are at risk because of their high sensitivity for the infection. All persons should follow good hygiene practices when keeping cats as pets.
Acknowledgments
We thank V. Eeckhaut, M. Foubert, and L. Winters for their excellent technical assistance.
Ivan Rychlik was supported by grant 1B44019 from the Ministry of Agriculture of the Czech Republic.
Biography
Dr. Van Immerseel is a postdoctoral researcher at Ghent University, Faculty of Veterinary Medicine, Department of Pathology, Bacteriology and Avian Diseases, where the work described in this study was performed. His research interests include bacterial pathogenesis and host-pathogen interactions, with a focus on Salmonella.
Footnotes
Suggested citation for this article: Van Immerseel F, Pasmans F, De Buck J, Rychlik I, Hradecka H, Collard J-M, et al. Cats as a risk for transmission of antimicrobial drug−resistant Salmonella. Emerg Infect Dis [serial on the Internet]. 2004 Dec [date cited]. http://dx.doi.org/10.3201/eid1012.040904
References
- 1.World Health Organization (WHO). WHO Surveillance programme for control of foodborne infections and intoxications in Europe. 7th Report 1993–1998. Berlin: Food and Agricultural Organization of the United Nations/WHO Collaborating Centre for Research and Training in Food Hygiene and Zoonoses; 2001.
- 2.Rabsch W, Tschape H, Baumler AJ. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 2001;3:237–47. 10.1016/S1286-4579(01)01375-2 [DOI] [PubMed] [Google Scholar]
- 3.Hald T, Vose D, Wegener HC, Koupeev T. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24:255–69. 10.1111/j.0272-4332.2004.00427.x [DOI] [PubMed] [Google Scholar]
- 4.Kimura AC, Reddy V, Marcus R, Cieslak PR, Mohle-Boetani JC, Knarreborg HD, et al. Chicken consumption is a newly identified risk for sporadic Salmonella enterica serotype Enteritidis infections in the United States: a case-control study in FoodNet sites. Clin Infect Dis. 2004;38:S244–52. 10.1086/381576 [DOI] [PubMed] [Google Scholar]
- 5.Stam F, Romkens TE, Hekker TA, Smulders YM. Turtle-associated human salmonellosis. Clin Infect Dis. 2003;37:167–9. 10.1086/379612 [DOI] [PubMed] [Google Scholar]
- 6.Stiver SL, Frazier KS, Mauel MJ, Styer EL. Septicemic salmonellosis in two cats fed a raw-meat diet. J Am Anim Hosp Assoc. 2003;39:538–42. [DOI] [PubMed] [Google Scholar]
- 7.Foley JE, Orgad U, Hirsh DC, Poland A, Pedersen NC. Outbreak of fatal salmonellosis in cats following use of a high titer modified-live panleukopenia virus vaccine. J Am Vet Med Assoc. 1999;214:67–70. [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved Standard M31-A. Wayne (PA): National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 9.Carlson SA, Bolton LF, Briggs CE, Hurd HS, Sharma VK, Fedorka-Cray PJ, et al. Detection of multiresistant Salmonella Typhimurium DT104 using multiplex and fluorogenic PCR. Mol Cell Probes. 1999;13:213–22. 10.1006/mcpr.1999.0240 [DOI] [PubMed] [Google Scholar]
- 10.Guerra B, Soto SM, Arguelles JM, Mendoza MC. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype 4,5,12:i:-. Antimicrob Agents Chemother. 2001;45:1305–8. 10.1128/AAC.45.4.1305-1308.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faldynova M, Pravcova M, Sisak F, Havlickova H, Kolackova I, Cizek A, et al. Evolution of antibiotic resistance in Salmonella enterica serovar Typhimurium strains isolated in the Czech Republic between 1984 and 2002. Antimicrob Agents Chemother. 2003;47:2002–5. 10.1128/AAC.47.6.2002-2005.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frana TS, Carlson SA, Griffith RW. Relative distribution and conservation of genes encoding aminoglycoside-modifying enzymes in Salmonella enterica serotype Typhimurium phage type DT104. Appl Environ Microbiol. 2001;67:445–8. 10.1128/AEM.67.1.445-448.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15:209–15. 10.1006/mcpr.2001.0363 [DOI] [PubMed] [Google Scholar]
- 14.Aminov RI, Chee-Sanford JC, Garrigues N, Teferedegne B, Krapac IJ, White BA, et al. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl Environ Microbiol. 2002;68:1786–93. 10.1128/AEM.68.4.1786-1793.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briggs CE, Fratamico PM. Molecular characterization of an antibiotic resistance gene cluster of Salmonella Typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebisch B, Schwarz S. Molecular typing of Salmonella enterica subsp. enterica serovar Enteritidis isolates. J Med Microbiol. 1996;44:52–9. 10.1099/00222615-44-1-52 [DOI] [PubMed] [Google Scholar]
- 17.Olsen JE, Skov MN, Threlfall EJ, Brown DJ. Clonal lines of Samonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J Med Microbiol. 1994;40:15–22. 10.1099/00222615-40-1-15 [DOI] [PubMed] [Google Scholar]
- 18.Hill SL, Cheney JM, Taton-Allen GF, Reif JS, Bruns C, Lappin MR. Prevalence of enteric zoonotic organisms in cats. J Am Vet Med Assoc. 2000;216:687–92. 10.2460/javma.2000.216.687 [DOI] [PubMed] [Google Scholar]
- 19.Spain CV, Scarlett JM, Wade SE, McDonough P. Prevalence of enteric zoonotic pathogens in cats less than 1 year of age in central New York State. J Vet Intern Med. 2001;15:33–8. 10.1111/j.1939-1676.2001.tb02294.x [DOI] [PubMed] [Google Scholar]
- 20.Liesegang A, Davos D, Balzer JC, Rabsch W, Prager R, Lightfoot D, et al. Phage typing and PFGE pattern analysis as tools for epidemiological surveillance of Salmonella enterica serovar Bovismorbificans infections. Epidemiol Infect. 2002;128:119–30. 10.1017/S0950268801006719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type three secretion system. Microbes Infect. 2001;3:1281–91. 10.1016/S1286-4579(01)01488-5 [DOI] [PubMed] [Google Scholar]
- 22.Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, Imberechts H, et al. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol. 2001;183:5725–32. 10.1128/JB.183.19.5725-5732.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]