Abstract
Background
Idiopathic Pulmonary Fibrosis (IPF) is characterized by profound changes in the lung phenotype including excessive extracellular matrix deposition, myofibroblast foci, alveolar epithelial cell hyperplasia and extensive remodeling. The role of epigenetic changes in determining the lung phenotype in IPF is unknown. In this study we determine whether IPF lungs exhibit an altered global methylation profile.
Methodology/Principal Findings
Immunoprecipitated methylated DNA from 12 IPF lungs, 10 lung adenocarcinomas and 10 normal histology lungs was hybridized to Agilent human CpG Islands Microarrays and data analysis was performed using BRB-Array Tools and DAVID Bioinformatics Resources software packages. Array results were validated using the EpiTYPER MassARRAY platform for 3 CpG islands. 625 CpG islands were differentially methylated between IPF and control lungs with an estimated False Discovery Rate less than 5%. The genes associated with the differentially methylated CpG islands are involved in regulation of apoptosis, morphogenesis and cellular biosynthetic processes. The expression of three genes (STK17B, STK3 and HIST1H2AH) with hypomethylated promoters was increased in IPF lungs. Comparison of IPF methylation patterns to lung cancer or control samples, revealed that IPF lungs display an intermediate methylation profile, partly similar to lung cancer and partly similar to control with 402 differentially methylated CpG islands overlapping between IPF and cancer. Despite their similarity to cancer, IPF lungs did not exhibit hypomethylation of long interspersed nuclear element 1 (LINE-1) retrotransposon while lung cancer samples did, suggesting that the global hypomethylation observed in cancer was not typical of IPF.
Conclusions/Significance
Our results provide evidence that epigenetic changes in IPF are widespread and potentially important. The partial similarity to cancer may signify similar pathogenetic mechanisms while the differences constitute IPF or cancer specific changes. Elucidating the role of these specific changes will potentially allow better understanding of the pathogenesis of IPF.
Introduction
Idiopathic pulmonary fibrosis (IPF) is a non-neoplastic pulmonary disease, characterized by extracellular matrix deposition, myofibroblasts foci formation and alveolar epithelial cell hyperplasia [1], [2], [3], [4]. The disease is progressive and in most cases unresponsive to corticosteroid and immunosuppressive therapy [5]. Although the exact etiology of the disease is still under investigation, several studies suggest that a combination of genetic and environmental factors may be the cause of IPF [6]. Exposure to wood, metal dust or stone/sand/silica as well as smoking, farming and handling livestock are associated with IPF in several independent studies [7]. A unique feature to the lung phenotype in IPF is the extent to which the lung is altered from normal. Alveolar epithelial cells and fibroblasts exhibit distinct and profound changes in their phenotypes with alveolar epithelial cells undergoing hyperplasia and potentially epithelial mesenchymal transdifferentiation and fibroblasts becoming activated and exhibiting myofibroblast features. Multiple studies demonstrated that the lung phenotype in IPF is dramatically different than that of the healthy lung with globally different patterns of mRNA and microRNA expression [8], [9], [10], [11], [12], [13], [14] and aberrations in multiple pathways such as coagulation [15], apoptosis [16], [17], oxidative stress [18], epithelial mesenchymal transition [19], [20], [21] and developmental pathways [21], [22], [23]. Usually it is assumed that multiple cycles of injury lead to this phenotype, however these injuries do not explain how those profound phenotypic changes are sustained and even progress years after the initial injury. Global epigenetic changes, traditionally defined in the context of heritable changes that are not coded by changes in DNA sequence, have rapidly emerged as a general mechanism by which cellular molecular phenotypes are stably altered during development, cellular differentiation, response to environmental stress and disease pathogenesis [24], [25], [26]. It is well established that nutritional, chemical and physical factors can have a profound effect on gene expression [27]. Not only can they cause mutations in the promoter and coding regions of genes but they can also orchestrate a variety of epigenetic changes [28]. Two of the best described mechanisms of epigenetic control are DNA methylation and chromatin remodeling. DNA methylation typically involves the addition of a methyl group to the 5 position of the cytosine pyrimidine ring of a CpG dinucleotide [29]. Clustered CpGs form CpG islands whose state of methylation is critical for the activity of transposable elements and the transcriptional regulation of genes through direct blockage of transcription factors or chromatin remodeling [30]. Alterations of CpG methylation have been implicated in many diseases where the hypermethylation of the promoter associated CpG islands results in transcriptional silencing [31] while the hypomethylation results in loss of imprinting and transcriptional activation [32]. Aberrant methylation of CpG dinucleotides is a well-recognized epigenetic hallmark of multiple diseases including lung cancers [33], [34], [35], [36]. So far, the extent and role of epigenetic changes has not been studied in IPF.
In this study, we analyze global methylation patterns of IPF using human CpG island microarrays. In addition to compiling a DNA methylation profile that differentiates IPF patients from normal individuals, we compared this profile to that of lung adenocarcinoma patients. Our results reveal an altered DNA methylation pattern in IPF which shows great similarity to the methylation pattern of lung cancer. Our work is the first step in understanding the role of DNA methylation in the pathogenesis of IPF. Furthermore, the similarity of IPF with cancer may reveal common underlying molecular mechanisms and offer therapeutic options for IPF patients adopted from cancer biology [37].
Methods
Sample Description
Lung tissue samples were obtained through the University of Pittsburgh Health Sciences Tissue Bank (Pittsburgh, PA). They included 12 frozen lung tissue samples from IPF patients, 10 frozen lung tissue samples from adenocarcinoma patients and 10 histological normal lung samples obtained from the same group of adenocarcinoma patients ( Table 1 ). The diagnosis of IPF was based on microscopic findings that were consistent with usual interstitial pneumonia [1], [3]. All adenocarcinoma tumors were obtained from patients staged as T1b-T2b N0 M0 [38]. All cancer patients were smokers and older than IPF patients. The lung samples that were removed from patients with lung cancer contained both adenocarcinoma tissues and normal histology tissues obtained from disease-free margins of the lung. The IPF patients fulfilled the diagnostic criteria of the American Thoracic Society and the European Respiratory Society [1]. Patients consented to the donation of the removed tissue to the tissue bank and the use of the archived tissue was approved by the University of Pittsburgh Institutional Review Board (IRB020123, IRB0506140, IRB0411036). (Pittsburgh, PA).
Table 1. Human subjects.
| Variable | Control/Cancer | IPF |
| Number of subjects | 10 | 12 |
| Average age, yr. | 71(±12.44) | 60(±5.51) |
| Male/Female | 6/4 | 8/4 |
MeDIP and Hybridization to Human CpG Island Microarrays
Genomic DNA was extracted from 10 mg of frozen lung tissues using the DNeasy Blood & Tissue kit (Qiagen Sciences, CA) following the manufacturer’s protocol. Five micrograms of genomic DNA were sonicated (Sonic Dismembrator, Fisher Scientific) to achieve fragment lengths between 300–600 bp. Methylated DNA was immunoprecipitated (IP) using 5-Methylcytidine monoclonal antibody (AbD Serotec, NC) as instructed in the Agilent Microarray Analysis of Methylated DNA Immunoprecipitation protocol (Agilent Technologies, Santa Clara, CA) [39], [40]. Immunoprecipitated DNA and total genomic DNA were labeled with Cy3 and Cy5 respectively, using Agilent Genomic Labeling kit PLUS (Agilent Technologies, Santa Clara, CA) and hybridized to the human CGI oligonucleotide microarrays (Agilent Technologies). The arrays were designed according to the University of California at Santa Cruz (UCSC) genome-browser CpG island list and contained 237,000 probes covering more than 90% of the 27,639 human CpG islands at a density of 1 probe per 100 bp as described in Straussman et al [41] along with quality control assays that assess the platform’s performance. Further validation of Agilent’s MeDIP microarray platform was achieved by Yamashita et al [42]. Following hybridization and washing, the arrays were scanned in an Agilent G2509C microarray scanner and raw data were obtained using Feature Extraction Ver.9.5.3.
Microarray Data Analysis
For our analysis we only included probes with a hybridization Tm value between 79°C and 93°C as these show higher quality signal [41]. We subsequently divided the probes according to their Tm into 14 groups/bins differing by 1°C. Probe signals in each bin were standardized to have an average of 0 and a standard deviation of 1. To work in a CpG island oriented manner we scored each island for its likelihood to be methylated. For that purpose, each probe was mapped to the genome and the signals of the probes that were mapped to a single CpG island were averaged to obtain the island’s methylation score [41]. The complete microarray data have been deposited in the Gene Expression Omnibus (GSE29895), are MIAME compliant and publicly available.
MassARRAY EpiTYPER Assay
CpG dinucleotide methylation was quantified by the MassArray EpiTYPER platform (Sequenom Inc, CA) [43]. The EpiTYPER assay is a MALDI TOF mass spectrometry based quantitative method for measuring CpG methylation down to a single dinucleotide resolution. 500 ng of fragmented DNA from each sample was modified by bisulfite treatment. Following PCR with specific primers and Shrimp Alkaline Phosphatase treatment, fragments were ligated to a T7 promoter segment, and then transcribed into RNA. The synthesized RNA was cleaved with RNase A and all cleavage products were analyzed by MassArray in the Genomics and Proteomics Core Laboratory (GPCL, University of Pittsburgh, Pittsburgh, PA) according to the manufacturer’s instructions. Primers were designed using the EpiDesigner Software (http://www.epidesigner.com/index.html) (Table S1 in Supporting Information).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from frozen lung tissue with miRNeasy mini kit (Qiagen Sciences,CA) following the manufacturer’s protocol [44]. 500 ng of the extracted RNA sample was used as a template for the reverse transcriptase reaction. 25 ng of the synthesized cDNA was amplified in a qPCR reaction using TaqMan universal PCR master mix (Applied Biosystems, Foster City,CA) and TaqMan gene expression assays for the following genes: STK17B (assay IDHs00177790_m1), STK3 (assay Hs00169491_m1), HIST1H2AH (assay Hs00544732_s1) and GUSB (assay Hs99999908_m1 ). All assays were done in triplicates and appropriate Non-Transcriptase and Non-Template control reactions were included. GUSB (encoding b-glucoronidase) was used as a housekeeping gene for normalization and the results were analyzed by the ΔΔCT method [45] after averaging the triplicates of each assay. Fold change was calculated by taking the average of all the control samples as the baseline.
Data Analysis
Differentially methylated CpG islands were identified by analyzing the CpG Island Microarray data with the Class Comparison feature of BRB-ArrayTools 3.7.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html). We controlled for multiple testing by setting the significance level at a False Discovery Rate (FDR) of less than 5% [46]. Data visualization was accomplished using the Genomica [47] and the JavaTreeView software packages. The Student’s t test was applied to for the EpiTYPER MassArray and qRT-PCR to test significance of the results. Significance of overlap of differentially methylated islands (DMI) between IPF and Cancer samples and enrichment of DMIs in promoter regions was calculated using the hypergeometric distribution. Pathway analysis was performed using DAVID Bioinformatics Resources 6.7 [48] and IPA Ingenuity Systems (http://www.ingenuity.com).
Results
The patterns of DNA methylation in lung samples of IPF, cancer patients and controls, were determined using Agilent Human CpG Islands microarrays. Overall, 12 IPF, 10 lung adenocarcinoma and 10 normal histology samples from the same adenocarcinoma patients were included in our study ( Table 1 ). The analysis of the microarray data was divided into two parts. In the first part, the IPF or the adenocarcinoma samples were compared to the control samples to compile two separate lists of differentially methylated CpG islands. In the second part, the two lists were compared to assess for differences or similarities between the methylation changes that are associated with each disease.
IPF Lung Samples Show a Different Methylation Profile when Compared to Normal Histology Lung Samples
The 25,406 out of 27,639 human CpG islands that had an acceptable Tm (see methods) were analyzed using the Class Comparison algorithm from BRB Array Tool software package. 625 CpG islands were found to be differentially methylated in IPF lung tissue samples when compared to control lung tissue samples ( Figure 1A , Table S2 in Supporting information). 91.2% of the 625 differentially methylated CpG islands were located in intronic, exonic or and intergenic areas and only 8.8% in promoters. Considering that 10,923 of the 25,406 (43%) CpG islands in our study localize to promoters, this result indicates that a significantly larger than expected (p < 10–79) proportion of changes in methylation, when comparing IPF and control samples, occurs in regions that are not annotated as promoters in the current genome build.
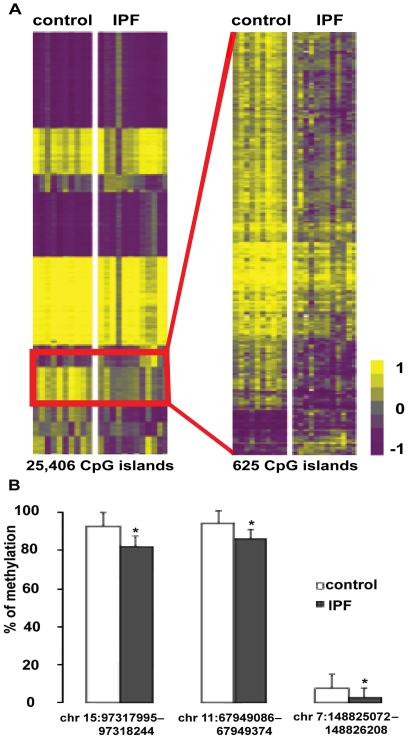
Figure 1. CpG islands are differentially methylated in IPF and control samples.
(A) Human CpG Island Microarray data: the heatmap on the left is the visual comparison of global methylation profiles between the 10 Control and the 12 IPF samples. The heatmap on the right consists only of the differentially methylated CpG probes highlighted by the red rectangle (p-value < 0.05, FDR<5%). Methylated CpG islands are shown in progressively brighter shades of yellow, depending on the fold difference, and hypomethylated CpG islands are shown in progressively brighter shades of purple. Grey stands for no difference between the two sample groups being compared. (B) EpiTYPER confirmation of differentially methylated CpG islands. Bars represent the average methylation of all the samples per study group. The X-axis shows the genomic location of each CpG island and Y-axis shows the percentage of methylation.
To validate the microarray results, 3 differentially methylated CpG islands showing various degrees of change in their methylation levels were picked and analyzed with the Sequenom’s MassArray EpiTYPER assay. The EpiTYPER assays showed decreased CpG island methylation in the IPF lung samples which was in agreement with the microarray data ( Figure 1B ). All differentially methylated CpG islands were mapped to the genome using the UCSC genome browser [49] and a list of genes that contain CpG islands showing significantly hyper- or hypomethylation in IPF lung samples was compiled (Table S2 in Supporting Information). A Functional Annotation Clustering of these genes using DAVID Bioinformatics Resources 6.7 revealed that a significant number of them are involved in apoptosis, cell morphogenesis, the regulation of cellular biosynthetic processes and histone acetylation ( Table 2 ). The modified Fisher Exact p-Value/EASE Score is calculated to measure gene-enrichment in any given annotation term. It ranges from 0 to 1 with 0 representing perfect enrichment. “Score” stands for Group Enrichment Score, which is calculated using the p-values of the individual members of each Functional Annotation Cluster. The higher the number is the higher the cluster ranks in biological significance [48].
Table 2. Functional annotation clustering of differentially methylated CpG islands.
| Cluster | Score | Count | p-Value | Genes |
| regulation ofapoptosis | 11.5 | 23 | 2.86E-12 | COL18A1,OBSCN,PRKCZ,MAEA,WFS1,BCAR1, STK17B,INTS1,ZBTB16,TNFRSF4,STK3,SRC, MCF2L,PROC,AKT1,IGF1R,NOTCH1,IGF2R, BIRC8, BCL6,DNAJB6, ARHGDIA,TERT |
| negative regulation of cellular biosynthetic process | 10.5 | 19 | 3.39E-11 | EIF2C2,CTBP1,CTBP2,NACC2,EHMT1.JARID2, RBM15B,ZEB2,CBX2,ZBTB16,PRDM16,TNFRSF, HDAC4,ZGPAT,STRA8,BCL6,NCOR2,DNAJB6, SMARCA4 |
| regulation of cell morphogenesis | 3.3 | 5 | 3.03E-03 | PLXNA3,PLXNB2,CDH4,ARHGDIA,MBP |
| histone acetylation | 2.8 | 4 | 1.36E-03 | KAT2A,BRD1,KAT2B,CREBBP |
Decrease in Promoter CpG Island Methylation is Associated with Increased Gene Expression
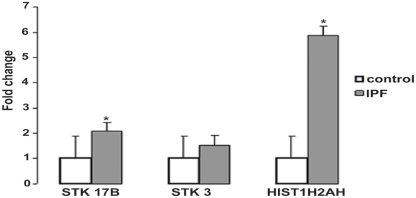
Typically, the methylation of promoter localized in CpG islands affects gene expression of the downstream genes [50]. All 625 differentially methylated CpG islands were checked for promoter localization and presence of a Trascriptional Start Site (TSS) using the UCSC Genome Browser [49]. 55 CpG islands were mapped in the promoter region of genes that had a well-characterized TSS ( Table 3 ). An IPA functional analysis showed that the genes with differentially methylated CpG islands in their promoters are associated with biological processes such as cellular assembly and organization, cellular growth and proliferation, cell morphology, cancer ,cell signaling, gene expression and cell death (Table S3 in Supporting Information). We analyzed by qRT-PCR three genes localized in differentially regulated regions. Serine/Threonine Kinase 17b (STK17B) and Serine/Threonine Kinase 3 (STK3) are involved in apoptosis while histone cluster 1 H2ah (HIST1H2AH) is essential is nucleosome formation. All three transcripts showed increased levels of expression in IPF samples compared to controls but only STK17B and HIST1H2AH have a p-value <0.05 while in the case of STK3 the p-value is 0.07 ( Figure 2 ).
Table 3. Differentially methylated promoters between IPF and control samples.
| CpG location | Gene | Eponine Predicted TSS | SwitchGear GenomicTSS | p-value | Fold change |
| chr2:242678973-242680235 | LOC728323 | chr2.35165-35173 | CHR2_P1372 | 5.70E-06 | 0.48 |
| chr7:148825072-148826208 | ZNF 746 | chr7.53295-53308 | CHR7_M1039 | 1.19E-03 | 0.52 |
| chr19:2653532-2653987 | GNG7 | chr19.27595-27597 | CHR19_M0064,CHR19_P0 | 1.72E-03 | 0.53 |
| chr22:37569114-37570371 | NPTXR | chr22.39508-39517 | CHR22_M0327 | 3.77E-05 | 0.56 |
| chr12:119290634-119291409 | MSI1 | chr12.12663-12672 | CHR12_M0815 | 4.79E-05 | 0.58 |
| chr17:53515753-53516493 | DYNLL2 | chr17.23937 | CHR17_P0699 | 1.10E-04 | 0.58 |
| chr8:99906637-99907112 | STK3 | chr8.55187 | CHR8_M0508 | 3.76E-04 | 0.58 |
| chr19:52614063-52614589 | MEIS3 | CHR19_M0732 | 3.60E-04 | 0.59 | |
| chr4:75122817-75123453 | CXCL3 | CHR4_M0336 | 3.54E-04 | 0.59 | |
| chrY:2537106-2537697 | NCRNA00103 | chrY.61288 | CHRY_P0007 | 2.22E-04 | 0.59 |
| chr9:126945496-126945768 | SCAI | CHR9_M0684 | 3.13E-04 | 0.59 | |
| chr13:31318630-31319274 | EEF1DP3 | chr13.13604-13608 | CHR13_P0091 | 2.02E-04 | 0.6 |
| chr1:46906261-46906982 | ATPAF1 | chr1.2564-2568 | 7.34E-04 | 0.6 | |
| chr1:76312735-76313241 | ST6GALNAC3 | chr1.2978,2979 | CHR1_P0726_R1 | 1.10E-04 | 0.61 |
| chr6:27222201-27223160 | HIST1H2AH | chr6.48519 | CHR6_M0196,CHR6_P0200 | 6.12E-04 | 0.61 |
| chr20:46971570-46972079 | ARFGEF2 | chr20.36249-36250 | CHR20_P0396 | 1.65E-04 | 0.62 |
| chr20:390530-391317 | TBC1D20 | chr20.35209-35211 | CHR20_M0004 | 3.33E-04 | 0.62 |
| chr3:20056462-20057430 | KAT2B | chr3.40742-40745 | CHR3_P0121 | 1.15E-04 | 0.63 |
| chr8:1908966-1910279 | KBTBD11 | chr8.53949-53953 | CHR8_P0014,CHR8_M0 | 3.49E-04 | 0.63 |
| chr9:21549133-21549816 | LOC554202 | chr9.56427 | CHR9_M0115 | 9.90E-04 | 0.63 |
| chr3:44493842-44494372 | ZNF445 | chr3.41072-41077 | CHR3_M0205 | 3.93E-05 | 0.64 |
| chr7:16427303-16427790 | ISPD | chr7.51105 | 4.83E-04 | 0.64 | |
| chr7:4889233-4890102 | RADIL | chr7.50913-50914 | CHR7_M0042 | 4.83E-04 | 0.64 |
| chr10:73203097-73203498 | C10orf54 | chr10.6463-6464 | 1.31E-03 | 0.64 | |
| chr17:35036840-35037445 | PPP1R1B | chr17.23254 | 9.02E-04 | 0.64 | |
| chr2:37237405-37237906 | EIF2AK2 | CHR2_M0214 | 7.71E-04 | 0.64 | |
| chr2:196743834-196744628 | STK17B | CHR2_M1078 | 4.38E-04 | 0.65 | |
| chr7:105539339-105540384 | SYPL1 | chr7.52701-52704 | CHR7_M0755 | 7.45E-04 | 0.65 |
| chr5:177949164-177950276 | COL23A1 | CHR5_M0978 | 5.10E-04 | 0.66 | |
| chr9:138236486-138236814 | LHX3 | CHR9_M0813 | 9.64E-05 | 0.66 | |
| chr6:38715755-38716126 | BTBD9 | chr6.48830 | 6.01E-04 | 0.67 | |
| chr22:37481403-37482422 | SUN2 | chr22.39495-39505 | CHR22_M0325.1 | 1.15E-03 | 9.68 |
| chr17:77421925-77423424 | ARHGDIA | chr17.24948-24955 | CHR17_M1049 | 1.00E-03 | 0.68 |
| chr1:219026639-219027226 | MOSC1 | chr1.4565 | CHR1_P1706_R1 | 5.74E-04 | 0.69 |
| chr16:23429074-23429400 | GGA2 | chr16.19779 | CHR16_M0269_R1 | 2.76E-04 | 0.71 |
| chr6:35996457-35997038 | SRPK1 | CHR6_M0389 | 8.45E-04 | 0.71 | |
| chr4:77391611-77392084 | FAM47D | chr4.44416-44418 | CHR4_P0385 | 2.44E-04 | 0.72 |
| chr3:52714577-52715466 | GLT8D1-SPCS1 | chr3.41472-41477 | CHR3_M0331,CHR3_P0373 | 3.65E-04 | 0.72 |
| chr19:40015371-40015792 | LOC400685 | chr19.29348 | CHR19_M0487 | 1.08E-03 | 1.36 |
| chr1:154808723-154809126 | IQGAP3 | CHR1_M1183_R1 | 3.07E-04 | 1.37 | |
| chr11:64645712-64646507 | FAU-MRPL49 | chr11.9471-9474 | CHR11_M0531_R1 | 8.27E-04 | 1.37 |
| chr22:36575287-36575627 | EIF3L | CHR22_P0282 | 1.21E-03 | 1.47 | |
| chr19:62972738-62973298 | ZNF 586 | CHR19_P0955.3 | 1.01E-03 | 1.49 | |
| chr2:10747175-10747692 | NOL10 | CHR2_M0064, CHR2_P0054 | 8.54E-04 | 1.51 | |
| chr22:29886124-29886466 | RNF185 | CHR22_P0227 | 2.63E-04 | 1.53 | |
| chr4:17225273-17225604 | MED28 | CHR4_P0145 | 1.32E-03 | 1.53 | |
| chr4:191142227-191143118 | TUBB4Q | chr4.45402 | 9.16E-05 | 1.53 | |
| chr17:24206105-24206445 | ERAL1 | CHR17_P0310 | 6.41E-04 | 1.56 | |
| chr17:34610012-34610471 | RPL19 | CHR17_P0434 | 3.92E-04 | 1.56 | |
| chr3:50357893-50358314 | ZMYND10 | chr3.41343 | CHR3_M0300 | 2.53E-04 | 1.56 |
| chr1:40008354-40009777 | BMP8B | chr1.2303 | 3.91E-04 | 1.61 | |
| chr1:68288824-68289061 | DIRAS3 | chr1.2955 | 1.58E-04 | 1.63 | |
| chr3:159844871-159845169 | GFM1 | CHR3_P0882 | 1.28E-03 | 1.66 | |
| chr3:139211681-139211893 | CLDN18 | CHR3_P0776 | 4.00E-06 | 1.69 | |
| chr5:169592408-169592807 | C5orf58 | CHR5_P0975 | 1.70E-06 | 1.81 |
Figure 2. Expression of genes with differentially methylated promoters.
qRT-PCR assay on 3 genes with hypomethylated promoter-associated CpG islands showed increase in the expression of the downstream gene. Y-axis shows fold change of detected transcripts in IPF samples when the expression in controls is set to baseline equal to 1. * denotes p-values <0.05. Error bars are based on Standard Deviation.
The Methylation Profiles of IPF and Adenocarcinoma Lung Samples Overlap
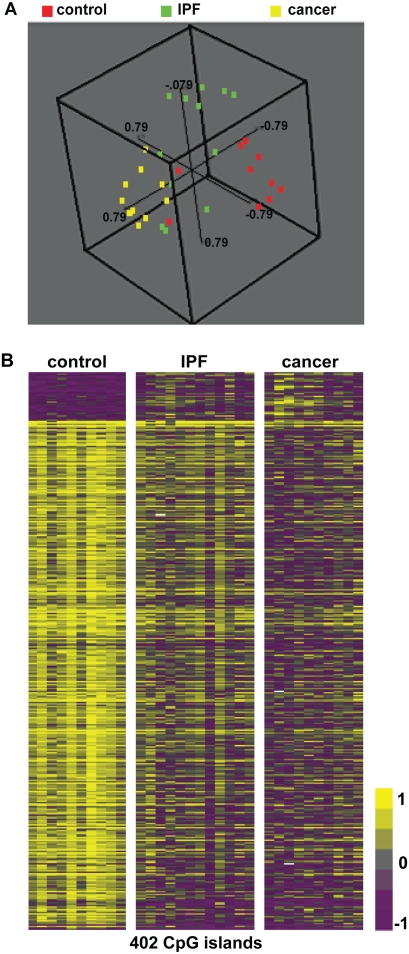
To determine the similarity of IPF samples to lung cancer we performed Principal Component Analysis (PCA) and Class Comparison using BRB-Array Tools on microarray data from 12 IPF patients and the 10 lung cancer patients (normal histology and adenocarcinoma samples included). PCA analysis demonstrated that IPF samples were positioned between the control and cancer samples suggesting that IPF samples had a methylation profile with partial similarity to both groups ( Figure 3A ). It is worth mentioning that IPF samples were more similar to cancer than control samples despite the fact that cancer and control tissue were obtained in pairs from the same patient. This observation may suggest that the majority of differences between IPF and controls may be related to differential environmental exposures or smoking effect because these differences persisted in the comparison of cancer to control despite the fact they came from the same subject. Class comparison analysis revealed that 2428 CpG islands were differentially methylated between cancer samples and normal histology controls. When compared to the 625 that are differentially methylated between IPF and Controls, 402 CpG islands overlapped. In other words, 65% of the CpG islands that have an altered methylation pattern in IPF lung samples are also modified in lung cancer samples ( Figure 3B and Table S4 in Supporting Information). This overlap is highly significant, as the probability of such an overlap to occur in random is very low (p<10−256). 45% of the 402 overlapped CpG islands are located in intronic and intergenic areas, 6% in promoters and 49% in exons.
Figure 3. Comparison of IPF and Adenocarcinoma to control samples.
(A) 3-D plot representation of the results after Principal Component Analysis of all 3 sample groups. Each color-coded dot represents a sample (red-control, green-IPF and blue-cancer) and each sample is positioned in the 3-D space according to its similarity or difference to the others. (B) Comparison of differentially methylated CpG islands that overlap between IPF and Lung Cancer. The heatmap consists of 402 differentially methylated CpG islands that are found to overlap between IPF and Lung Cancer. High methylation levels of a CpG islands are shown in yellow while low methylation levels of methylation are shown in purple. Grey stands for no difference between the two groups being compared.
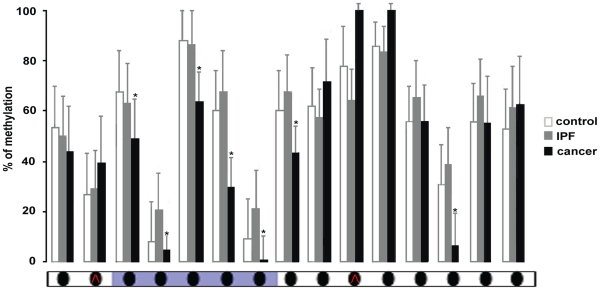
To determine whether similar methylation patterns in IPF and cancer result from a global change in methylation we assessed LINE-1 methylation. LINE-1 retrotransposons are abundantly and equally distributed across the genome and their methylation pattern is often used as an indicator of global methylation levels [51]. The methylation status of LINE-1 (GenBank: X58075.1) was defined in all three study groups (IPF, Cancer and Control) using the EpiTYPER MassArray assay. The PCR primers were designed to encompass the 15 CpG sites or units including the possible intrinsic LINE-1 promoter (Table S1 in Supporting Information). Although LINE-1 elements were found to be hypomethylated in the adenocarcinoma samples no significant change of the methylation levels was detected the in IPF samples ( Figure 4 ) suggesting that methylation changes in IPF were specific to regions.
Figure 4. Methylation profile of LINE-1 retrotransposon.
Global methylation levels of the LINE-1 retrotransposons were defined using the EpiTYPER mass array assay. Each bar represents the methylation in one of the 15 CpG dinucleotides or CpG units that span the LINE-1 sequence. Methylation levels are calculated as the average of all samples in each group (Control, IPF, Cancer) and standard error bars are included. The X axis shows the CpG dinucleotide or the CpG unit and the Y axis shows the percentage of methylation. The LINE-1 promoter is indicated in purple. The red arrowheads indicate units of 2-3 CpG dinucleotides.
Discussion
In this study, we used human CpG island microarrays to identify the differentially methylated CpG islands in the lung tissue of IPF patients. Our results indicate that the CpG island methylation profile of the IPF lung samples is very different from that of control samples and greatly overlaps with methylation changes observed in lung adenocarcinoma samples. Despite the observed similarity in CpG methylation between IPF and lung cancer, the lack of LINE-1 methylation in IPF suggests a more specific DNA methylation, which is confined to certain regions of the genome.
One of the most impressive results of our study is the extent of differentially methylated regions in the IPF lungs. Interestingly the majority of the differentially methylated CpG islands rest in promoter-distal sites or intragenic regions and only 8.8% of them are localized in gene promoters. Whereas the methylation status of promoter associated CpG islands can directly affect transcription, the role of the CpG methylation outside the immediate promoter region remains somewhat unclear. It is proposed that methylation of CpG island shores outside the promoter could also control transcription of downstream genes [52] or lead to histone modifications [53]. Methylation changes that occur in intragenic regions could impact RNA splicing [54]. In addition, methylation changes may affect the expression of non-coding RNAs [55] and thus indirectly affect global changes in gene expression. The biological impact of modest changes in the degree of CpG methylation is in fact unpredictable. As an example in the case of prostate cancer, a gradual increase in methylation from 12.6% to 19.3% or 21.8% signified a transition from a benign state to a localized or metastatic cancer, respectively [56]. However, regardless of the direct downstream effects, the extent of the methylation changes we found, supports previous observations about the degree and profundity of molecular changes in the IPF lung [8], [9], [10], [11], [12], [13], [14].
Naturally, the motivation to assemble methylation profiles is to find the underlying mechanisms that drive changes in gene expression. The detailed characterization of each one of the differentially methylated CpG islands in IPF patients is beyond the scope of this study. Globally, genes with differentially methylated CpG islands in their promoters were involved in biological processes such as cellular assembly and organization, cellular growth and proliferation, cell morphology, cancer, cell signaling, gene expression and cell death. All of these processes could be implicated in IPF pathogenesis. In our validation we focused on genes with differentially methylated promoters. We selected the Serine/Threonine Kinase 17b (STK17B) and Serine/Threonine Kinase 3 (STK3) because of their role in apoptosis [57] and the histone cluster 1 H2ah (HIST1H2AH) because of the recent interest in histone modifications in fibrosis[58]. STK17B and HIST1H2AH were significantly up-regulated in our IPF samples which is in agreement with the hypo-methylated state of their promoter associated CpG islands. Interestingly, the majority of the differentially methylated islands that were within or close to known genes were outside promoter regions. Some of these methylation changes were in genes that were previously reported to be increased in IPF such as COL18A1 [11], genes that are implicated in myofibroblast differentiation such as NOTCH1 [59] or markers of progressive IPF like SMARCA4 [60]. In addition, the promoter of CXCL3, a gene which is found to be up-regulated in the lung of bleomycin treated mice [61], was also hypomethylated in our IPF samples. When we looked for the overlap of differentially expressed genes in IPF in our previously published gene expression datasets [13], [14] we found that there were 46 genes that had both differentially methylated gene related CpG islands and gene expression changes. While a detailed analysis of methylation and expression changes in the same tissue would be better suited to address the correlation of methylation and gene expression changes our findings suggest that at least some of the methylation changes that we observed do have an effect on lung gene expression and thus may contribute to the lung phenotype in IPF.
A remarkable finding of our study is the similarity in DNA methylation patterns between IPF and lung adenocarcinoma. Recently, Vancheri et al compared IPF to cancer and described the pathogenic similarities between the two diseases. More specifically, they referred to common genetic and epigenetic alterations, uncontrolled proliferation, tissue invasion and perturbation of signal transduction pathways [37]. The similarity between cancer and IPF spreads to microRNA expression such as in the case of let-7d and hsa-miR-21, which are found to be down-regulated or up-regulated respectively in both diseases [8], [9]. All of these observations are in accord with published studies reporting high incidence of cancer in IPF patients when compared to healthy individuals [62], [63]. DNA hypomethylation is a hallmark of cancer [64] and in many types of cancer including lung carcinomas it is accompanied with lower levels of methylation in repetitive DNA elements [34], [65]. While the similarity in the differentially methylated CpG islands suggests common epigenetic mechanisms between IPF and cancer, our analysis of LINE-1 methylation indicates that this similarity is limited. LINE-1 repeats comprise about 20% of the human genome [66]. LINE-1 elements are usually methylated in somatic tissues but they are often hypomethylated in tumors [65], [67] resulting in increased mobility, which in turn leads to gene disruptions [68] and chromosomal instability [69]. While LINE-1 retrotransposons were hypomethylated in our cancer samples they were not in IPF samples leading to the conclusion that CpG island methylation changes in IPF are somewhat parallel to cancer but are not as extensive and do not involve global changes in LINE-1 methylation. This suggests that despite the similarities between the DNA methylation profiles of IPF and cancer, there are different mechanisms that cause and sustain these changes.
One of the major concerns in our global profiling approach is tissue heterogeneity. The IPF lungs contain mixed areas of normal tissue, myofibroblast foci and honeycombing [1]. The IPF lung is also highly variable in its cellular content as it contains normal cells like epithelial, endothelial cells and fibroblasts as well as abnormal ones like hyperplastic type II alveolar epithelial cells, myofibroblasts, potentially altered endothelial cells and varying degrees of inflammatory cells. Thus it is possible that the signal we obtained is only an under-estimation of the real epigenomic changes caused by an admixture of normal and abnormal regions, microenvironments and cell types. Naturally, it is impossible based on our analysis to determine whether the observed DNA methylation changes are cell type specific. In this context, our strategy of averaging signals across an island could also lead to loss of information and underestimation of epigenetic changes. However, we chose this approach because although it is less sensitive, we felt it provided us with global results, reduced the need to deal with probe variability and provided a good approximation of differentially methylated CpG islands. In the future it may make sense to refine both the measurement approach and data analysis to obtain more detailed results. The heterogeneity of our samples as well as the different methodologies used to identify the differences in CpG methylation could also explain the absence of PTGER2 and Thy-1 from our list of significantly methylated genes. The promoters of PTGER2[70] and Thy-1[71] were found to be hypermethylated in fibrotic lung fibroblasts and fibrotic tissue from IPF patients resulting in low levels of the coded proteins. In fact Thy-1 it is shown that the downregulation occurs only in areas of dense fibrosis and fibrotic foci while the rest of the tissue remains unaffected [70], [71]. However, the demonstration of significant global methylation changes despite the limitations of our methods, may be indicative of the importance of epigenomic regulation in IPF and lead to many more detailed discoveries and insights.
To the best of our knowledge our study is the first one to describe global DNA methylation changes in IPF lungs. Taken together with the extensive changes in gene histology, gene expression and microRNA profiles our results highlight the profundity and complexity of events underlying the phenotypic changes in IPF and to some extent suggest that interfering with one pathway may not be sufficient to reverse these changes. The differentially methylated CpG islands we identified should be further studied as their regulation could provide insights about how genotype and the environment interact to determine the lung phenotype in IPF. Based on our results, we believe that epigenetic modifications play a key role in the pathogenesis of IPF and thus could serve as disease biomarkers and therapeutic targets.
Supporting Information
The sequence of EpiTYPER MassArray primers.
(DOC)
Differentially methylated CpG islands distinguishing IPF from controls.
(DOC)
Functional Analysis of the 55 Differentialy methylated promoters in IPF.
(DOC)
Differentially methylated CpG islands overlapping between IPF and cancer.
(DOC)
Acknowledgments
The authors would like to thank the members of Dr. Kaminski’s Lab for their support and constructive criticism.
Footnotes
Competing Interests: The authors have read the journal’s policy and have the following conflicts: Dr. Yakhini is employed at Agilent Laboratories and his work on the manuscript was funded by Agilent. There are no patents, products in development or marketed products to declare. Dr Yakhini’s employment at Agilent had no impact on the design of the study and does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: NK, EIR, MK, KFG, KPV and GY were funded by NIH grants R01HL095397, R01LM009657, RC2HL101715 and the Dorothy P. and Richard P. Simmons Chair for Pulmonary Research. EIR was funded by American Lung Association Senior Research Training Fellowship Grand. The above funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS). Am J Respir Crit Care Med. 2000;161:646–664. doi: 10.1164/ajrccm.161.2.ats3-00. [DOI] [PubMed] [Google Scholar]
- 2.Gross TJ, Hunninghake GW. Idiopathic pulmonary fibrosis. N Engl J Med. 2001;345:517–525. doi: 10.1056/NEJMra003200. [DOI] [PubMed] [Google Scholar]
- 3.Katzenstein AL, Myers JL. Idiopathic pulmonary fibrosis: clinical relevance of pathologic classification. Am J Respir Crit Care Med. 1998;157:1301–1315. doi: 10.1164/ajrccm.157.4.9707039. [DOI] [PubMed] [Google Scholar]
- 4.Selman M, King TE, Pardo A. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med. 2001;134:136–151. doi: 10.7326/0003-4819-134-2-200101160-00015. [DOI] [PubMed] [Google Scholar]
- 5.Collard HR, King TE Treatment of idiopathic pulmonary fibrosis: the rise and fall of corticosteroids. Am J Med. 2001;110:326–328. doi: 10.1016/s0002-9343(01)00622-2. [DOI] [PubMed] [Google Scholar]
- 6.Taskar VS, Coultas DB. Is idiopathic pulmonary fibrosis an environmental disease? Proc Am Thorac Soc. 2006;3:293–298. doi: 10.1513/pats.200512-131TK. [DOI] [PubMed] [Google Scholar]
- 7.Taskar V, Coultas D. Exposures and idiopathic lung disease. Semin Respir Crit Care Med. 2008;29:670–679. doi: 10.1055/s-0028-1101277. [DOI] [PubMed] [Google Scholar]
- 8.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, et al. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol. 2010;298:H1235–1248. doi: 10.1152/ajpheart.00254.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, et al. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med. 2006;173:188–198. doi: 10.1164/rccm.200504-644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selman M, Carrillo G, Estrada A, Mejia M, Becerril C, et al. Accelerated variant of idiopathic pulmonary fibrosis: clinical behavior and gene expression pattern. PLoS One. 2007;2:e482. doi: 10.1371/journal.pone.0000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med. 2008;5:e93. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;180:167–175. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scotton CJ, Krupiczojc MA, Konigshoff M, Mercer PF, Lee YC, et al. Increased local expression of coagulation factor X contributes to the fibrotic response in human and murine lung injury. J Clin Invest. 2009;119:2550–2563. doi: 10.1172/JCI33288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thannickal VJ, Horowitz JC. Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proc Am Thorac Soc. 2006;3:350–356. doi: 10.1513/pats.200601-001TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fattman CL. Apoptosis in pulmonary fibrosis: too much or not enough? Antioxid Redox Signal. 2008;10:379–385. doi: 10.1089/ars.2007.1907. [DOI] [PubMed] [Google Scholar]
- 18.Hecker L, Vittal R, Jones T, Jagirdar R, Luckhardt TR, et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med. 2009;15:1077–1081. doi: 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 2006;103:13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis BC, Borok Z. TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol. 2007;293:L525–534. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 21.Kim KK, Wei Y, Szekeres C, Kugler MC, Wolters PJ, et al. Epithelial cell alpha3beta1 integrin links beta-catenin and Smad signaling to promote myofibroblast formation and pulmonary fibrosis. J Clin Invest. 2009;119:213–224. doi: 10.1172/JCI36940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med. 2008;5:e62. doi: 10.1371/journal.pmed.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vuga LJ, Ben-Yehudah A, Kovkarova-Naumovski E, Oriss T, Gibson KF, et al. WNT5A is a regulator of fibroblast proliferation and resistance to apoptosis. Am J Respir Cell Mol Biol. 2009;41:583–589. doi: 10.1165/rcmb.2008-0201OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz DA. Epigenetics and environmental lung disease. Proc Am Thorac. 2010;7(2):123–125. doi: 10.1513/pats.200908-084RM. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 26.Toyota M, Suzuki H, Yamashita T, Hirata K, Imai K, et al. Cancer epigenomics: implications of DNA methylation in personalized cancer therapy. Cancer Sci. 2009;100:787–791. doi: 10.1111/j.1349-7006.2009.01095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 2008;49:4–8. doi: 10.1002/em.20366. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 30.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 31.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 32.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 33.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome–components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 34.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009;19:181–187. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vancheri C, Failla M, Crimi N, Raghu G. Idiopathic pulmonary fibrosis: a disease with similarities and links to cancer biology. Eur Respir J. 2010;35:496–504. doi: 10.1183/09031936.00077309. [DOI] [PubMed] [Google Scholar]
- 38.Lababede O, Meziane MA, Rice TW. TNM staging of lung cancer: a quick reference chart. Chest. 1999;115:233–235. doi: 10.1378/chest.115.1.233. [DOI] [PubMed] [Google Scholar]
- 39.Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP). Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 40.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 41.Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita S, Hosoya K, Gyobu K, Takeshima H, Ushijima T. Development of a novel output value for quantitative assessment in methylated DNA immunoprecipitation-CpG island microarray analysis. DNA Res. 2009;16:275–286. doi: 10.1093/dnares/dsp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 47.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaenisch R, Bird A. Nat Genet 33 Suppl; 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. pp. 245–254. [DOI] [PubMed] [Google Scholar]
- 51.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klug M, Heinz S, Gebhard C, Schwarzfischer L, Krause SW, et al. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010;11:R63. doi: 10.1186/gb-2010-11-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi JK. Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol. 2010;11:R70. doi: 10.1186/gb-2010-11-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dudziec E, Miah S, Choudhry HM, Owen HC, Blizard S, et al. Hypermethylation of CpG Islands and Shores around Specific MicroRNAs and Mirtrons Is Associated with the Phenotype and Presence of Bladder Cancer. Clin Cancer Res. 2011;17:1287–1296. doi: 10.1158/1078-0432.CCR-10-2017. [DOI] [PubMed] [Google Scholar]
- 56.Kim JH, Dhanasekaran SM, Prensner JR, Cao X, Robinson D, et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 2011;21:1028–1041. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanjo H, Kawai T, Akira S. DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J Biol Chem. 1998;273:29066–29071. doi: 10.1074/jbc.273.44.29066. [DOI] [PubMed] [Google Scholar]
- 58.Pang M, Zhuang S. Histone deacetylase: a potential therapeutic target for fibrotic disorders. J Pharmacol Exp Ther. 2010;335:266–272. doi: 10.1124/jpet.110.168385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu T, Hu B, Choi YY, Chung M, Ullenbruch M, et al. Notch1 signaling in FIZZ1 induction of myofibroblast differentiation. Am J Pathol. 2009;174:1745–1755. doi: 10.2353/ajpath.2009.080618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boon K, Bailey NW, Yang J, Steel MP, Groshong S, et al. Molecular phenotypes distinguish patients with relatively stable from progressive idiopathic pulmonary fibrosis (IPF). PLoS One. 2009;4:e5134. doi: 10.1371/journal.pone.0005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keane MP, Belperio JA, Moore TA, Moore BB, Arenberg DA, et al. Neutralization of the CXC chemokine, macrophage inflammatory protein-2, attenuates bleomycin-induced pulmonary fibrosis. J Immunol. 1999;162:5511–5518. [PubMed] [Google Scholar]
- 62.Ozawa Y, Suda T, Naito T, Enomoto N, Hashimoto D, et al. Cumulative incidence of and predictive factors for lung cancer in IPF. Respirology. 2009;14:723–728. doi: 10.1111/j.1440-1843.2009.01547.x. [DOI] [PubMed] [Google Scholar]
- 63.Le Jeune I, Gribbin J, West J, Smith C, Cullinan P, et al. The incidence of cancer in patients with idiopathic pulmonary fibrosis and sarcoidosis in the UK. Respiratory medicine. 2007;101:2534–2540. doi: 10.1016/j.rmed.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 65.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 66.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 67.Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 69.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 70.Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, et al. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am J Pathol. 2010;177:2245–2255. doi: 10.2353/ajpath.2010.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequence of EpiTYPER MassArray primers.
(DOC)
Differentially methylated CpG islands distinguishing IPF from controls.
(DOC)
Functional Analysis of the 55 Differentialy methylated promoters in IPF.
(DOC)
Differentially methylated CpG islands overlapping between IPF and cancer.
(DOC)