Abstract
Drosophila responds to Gram-negative bacterial infection by activating the immune deficiency (IMD) pathway, leading to production of antimicrobial peptides (AMPs). As a receptor for the IMD pathway, peptidoglycan-recognition protein (PGRP), PGRP-LC is known to recognize and bind monomeric peptidoglycan (DAP-type PGN) through its PGRP ectodomain and in turn activate the IMD pathway. The questions remain how PGRP-LC is activated in response to pathogen infection to initiate the IMD signal transduction in Drosophila. Here we present evidence to show that proteases such as elastase and Mmp2 can also activate the IMD pathway but not the TOLL pathway. The elastase-dependent IMD activation requires the receptor PGRP-LC. Importantly, we find that live Salmonella/E. coli infection modulates PGRP-LC expression/receptor integrity and activates the IMD pathway while dead Salmonella/E. coli or protease-deficient E. coli do neither. Our results suggest an interesting possibility that Gram-negative pathogen infection may be partially monitored through the structural integrity of the receptor PGRP-LC via an infection-induced enzyme-based cleavage-mediated activation mechanism.
Key words: signal transduction, Drosophila innate immune activation, immune deficiency (IMD) signaling pathway, pgrp-lc, elastase, matrix metalloproteinase
Introduction
Drosophila melanogaster relies solely on innate immunity for defense against pathogen infection.1–3 The humoral immunity response of Drosophila is stimulated by the signals from the two evolutionarily conserved signal transduction pathways, TOLL and immune deficiency (IMD), which are differentially activated in response to distinct classes of pathogens.1 The TOLL pathway, which responds to either fungi or Gram-positive bacteria, is activated upon pattern recognition by extracellular signaling components that act upstream of TOLL such as PGRP-SA (Semmelweis), Gram-negative binding protein 1 (GNBP1), PGRP-SD, a serine protease (Persephone) and a serine protease inhibitor (Serpin).4–9 Through a mechanism that is yet to be fully understood, multiple serine proteases are activated post pattern recognition and multiple protease-dependent recognition cascades function upstream of TOLL and converge at the step of proteolytic cleavage of the TOLL ligand, Spätzle (Spz).10–13 The cleaved Spz then binds to TOLL and activates the downstream NFκB signaling pathway.14,15 Thus, the mechanism of TOLL activation sets a precedent for a protease-dependent activation mechanism of innate immunity in Drosophila.
Gram-negative bacterial infection, on the other hand, activates the Drosophila IMD pathway, leading to the production of AMPs like Diptericin and other immunity factors, which then neutralizes the invading pathogens (reviewed in ref. 1). Activation of the IMD pathway requires the type II transmembrane receptor PGRP-LC and its co-receptors.16–19 The PGRP-LC gene has three isoforms with identical intracellular and transmembrane domains but diverse extracellular PGRP domains (PGRP-LCa, -LCx and -LCy).16–18,20 It has been reported that PGRP-LC recognizes Gram-negative bacteria through binding of unique microbial pattern recognition molecules such as diaminopimelic acid peptidoglycan (DAP-PGN) with its ectodomain PGRP.21–23 The exact mechanism of PGRP-LC activation is not yet clear, although the injection of PAMPs such as Gram-negative PGN is known to activate the IMD pathway in vivo21–23; and the binding of DAP-PGN to PGRP-LC and dimerization of PGRP-LC are known to activate the IMD pathway in S2 cells.22–25 However, the questions of how these common PAMPs such as DAP-PGN are generated and whether these immune agonists are present in cell wall metabolites of commensal bacteria that live in the host animals remain to be determined. With limited innate immunity receptor diversity, how the pattern recognition receptors such as PGRPs recognize specific pathogens amidst millions of commensal microbes, and elicit an appropriate immune response is an intriguing question. The gap in our understanding of how innate immune system is activated in response to pathogen infection, tissue injury and inflammation suggest that our current model of PAMP recognition may admit expansion. To begin to address this question, we first ask whether an infection/injury-induced protease release plays a role in the activation of PGRP-LC similar to Spz/TOLL activation.
Activation of protease cascades commonly occurs in host defense, inflammation, tissue injury and wound repair in vertebrates and invertebrates.26–40 To ask whether protease release during host-pathogen antagonism may provide one of additional “danger” cues and signals for activating host immunity and for host cells to discriminate pathogens vs. nonpathogens,41 we examined whether proteases could activate the IMD pathway and whether live Gram-negative bacterial infection could modulate the receptor structural integrity of PGRP-LC. Here we report that the Drosophila IMD pathway can be activated by elastase and Mmp2 in vivo. We show that elastase, which induces IMD but not TOLL activation in flies, does cleave PGRP-LC in vitro. Moreover, cleavage intermediates of PGRP-LC can be detected in cultured Drosophila cells upon live Salmonella/E. coli infection whereas no cleavage of PGRP-LC occurs following challenge by 10-fold more dead or protease-deficient bacteria. Thus, we hypothesize that PGRP-LC may not only recognize exogenous microbial molecular patterns such as DAP-PGN through its extracellular PGRP domain, but also activate the IMD pathway in response to infection/injury-induced receptor cleavage/proteolysis during pathogen-host antagonism and tissue damage through the modulation of PGRP-LC structural integrity. Whether such a protease-cleavage dependent PGRP-LC activation mechanism is operating in response to Gram-negative bacterial infection in vivo remains to be further defined in Drosophila.
Materials and Methods
Reagents.
Ultrapure heparan sulfate (HS), hyaluronic acid (HA), chondroitin sulfate B (CS), and endotoxin removal resin (END-X B15) were purchased from Seikagaku (Falmouth, MA). Purified neutrophil elastase (24 U/ml) was purchased from Calbiochem (La Jolla, CA). All materials listed above were certified endotoxin-free or were treated with endotoxin removal resin and tested to be endotoxin-free by Limulus amebocyte lysate assay gel clot method (Seikagaku).42 Lipopolysaccharide (LPS) derived from Escherichia coli was purchased from Sigma-Aldrich (St. Louis, MO). Glutathione sepharose 4B beads, anti-FLAG-M5 and anti-Actin antibodies were purchased from Sigma-Aldrich and were used at 1:2000 dilutions for western blot analysis.
Administration of proteases and endogenous substances.
Drosophila wild-type flies (Oregon-R, OR) were maintained on standard cornmeal agar medium at 25°C. Genetic crosses were performed according to standard procedures. Investigators have for decades injected flies with test substances like PGN and LPS as there is no better way to deliver them in this “water-repellent” insect model system. The common practice of using an entomology needle to inject/puncture Drosophila with microorganisms/immune agonists for infection or agonist delivery actually elicits two distinct biological responses, i.e., tissue injury and pathogen infection. To avoid tissue injury, we chose to gently coat flies with protease/saccharides. This unconventional method was used due to the following considerations: (1) To avoid physical damage to the animals in our delivery method, we favor non-invasive methods of gentle coating or the UAS-GAL4 expression system. (2) For our experimental design, we strenuously tried to avoid mechanically inducing tissue damage. We used a gentle way of shaking and coating flies using the method as described below. Triplicate groups of young adult flies (33 females and 33 males) were treated with 1X PBS control, or 1X PBS containing elastase (3 Unit/ml), LPS (40 µg/ml), HS (5 mg/ml), HA (20 mg/ml) or CS (20 mg/ml) for 12 h. Large fly vials (50 ml volume) were coated with 200 µl of the sterile solutions listed above. The solution was shaken into tiny droplets (size: ∼1/20 of volume of a fly) that coated the inner surface of the vials. There is no chance that flies could drown in such a small amount of fluids. Anesthetized flies were added to the vials and gently rolled round for 20–30 sec in these small droplets of solutions to coat their bodies with the elastase and saccharide solutions. Flies recovered from the gentle coating technique immediately and shook off the majority of the coated solution excepting a small amount entrapped between the wing and body. After 20–30 min, all the solution evaporated and flies resumed their normal behaviors in the vials. The treated flies were then incubated in an Environmental Chamber with humidity (60%) and temperature (25°C) control overnight. (3) In addition to use this method of coating flies with proteases, we also examine whether expression of Drosophila matrix metalloproteinases (MMPs) in the fat body induces IMD activation by using conventional the GAL4-UAS gene delivery system.
Histology and immunohistochemistry.
Immunohistochemical staining was performed on formalin/DMSO-fixed and paraffin-embedded whole-mount Drosophila adults. Our fixation and embedding protocol was modified based on the standard histology protocols commonly used for processing human tumor tissues. Five micrometer-cut sections were deparaffinized, rehydrated in graded ethanol and stained with Hematoxylin and Eosin (H&E) and Gram-staining as described.43
Mmp transgenic flies.
Transgenic lines were generated carrying one of the two splicing forms (F1 or F2) of Mmp1 mutants that were predicted to be constitutively active forms of Mmp1 (UAS-Mmp1.F1C93A and UAS-Mmp1.F2C93A). The mutation C93A was engineered to be a constitutively active form of Mmp1. Cys93 of the pro-domain coordinates with the zinc atom at the active site; its substitution with Ala is expected to cause the pro-domain to be unable to inhibit the active site, eliminating the zymogen form of the enzyme and causing it to become constitutively active. UAS-Mmp1.F1, UAS-Mmp1.F2 and UAS-Mmp2 were previously reported in Page-McCaw et al.63 Two independent lines of each UAS-Mmp transgene inserted on either chromosome II or III were tested in this study. MMP was expressed in the female fat bodies using the fat body-specific driver, YP1.44,45 YP-1-Gal4-UAS-Mmp2 females are semi-lethal, infertile with significantly shortened life span, and genetic cross was set up en masse in order to obtain enough female offspring for northern blot analysis. Cg-GAL4 directs transcription of UAS-transgene in larval fat body, anterior lobe of the lymph gland and circulating hemocytes.46 Cg-GAL4-UAS-MMP animals were embryonic lethal while Cg-GAL4-UAS-Mmp1C93A animals were larval lethal and exhibited fat-body histolysis phenotypes.
RNA extraction and northern blot.
RNA extraction was performed as described.47 Activation of the innate immune response was assayed by determining the expression of Drosomycin, Diptericin, Cecropin A, Defensin and Attacin mRNA using total RNA extracted from mock-treated flies or flies treated with either E. coli, protease, LPS, HS, CS or HA. Ribosomal protein rp49 mRNA was used as an internal control to monitor RNA loading.
Bacterial infection in S2 cells.
The FLAG-tagged PGRP-LCa/x open-reading frames (ORFs) were cloned into the pMT-V5-His vector and transfected into S2 cells using Cellfectin (Invitrogen, San Diego, CA). Stable cell lines expressing PGRP-LC were cloned and the receptor expression was verified by western blot analysis. The PGRP-LC expression was induced by Cu2+ overnight, and then cells were washed three times using serum-free S2 cell medium. E. coli [XL1-Blue and BL21(DE3) (Stratagene)], Salmonella Typhimurium and Staphylococcus carnosus were grown in liquid media, and electro-competent bacterial stocks were prepared. Concentrated E. coli, Salmonella and Staph were fixed in 4% paraformaldehyde or 50–75%–100% EM grade ethanol overnight at 4°C. The fixed bacteria were washed six times in sterile PBS and tested for complete sterility on LB plates overnight at 37°C before adding to S2 cells for the subsequent infection experiments. To ensure that absolutely equal amounts of live and dead bacteria were added under the same experimental conditions, we normalized the amount of bacteria by bacterial mass (pellet size) after centrifugation and followed by the precise serial dilution to add exact and known amounts of bacteria into S2 cells in the well-defined infection ratios. Since we cannot quantify the amount of dead bacteria by performing colony formation assays, we used the dilution series starting from the absolute bacterial mass to quantify both the live and dead bacteria using the same dilution method. The absolute bacterial mass/pellet size gives us an accurate way to calculate the amount of live bacteria being added to the S2 cells. The absolute amount of live bacteria added was counted, confirmed and verified on LB plates. The concentration of our electrocompetent E. coli stock is 6 × 1010/ml and that of the Salmonella stock is 5 × 1010/ml. To ensure that each experiment has the same bacteria to S2 cell infection ratio, the volume of bacterial stock added to each 6-well plate of S2 cells corresponded to pre-set increments between 10,000 and 100 million bacteria. S2 cells were seeded at a uniform density of 2 million cells per 6-well plate, resulting in infection ratios of 50:1; 20:1, 10:1, 5:1, 2:1, 1:1, 1:10, 1:100 and 1:200 as shown in Figures 3 and 4. We avoided adding the freshly grown bacterial culture into the S2 cells since the LB medium contains yeast extract that has many potential immune agonists. We prepared electrocompetent bacterial cells so that the bacterial cells were washed excessively with a large amount of sterile water to rid them of as much impurity/bacterial metabolites/yeast contaminants as possible before adding them to infect S2 cells expressing PGRP-LC. Finally, we used 10–50 times more dead bacteria than live ones into the S2 cell culture to unambiguously demonstrate that dead bacteria did not induce any cleavage of PGRP-LC while live Gram-negative bacteria readily induced the receptor cleavage at the surface of the S2 cells. Thus, the stable S2 cell lines expressing the receptor PGRP-LC were treated with similarly washed wild type and protease-deficient E. coli, live and dead Salmonella, E. coli or Staph. Finally, the S2 cell medium contains potent antibiotics (Penicillin-Streptomycin) that should be sufficient to suppress significant live bacterial growth in the duration of our experiments (12–16 h). Post infection, the stable S2 cells expressing PGRP-LC were collected, lysed in 1X Beach buffer containing 1X Roche complete protease inhibitors. The PGRP-LC expression level was analyzed by western blot using the anti-FLAG-M5 antibody. The Actin expression was used as an internal loading control in the cell lysates.
Figure 3.
Loss of PGRP-LC integrity in response to live E. coli/Salmonella infection in S2 cells. (A) A schematic illustration of the full-length receptors, PGRP-LCa/x, is shown. Identical intracellular and transmembrane (TM) domains are depicted as gray boxes and divergent extracellular PGRPa/x domains are depicted as pink and blue boxes. The FLAG tag (black bar) is added to the intracellular N-terminus of PGRP-LCa/x. This intracellular FLAG-tagged PGRP-LCa/x should be protected from any extracellular cleavage events and thus facilitate the detection of any receptor cleavage intermediates in response to pathogen infection by the western blot analyses. (B) PGRP-LC is a membrane receptor that is prominently expressed on the cell surface. (Panels 1–3) Stable S2 cell lines expressing FLAG-tagged PGRP-LC were established under the control of the inducible metallothionein promoter. Immunofluorescent staining shows that PGRP-LC is expressed on the membrane surface (panel 1). DAP I and Phalloidin were used as controls to stain nuclei (panel 2) and F-actin (panel 3). (Panels 4–6) Human cancer cells expressing EGFP-tagged PGRP-LC were established under the control of the CMV promoter. Three representative confocal images of the transfected cells are shown. PGRP-LC is clearly a membrane protein with some vesicles, ER/Golgi staining in the cytoplasm. (C)–(H) The expression of intracellular FLAG-tagged PGRP-LCx/a in S2 cells in response to live E. coli/Salmonella/Staph infection was determined by western blot analysis. (C) Expression of PGRP-LCx/a was established in stable S2 cell lines (two black arrows). S2 cells were used as negative control. A minimal amount of anti-FLAG-M5 cross-reactivity was detected with bacterial proteins [E. coli/Salmonella/Staph/BL21(DE3)] used in this study. A nonspecific band that cross-reacted with the anti-FLAG-M5 mAb was occasionally detected in untreated S2 cells. No receptor cleavage was observed under sterile condition. (D) Dosage-dependent cleavage of PGRP-LCx by live E. coli was examined after the cells were inoculated overnight with increasing amounts of E. coli. At the infection ratio of 5:1 (bacteria to S2 cells), PGRP-LC is cleaved. (E) No PGRP-LCa cleavage was detected under similar and higher bacterial conditions upon Staph (Gram-positive bacteria) infection overnight. (F) and (G) In order to capture the receptor cleavage intermediates, the S2 cells expressing PGRP-LC were subjected to a short time course of E. coli or Salmonella infection at the infection ratio of 10:1 (bacteria to S2 cells) for 0–10 h. Some PGRP-LC cleavage intermediates were readily detected (as marked by the red arrows). Anti-FLAG-M5 mAb was used to detect PGRP-LCa/x expression and Actin was used as a loading control.
Figure 4.
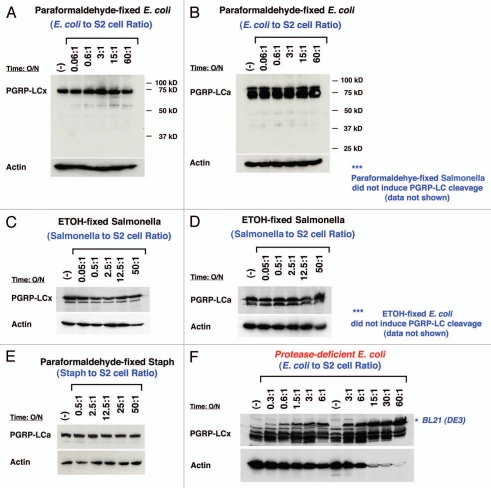
PGRP-LC expression is not affected by the presence of high concentration of dead bacteria or protease-deficient E. coli. The expression of FLAG-tagged PGRP-LCa/x in S2 cells in response to dead E. coli/Salmonella/Staph and protease-deficient BL21(DE3) infection was determined by western blot analysis. (A), (B), (C), (D) and (E) None of the structurally intact but dead bacteria (either paraformaldehyde-fixed or ethanol-fixed E. coli, Salmonella or Staph) triggered any PGRP-LC cleavage at very high infection ratio of 50:1 (10-fold higher concentration than live bacterial infection). (F) A protease-deficient E. coli strain, BL21(DE3), was used to infect S2 cells stably expressing FLAG-tagged PGRP-LCa/x proteins. No significant receptor cleavage was detected at a high infection ratio of 60:1. Anti-FLAG-M5 mAb was used to detect PGRP-LCa/x expression and Actin was used as a loading control.
Affinity purification of GST-PGRP-LCa/GST-PGRP-ECa fusion proteins and biochemical cleavage of PGRP-LC/PGRP-EC by elastase in vitro.
The full-length (FL) and extra-cellular (EC) fragments of PGRP-LCa were amplified using high-fidelity PCR. The following primers were used (restriction enzyme sites are underlined, start codon and stop codon are boldfaced). The PGRP-LCa-FL N-terminal forward primer (EcoRI): 5′-GGG AAT TCA TGC CTT TTA GCA ATG AAA CGG AAA TGA G-3′;
PGRP-LCa-EC N-terminal forward primer (EcoRI): 5′-CGG GAA TTC ATG ACA AAT CTC TTC GGA AAG ACG TTG-3′; PGRP-LCa-FL C-terminal reverse primer (Not1): 5′-CAA CGC CGG CGT CAC GAC CAA TGA GTC CAG TTG GC-3′. The PGRP-LC full-length and ectodomain fragments were cloned into the pGEX vector and sequenced to be error-free. The expression and affinity purification of these GST-PGRP-LC/GST-PGRP-EC fusion proteins were performed as previously described.48
Aliquots (20 µl of a 50% slurry) of the GSH-affinity-purified GST-PGRP-LCa-FL, GST-PGRP-LCa-EC and GST were treated with 1 µl of elastase (1 Unit/ml) at room temperature for 1–60 min and 1–6 h respectively. The protease cleavage reactions were stopped by adding an equal volume of 1X SDS loading buffer containing Roche complete protease inhibitors and freezing the samples immediately at −20°C. The protein samples were boiled at 100°C for 10 min and spun down at 4°C. The PGRP-LC cleavage products were separated by SDS-PAGE and gels were stained with Coomassie Blue.
Protease cleavage of PGRP-LC in S2 cells.
Two million of the stable S2 cells expressing PGRP-LC were seeded in 6-well plates for overnight, and were gently washed three times using serum-free S2 cell medium purchased from Gibco-BRL (Invitrogen) to rid of many protease inhibitors that are enriched in the fetal bovine serum (FBS) in the tissue culture medium. The S2 cells were treated with elastase (24U/ml) and thermolysin (1 µg/µl) isolated from Bacillus thermoproteolyticus rokko (Sigma-Aldrich, a thermophilic extracellular metalloproteinase) in 2 ml serum-free S2 cell medium for two hours. Post protease cleavage, the S2 cells were collected, lysed in 1X Beach buffer containing 1X Roche complete protease inhibitors. The PGRP-LC expression level was analyzed by western blot using the anti-FLAG-M5 antibody and Actin expression was used as an internal loading control.
Proteomic ID of PGRP-LC cleavage products.
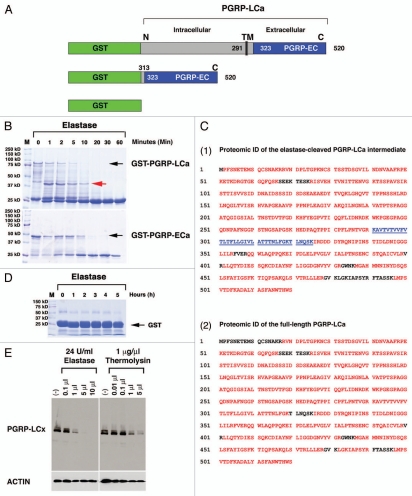
Both GST-PGRP-LCa-FL and GST-PGRP-LCa-EC could be rapidly cleaved by elastase in vitro (Fig. 5B). The elastase-cleaved GST-PGRP-LCa intermediate fragment marked by a red arrow was isolated and sent for proteomic identification (ID) at the Mayo Clinic Proteomics Core. In parallel, the full-length PGRP-LCa as marked by a black arrow was isolated and used as a control. Protein ID was made via in-gel trypsin digest and nano-LC-MS/MS with linear ion trap mass spectrometry.
Figure 5.
PGRP-LCa is an elastase substrate in vitro and a membrane-expressing PGRP-LCx can be cleaved by mammalian elastase and bacterial MMP in S2 cells. GSH-sepharose beads were used to affinity-purify full-length (FL) GST-tagged PGRP-LCa (named GST-PGRP-LCa), GST-tagged extracellular (EC) PGRPa domain (named GST-PGRP-LCa-EC) and GST proteins. GST protein is used as a negative control. (A) A schematic illustration of GST-tagged-PGRP-LCaFL/EC is shown. (B) The full-length GST-PGRP-LCaFL/EC fusion proteins are marked by black arrows. Both the full-length and extracellular fragment of PGRP-LCa can be readily cleaved by elastase in vitro. Equal aliquots (20 µl) of the GST-PGRP-LC fusion proteins were treated with 1 µl of elastase at different time points (1–60 min). The cleaved GST-PGRP-LCFL/EC intermediates were separated by SDS-PA GE and visualized by Coomassie Blue staining. A cleaved PGRP-LCa intermediate marked by the red arrow was sent for proteomic identification (ID) of the putative cleavage sites on PGRP-LC by elastase. (C) Proteomic ID results showed that the cleavage of PGRP-LCaFL/EC by elastase is rather nonspecific and elastase can completely cleave the extracellular PGRPa domain in vitro. (Panels 1 and 2) The proteomic identification results of PGRP-LCa cleavage intermediate as marked by the red arrow in Figure 5B (panel 1) and the full length PGRP-LCa (panel 2) are shown. The peptides identified and matched to the published PGRP-LCa amino acid sequence from the MS data are depicted in red. The near complete MS peptide coverage of the entire PGPLC-LCa coding sequence was detected in both the cleaved intermediate and the full-length receptor, indicating that the cleavage sites are rather nonspecific. (Panel 1) The transmembrane domain of PGRP-LCa that is located between 291–325 amino acids is not detected in the elastase-cleaved PGRP-LCa intermediate (marked by the underlined blue color). One possibility is that elastase may cut PGRP-LC near the TM domain between 291–325 Lysine (K) positions. Another possibility is that this hydrophobic TM peptide may be lost during sample preparation for MS ID in the elastase cleaved PGRP-LC intermediate. (D) GST protein was used as a negative control and treated with elastase for an extended period (6 h). No cleavage of GST protein was observed. (E) Stable S2 cell lines expressing intracellular FLAG-tagged PGRP-LCx were established under the control of the inducible metallothionein promoter. Dosage-dependent cleavage of PGRP-LCx by elastase (24 U/ml) or bacterial MMP, thermolysin (1 µg/ml) was examined after the cells were inoculated for 2 h with increasing amounts of proteases in serum-free medium under sterile conditions. The results showed that PGRP-LCx was readily accessible to elastase/thermolysin-mediated proteolysis. Anti-FLAG-M5 mAb was used to detect PGRP-LC expression and Actin was used as a loading control.
The Coomassie Blue stained SDS-PAGE gel bands are prepared for mass spectrometry analysis using the following procedures. Prior to trypsin digestion, the gel pieces were destained with 50% acetonitrile/50 mM Tris pH 8.1 until clear then reduced with 25 mM DTT/50 mM Tris pH 8.1 for 40 min at 55°C, followed by alkylation with 40 mM iodoacetamide/50 mM Tris pH 8.1 for 40 min at room temperature. Proteins are digested overnight with 30 µl (0.004 µg/µl) trypsin (Promega Corporation, Madison, WI) in 25 mM Tris pH 8.1/0.0001% Zwittergent 3–16 at 37°C. The peptides are extracted with 60 µl of 2% trifluoroacetic acid for 45 min followed by 80 µl of acetonitrile. The pooled extracts are concentrated to less than 5 µl on a SpeedVac spinning concentrator (Savant Instruments, Holbrook, NY), then brought up in 0.1% formic acid/0.05% trifluoroacetic acid for protein identification by nano-flow liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) analysis using a ThermoFinnigan LTQ Linear Ion Trap mass spectrometer (ThermoElectron, San Jose, CA) coupled with a Michrom Paradigm MS4 (Michrom BioResources Inc., Auburn, CA). The peptide mixture is loaded into an OPTI-PAK inline trap cartridge (Optimize Technologies Inc., Oregon City, OR), custom packed with Michrom Magic C8 media, then injected onto a 75 um × 10 cm ProteoPepII C18 PicoFrit nanoflow column (New Objective Inc., Woburn, MA) and eluted using a gradient of 0.1% formic acid/5% acetonitrile to 0.1% formic acid/50% acetonitrile in 50 min. The linear ion trap experiment was set as data dependent triple play consisting of a full scan for ions in mass range of 400–1400 m/z, triggering to 10 amu profile mode zoom scan then MS/MS mode on the full scan ions with intensities exceeding a preset threshold. The MS/MS raw data were converted to DTA files using ThermoFinnigan's Bioworks 3.1 and correlated with theoretical fragmentation patterns of tryptic peptide sequences from the Swissprot databases using and Mascot™1 (Matrix Sciences, London, UK) search algorithm running on a 10 node cluster. All searches were conducted with variable modifications allowing +16 for methione sulphoxide, and +57 for carboxamidomethyl-cysteines. The search was restricted to trypsin generated peptides allowing for 2 missed cleavages and was left open to all species. Peptide mass tolerances were ± 1.5 Daltons and fragment mass tolerance was set to ± 0.8 Daltons. Protein identifications are considered when Mascot searches give at least two consensus peptides with individual probability scores exceeding 95% and ranking in the top five of all the hits for their respective MS/MS spectra.49
Results
Activation of the IMD pathway by exogenous serine protease, elastase, in vivo.
The Drosophila genome contains 10 elastaselike serine proteinases, some of which are strongly upregulated in response to immune challenge.50 To test if elastase, a model serine protease released by mammalian neutrophils, could elicit IMD activation in Drosophila, wild-type OR flies were gently coated with exogenous elastase free of any bacterial cell wall contaminants.42 This method was used to avoid puncture or physical wounding to the animals during the delivery of this enzyme. As shown by the northern blot of AMP production, wild-type flies treated with elastase produced high levels of Diptericin mRNA, but not Drosomycin mRNA, indicating the IMD pathway was activated by elastase while TOLL was not (Fig. 1A). This response appeared to be mediated by the enzymatic activity of elastase (not by any contaminating microbial substances in the elastase solution), as the IMD activation did not occur in wild-type flies treated with heat-inactivated elastase (Fig. 1B). Together, these observations suggest that the elastase treatment can specifically activate the IMD pathway but not the TOLL pathway.
Figure 1.
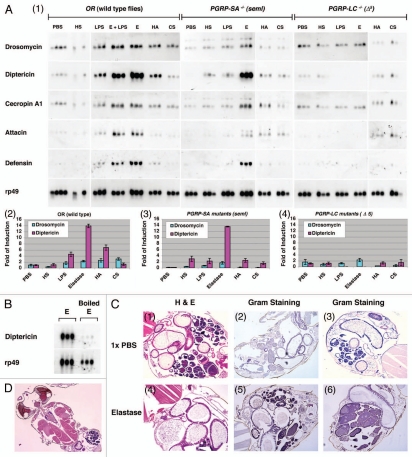
Elastase activates the IMD pathway through the receptor PGRP-LC. (A) Expression of antimicrobial peptides (AMP) was examined in triplicate in wild-type OR flies and mutant flies, PGRP-LC (Δ5 null) and PGRP-SA (seml), after treatment with sterile 1X PBS, elastase (3 Unit/ml), LPS (40 µg/ml), heparan sulfate (HS, 5 mg/ml), hyaluronic acid (HA , 20 mg/ml) or chondroitin sulfate (CS, 20 mg/ml) overnight. Northern blot analysis was performed using 50 µg total RNA extracted from the treated flies, and the blots were hybridized with 32P-labeled cDNA probes to antimicrobial peptides including Drosomycin, Diptericin, Cecropin A1, Attacin, and Defensin. rp49 mRNA was used as a control. No bacteria were added in these experiments to induce IMD activation. Film exposure is in the linear range. AMP production was examined by northern blots (panel 1). The relative levels of AMP expression, Diptericin/rp49 and Drosomycin/rp49, are shown in bar graphs in wild-type and mutant flies (panels 2, 3 and 4). Note that elastase was sufficient to induce robust IMD activation, whereas the saccharides did not. (B) AMP production in OR flies treated with elastase (3 U/ml) and heat-inactivated elastase (boiled at 100°C for 5 min) was examined. Note that the loss of elastase enzymatic activity is associated with loss of IMD activation. (C) PBS- and elastase-treated flies were embedded in paraffin blots. The intestinal morphologies of 1X PBS- and elastase-coated adult flies are shown. Hematoxylin and Eosin staining (H&E) is used to view adult abdominal morphology/cellular histology using serial paraffin sections (panels 1 and 4). Gram staining was used to detect for the loss of structural integrity of elastase-treated flies and the presence of commensal bacteria in internal organs post elastase treatment (panels 2, 3, 5 and 6). Note that there is no bacterial presence outside of the intestines of elastase-treated flies when compared with PBS-treated flies, demonstrating that the intestinal integrity remains intact at the microscopic level and no commensal bacteria leaked into the internal organs upon elastase treatment. (D) Section shows the general morphology of adult retina, brain, heart and muscles, demonstrating that paraffin section can be easily adapted for Drosophila research of host-pathogen interaction.
It is known that septic injury and tissue damage can lead to TOLL activation.1,10,51 Since it is well known that serine proteases are necessary for Spz cleavage and the proteolytic cleaved Spz activates the TOLL pathway in Drosophila,1,14,52 the observation that the TOLL pathway is not affected by elastase, a serine protease, is intriguing, suggesting that distinct serine proteinases may be specifically required for either TOLL or IMD activation in Drosophila. The elastase-dependent IMD activation was highly specific. First, the fact that TOLL pathway was not activated in these elastase-treated flies supported the conclusion that the gentle coating technique used has not disrupted the general integrity of the cuticle structure and led to global tissue damage that would result in TOLL activation. Second, no microscopic melanization was observed in elastase-treated flies further supported this conclusion since the known tissue damage will activate the melanization cascades. Third, to ensure that the elastase treatment did not activate the IMD pathway indirectly by causing tissue damage that results in the leakage of commensal bacteria into the internal organs and subsequent secondary infection in the fly bodies, we embedded 1X PBS buffer- and elastase-treated adult flies in paraffin blots and performed H&E and Gram staining. The serial section and histology study confirmed that the major organs were intact and no commensal bacteria had leaked into the fly internal organs post elastase treatment under high and low magnification (Fig. 1C and D, data not shown). Lastly, the fly intestines were not damaged by elastase and no commensal bacteria were detected in the sterile interiors and internal organs of the animals (Fig. 1C, panels 5 and 6) when compared with the controls (Fig. 1C, panels 2 and 3). TOLL pathway is not activated in these elastase-treated flies clearly indicated that the “tissue damage” induced by this serine protease is not sufficient to activate the TOLL pathway. Based on the evidence, we conclude that elastase treatment is rather specific in activating the IMD pathway.
In mammalian systems, elastase is known to release TLR4 agonists42,53 and therefore, it is possible that the elastase treatment activates IMD pathway by releasing conserved sacharides such as the heparan sulfate (HS), hyaluronic acid (HA) or chondroitin sulfate (CS). These endogenous saccharides are the core and conserved components of extracelular matrix (ECM), they are ubiquitously present on the surface of all mammalian and insect cells, they decorate cell surface proteins, and are required for modulating many membrane receptor signaling pathways during development.54–56 To test this, flies were coated with endogenous saccharides (HS, HA and CS) and an exogenous microbial saccharides, lipopolysaccharide (LPS) and AMP production was analyzed by northern blot. In contrast to treatment with elastase, none of the saccharides tested induced any substantial activation of either the IMD or TOLL pathway (Fig. 1A). Thus, while the protease activity of elastase appears to be required for IMD activation, these known agonists of mammalian TLR4 do not appear to activate the IMD pathway by gentle coating without needle puncturing. These results suggest that elastase treatment may activate the Drosophila IMD pathway through a different mechanism from that described for mice and humans.42,57,58
Elastase-mediated IMD activation requires PGRP-LC.
Members of the PGRP family detect different types of microbial invasion and activate the appropriate immune signaling pathways in Drosophila.1,59,60 To determine whether any of the infection-sensing PGRP receptors were required for elastasedependent Diptericin mRNA induction, we tested mutant flies with complete loss-of-function mutations in PGRP-LC-/- (Δ5) and PGRP-SA-/- (seml) for responses to IMD agonists. Similar to wild-types flies (OR), elastase treatment of seml mutant flies activated the IMD pathway, as indicated by expression of Diptericin, Attacin and Cecropin A mRNA (Fig. 1A, panels 1, 2 and 3). In contrast, elastase-mediated activation of the IMD pathway did not occur in PGRP-LC-/- (Δ5) mutant flies (Fig. 1A, panels 1 and 4). Similar to the OR flies, HS, HA, CS saccharide treatment did not activate either the TOLL or IMD pathway in these mutant flies, suggesting that these saccharide substances do not by themselves elicit an immune response by gentle coating the surface of adult flies with no puncturing. Collectively, these results show that activation of the IMD pathway by elastase requires functional PGRP-LC. It is conceivable that elastase may act directly or indirectly on this receptor.
Activation of the IMD pathway by matrix metalloproteinase (Mmp2).
As many serine proteases have been implicated in fly immunity (TOLL activation and melanization), we asked whether the observed IMD activation was specific to elastase or reflected a broader mechanism of specific protease-mediated activation of the IMD pathway. To address this question, we ectopically expressed matrix metalloproteinases (MMPs), which are known to induce the breakdown of extracellular matrix and induce tissue damage as well as elicit host immune responses.61,62 We employed the GAL-UAS system to express MMP in fat bodies, a major organ responsible for Drosophila humoral immunity, and we then measured AMP production in transgenic flies expressing MMP in the internal organs.
Drosophila has two MMP genes: Mmp1 and Mmp2.63 Mmp2 is a membrane-associated MMP with a glycosylphosphatidylinositol (GPI) anchor while Mmp1 is a secreted MMP.64 To evaluate the potential function of these proteases in IMD activation, we expressed Mmp1, Mmp1C93A (a constitutively-active mutant), or Mmp2 in adult female fat bodies using the yolk protein 1 promoter (YP1-GAL4)15,44,45 and then determined the immune responses in the transheterozygous females. We chose the fat body-specific driver (YP1-GAL4) because fat bodies are a major organ responsible for Drosophila humoral immunity. In addition, we reasoned that the expression of MMP in an internal environment (away from the Drosophila external cuticle and gut/intestinal system) should minimize the complication of secondary infection by external or gut commensal bacteria.
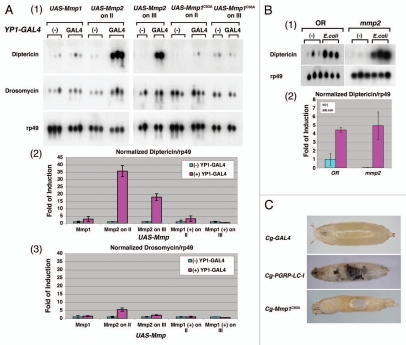
Ectopic expression of Mmp2 in the resultant transgenic flies led to marked activation of the IMD pathway while ectopic expression of either Mmp1 or activated Mmp1C93A did not (Fig. 2A, panels 1, 2 and 3). To ensure that Mmp1 and Mmp1C93A is expressed and active in these transgenic flies, MMP was expressed under the control of Cg-GAL4, which directs transcription in the larval fat body, the anterior lobe of the lymph gland and the circulating hemocytes.46 As shown in Figure 2C, Mmp1C93A is clearly an active and functional MMP whose expression under the control of Cg-GAL4 leads to severe internal tissue damage and histolysis (Fig. 2C). The Cg-GAL4-UAS-Mmp1 and Cg-GAL4-UAS-Mmp2 animals are embryonic lethal. The membrane-spanning and ectodomain-deleted PGRP-LC-I fragment was used as a positive control to show the expression of the active receptor under Cg-GAL4 led to massive melanization phenotypes.43 Interestingly, no melanization phenotype was observed in these Cg-GAL4-UAS-Mmp1C93A animals with massive internal tissue injury (Fig. 2C). These results suggest that internal tissue damage per se may not necessarily lead to melanization. External puncture/tissue damage is known to activate melanization cascades in Drosophila.34,65,66 The finding that the melanization cascades are not activated in response to internal tissue damage by Mmp1C93A is intriguing. The question of whether external and internal tissue damages are perceived differently in Drosophila remains to be further defined.
Figure 2.
Expression of Drosophila matrix metalloproteinase, Mmp2, in fat bodies is specific in activating the IMD pathway in vivo. (A) Expression of Mmp2 in female adult fat bodies (YP1-GAL4) resulted in a marked increase in Diptericin mRNA production in the absence of bacterial infection. Northern blot was performed as described above. Two independent transgenic lines carrying Mmp1, Mmp1C93A or Mmp2 transgene inserted on either chromosome II or III were tested. No bacteria were used in these experiments to induce IMD activation. AMP production was examined by northern blots (panel 1). The relative levels of AMP expression, Diptericin/rp49 and Drosomycin/rp49, are quantified by bar graphs in MMP-expressing flies (panels 2 and 3). Note that Mmp2 expression induced robust IMD activation whereas the Mmp1 did not. (B) Mmp2 mutant larvae have a normal IMD response in response to E. coli infection. Two hundred wild-type and Mmp2 mutant larvae at the first and second instar stage were treated with or without E. coli for six hours. The northern analysis was performed as described (panel 1). The relative levels of AMP expression, Diptericin/rp49, in response to infection are quantified by bar graph. (C) Expression of Mmp1C93A under the control of Cg-GAL4 resulted in tissue histolysis (holes), internal tissue damage and subsequent lethality, suggesting that Mmp1C93A is an active MMP. However, no melanization reaction was observed. The expression of extracellular PGRP domain-deleted receptor, PGRP-LC-I, is known to drive melanization reaction in the absence of an infection, and it was used as a control.
Since the ectopic expression of Mmp2 can activate the IMD pathway, it is possible that Mmp2 is a bona fide signaling component of the IMD pathway that acts upstream of PGRP-LC. To test this idea, we examined the IMD activation in null mmp2 mutant larvae treated with or without E. coli. The mmp2 mutant larvae mounted as robust an immune response against E. coli as the wild-type larvae, indicating that Mmp2 is not a bona fide signaling component required for the IMD activation (Fig. 2B, panels 1 and 2). However, the result does not exclude the possibility that Mmp2 functions in a genetically redundant fashion, directly or indirectly, in the proteolytic activation of other yet-to-be-identified upstream signaling components in the PGRP-LC pathway.
Since the YP1-GAL4-UAS-Mmp2 transgenic flies are infertile, semi-lethal, have significantly shortened life span and die after a few days post eclosion at 18–20°C (pupal lethal at 25°C), it precludes our ability to experimentally examine the genetic epistasis between Mmp2 and PGRP-LC. Nevertheless, the results show that expression of Mmp2 (but not Mmp1), a different class of protease from elastase with distinct enzymatic mechanism and substrate specificity, activates the IMD pathway in a manner similar to that of elastase. Thus, the IMD pathway can be activated by two proteases, elastase and Mmp2, in Drosophila.
PGRP-LC can be cleaved in response to live Salmonella/E. coli infection in S2 cells.
We have thus far provided evidences showing that the elastase-dependent IMD activation requires a functional receptor, PGRP-LC. These observations raise an interesting possibility that PGRP-LC may be a substrate of a yet-to-be-identified protease(s) released during active pathogenic infection. To determine whether the infection-mediated and protease-mediated IMD activation are linked events, we examined whether live bacterial infection activates the IMD pathway by cleaving PGRP-LC in the extracellular domain. To address this question, we tested whether exposure to Gram-negative bacterial infection modulates PGRP-LC expression in Drosophila S2 cells. A FLAG tag was added to the intracellular N-terminus of PGRP-LCa/x (Fig. 3A). We designed the reporter construct so that the FLAG-tagged intracellular domain of PGRP-LC should be protected from any protease-dependent cleavage event extracellularly in response to pathogen infection and we hope to use this construct to capture cleavage intermediates of the receptor. Drosophila S2 cells stably expressing one of the two full-length receptors—PGRP-LCx and PGRP-LCa—under the control of an inducible metallothionein promoter was generated. Immunochemical staining revealed that the subcellular localization of both PGRP-LC receptors was predominantly at the membrane, consistent with the published results (Fig. 3B; data not shown; ref. 25). Importantly, the induced PGRP-LC expression was stably maintained in serum-free medium, and no receptor cleavage was detected by western blots in the absence of live pathogenic bacterial infection (Fig. 3C).
Having established PGRP-LC-expressing stable S2 cell lines, we next analyzed the effects of live pathogen infection on the expression and structural integrity of PGRP-LC. The PGRP-LC expressing cells were treated with increasing amounts of live E. coli, Salmonella Typhimurium or Staphylococcus carnosus and alterations in PGRP-LC expression were examined by western blot analysis. As a control, minimal cross-reactivity was detected between bacterial proteins and the anti-FLAG-M5 antibody used (Fig. 3C). To infect these S2 cells expressing the PGRP-LC with live bacteria, we did a serial dilution that exposed the S2 cells with increased amount of bacteria until the infection ratio reached 1:10 ratio (S2 cells: bacterial cells). Interestingly, we found that PGRP-LCx/a expression disappeared following infection by live E. coli and Salmonella (Gram-negative) at ratio of 1:5 for overnight, while Gram-positive Staphylococcus infection did not result in any changes in PGRP-LC expression using the same infection ratio (Fig. 3D and E). Importantly, increasing amounts of a possible cleavage intermediate of N-terminal FLAG-tagged PGRP-LC (marked by the red arrows) were detected concomitant with the decreasing amounts of the full-length receptor during live E. coli or Salmonella infection at an infection ratio of 1:10 (Fig. 3F and G). Our results suggest that the receptor PGRP-LC might be degraded or cleaved in response to the presence of live Salmonella/E. coli infection in S2 cells.
Protease-deficient E. coli or dead bacteria do not alter PGRP-LC expression in S2 cells.
To determine whether this cleavage of the receptor PGRP-LC during live infection specifically reflects the action of infection-induced virulence factor or bacterial digestive enzymes for invasion, or can be achieved by the recognition of bacterial surface antigens, we asked whether the structurally intact but dead bacteria would also induce similar PGRP-LC cleavage/degradation in Drosophila S2 cells. To minimize the possibility that a specific bacterial antigen could be destroyed during the specimen processing, two independent chemical fixatives (4% paraformaldehyde and 100% ethanol) were used to preserve the structural integrity and surface patterns of the bacterial cells following the electron microscope (EM) procedure. We chose not to use the heat-inactivating method to avoid any structural and integrity damages to bacterial cells. The results showed that treating the PGRP-LC-expressing S2 cells with chemically-fixed and thus dead E. coli/Salmonella, or Staphylococcus at 50-fold or 10-fold higher concentrations than the live bacteria did alter neither PGRP-LCx nor PGRP-LCa expression or receptor integrity (Figs. 4A–E and data not shown), suggesting that live Gram-negative bacterial infection can induce the PGRP-LC cleavage while dead but structurally intact bacteria cannot do so.
As the accumulated evidence suggests that proteolysis may play a role in the IMD activation, we reasoned that the factor required for bacteria to elicit PGRP-LC degradation might be a protease. To test this idea, we treated the stable S2 cells expressing PGRP-LC with live BL21(DE3), a protease-deficient E. coli strain with a mutated outer membrane serine protease (OmpT). OmpT is a surface protease that is responsible for conferring resistance to antimicrobial peptides and is known to be physiologically relevant for the virulence of Yersinia pestis and clinical E. coli isolates.67–73 The result showed that massive BL21(DE3) infection was indeed defective in inducing modulation of PGRP-LC expression when compared with wild-type live Salmonella/E. coli infection (Fig. 4F). Thus, live E. coli/Salmonella infection (at an infection ratio of 1:5) induced rapid PGRP-LC cleavage/degradation while none of the dead bacteria (at an infection ratio of 1:50), live protease-deficient E. coli (at an infection ratio of 1:60) or live Gram (+) bacteria, Staphylococcus, (at an infection ratio of 1:5) modulated PGRP-LC receptor intergrity in S2 cells nor did they induce IMD activation in vivo (Figs. 3 and 4; ref. 43]. These results suggest an interesting possibility that active infection and specific pathogen-host antagonism may secret a virulence factor(s) that modulates structural integrity of PGRP-LC in the presence of pathogenic bacteria. They may also point to a potential role for the bacterial surface serine protease OmpT in triggering PGRP-LC signaling during Gram-negative bacterial infection in vivo. The role of the bacterial OmpT in the IMD activation will be elucidated in the transgenic fly models in the future.
PGRP-LC can be cleaved by elastase in vitro and in vivo.
PGRP-LC cleavage/degradation during a live bacterial infection raises the question of whether PGRP-LC can be cleaved directly by protease release induced by active infection. Since the elastase-induced IMD activation depends on a functional PGRP-LC, we examined whether PGRP-LC could be an elastase substrate in vitro. The full-length (FL) and extracellular (EC) fragments of PGRP-LCa were GST-tagged and affinity purified (Fig. 5A). The two GST-PGRP-LCFL/EC fusion proteins were treated with elastase, a model serine protease. Following a similar time course, both the full-length and ectodomain of PGRP-LC were rapidly cleaved by elastase in 10–20 min in vitro (Fig. 5B). PGRP-LCEC was completely degraded by elastase while a LC intermediate product was detected in PGRP-LCFL cleavage (marked by the red arrow) (Fig. 5B). Comparative proteomic analysis of the elastase-cleaved LC intermediate and the full-length PGRP-LCa indicated that the elastase-mediated proteolytic cleavage of PGRP-LC was rather nonspecific, because a near complete MS peptide coverage was observed from the LC intermediate spanning the entire PGRP-LC coding region (Fig. 5C). Affinity-purified GST protein, used as a negative control, was not cleaved by elastase after a 6-h prolonged incubation (Fig. 5D). Thus, these results show that PGRP-LCFL/EC can be rapidly cleaved by elastase within 10 min in vitro, suggesting an intriguing possibility that the extra-cellular fragments of PGRP-LC may be readily cleaved by certain pathogen/injury-induced digestive enzymes while its intracellular domain may be protected by plasma membrane to signal the presence of active pathogen infection and invasion to the host cells and enable them to mount an effective innate immune and host defense response.
We next examined whether PGRP-LC stably expressed on the surface of Drosophila S2 cells could be similarly cleaved by elastase and active MMP, both of which trigger the IMD activation in flies in vivo (Figs. 1 and 2). Elastase and thermolysin (bacterial MMP) were added to S2 cells stably expressing PGRP-LCx in serum-free medium under sterile condition and the PGRP-LC expression and integrity was monitored by western blot. In agreement with the in vitro biochemical cleavage results with GST-tagged PGRP-LCaFL/EC (Fig. 5B), PGRP-LCx expressed in the S2 cells was readily cleaved by elastase or thermolysin (Fig. 5E). Thus, these results provide additional evidence to support that PGRP-LC may be a bona fide elastase substrate in vitro and in S2 cells. Nevertheless, the endogenous proteases that could cleave PGRP-LC in vivo in response to live Gram-negative infection remain to be identified.
Discussion
While the activation of serine protease cascades and Spz proteolysis are known to play a pivotal role in TOLL activation in Drosophila,11–13,74 the involvement of proteases in PGRP-LC/IMD activation, the other major signaling pathway of Drosophila humoral immunity, has not been reported. Here we provide first evidence to show that proteases such as elastase and Mmp2 can specifically activate the Drosophila IMD pathway in vivo. Moreover, the elastase-induced activation of the IMD pathway requires a functional PGRP-LC and the extent of PGRP-LC proteolysis appears to be correlated with live bacterial infection. Taken together, these data suggest that, similar to the TOLL pathway, protease activity may play a role in the IMD activation.
Even though the physiological relevance of this proteasedependent IMD activation remain to be elucidated, it nevertheless seems possible that these proteases may mimic some aspects of pathogenic bacterial infection, e.g., the activity of yet-to-be-identified proteases perhaps emanating from invading microbes as well as activated host immune cells during pathogen-host antagonism. Indeed, several evidences suggest that this may be the case. First of all, infection of Drosophila S2 cells or transgenic animals with live Salmonella/E. coli, but not dead Salmonella/E. coli at a 10-fold higher concentration, results in the cleavage of the receptor PGRP-LC. Second, the receptor PGRP-LC can be directly cleaved by elastase in vitro and S2 cells, suggesting that elastase may mimic the action of an infection-induced virulent factor(s) that lead to the cleavage of the receptor PGRP-LC and in turn activate the IMD pathway. Third, an ectodomain (PGRP)-deleted transmembrane receptor that is common among all three PGRP-LC isoforms, PGRP-LC-I, when overexpressed, function as a constitutively active receptor in the absence of pathogen infection.43 Together, these observations suggest that a bacterial protease may trigger the cleavage of the extracellular (PGRP) domain of PGRP-LC, rendering the receptor active to activate the downstream IMD pathway to combat the invading Gram-negative bacteria and repair the invasion-dependent tissue damage.
It has been reported that extracellular serine proteases can activate mammalian MMPs.75 It is also known that an exogenous fungal protease PR1 can cleave an endogenous Persephone protease that in turn activates TOLL pathway, suggesting that host immune sensor can be modulated by virulence factors released during pathogen-host interaction.13 It remains to be determined whether infection/injury-induced proteases are released during pathogen-host antagonism, what the identities of these proteases are and where they come from, whether they cleave PGRP-LC extracellularly and whether a damaged receptor PGRP-LC sends a “danger” signal to the host cells to activate the host defense responses in vivo. Despite the limitations in our experiments, our data have provided important cues in explaining how innate immune pathways are activated in response to pathogen infection in vivo. Thus, we would like to offer the following hypothesis.
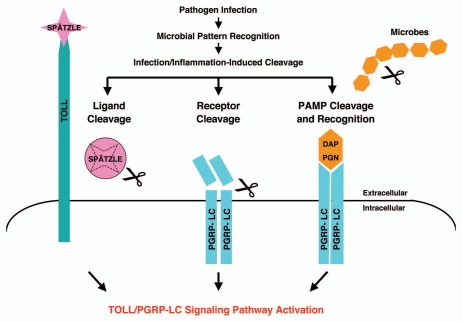
We speculate that the modulation of host innate immunity receptors/sensors may be a second-phase interaction for host cells to differentiate pathogenic from non-pathogenic microbes after initial-phase of PAMP recognition and host-microbe interaction. The infection-dependent modulation of host surveillance receptors/sensors may lead to irreversible activation of innate immunity pathways and in turn allow for propagation of a localized host response against the invading pathogens. Thus, pathogenic microbes may engage host cells, release virulence factors, digestive enzymes/proteases for invasion and infection which could in turn cleave the innate immunity receptors/sensors while nonpathogenic microbes will engage host cells without stimulating the release of any harmful substances that may compromise the integrity of the host sentinel receptors/sensors. A working model is proposed in which PGRP-LC acts as a sentinel receptor for the IMD pathway that can be activated by PAMP recognition via DAP-PGN binding as well as a proteolysis-dependent mechanism (Fig. 6). We hypothesize that the structural integrity of PGRP-LC may thereby constitute a tissue well-being signal and the cleavage of the PGRP ectodomain may signal the onset of pathogenic infection and tissue damage.
Figure 6.
A model summarizing the infection-induced protease-dependent cleavage of innate immunity sensors/receptors in response to pathogenic infection and tissue damage in Drosophila A schematic illustration of the protease-dependent activation of Drosophila IMD and TOLL pathways is shown. It is well established that the TOLL ligand Spätzle is processed by an infection-activated serine protease cascade and that the cleaved Spätzle binds to TOLL and thereby activates the TOLL pathway.1 The receptor PGRP-LC can be activated by binding to bacterial elicitors (monomeric or polymeric DAP -PGN).1,21,23 To complement the well-established mechanism of innate immune activation via microbial pattern recognition (PAMP), we hypothesize that PGRP-LC may also be cleaved by infection-induced proteases released during pathogen-host antagonism. The structural integrity of the sentinel receptor PGRP-LC may constitute a “tissue well-being” signal. The infection-induced protease release may be a “danger/damage” signal that may help the host cells to detect pathogen infection and tissue injury.
PGRP-LC is known to recognize and bind monomeric peptidoglycan (DAP-type PGN) through its PGRP ectodomain and in turn to activate the IMD pathway.21–23 These elegant studies clearly demonstrate the PGRP-LC is a pattern recognition receptor that can be activated by specific pathogen-associated molecular pattern like DAP-PGN. Our infection-induced and protease-mediated IMD activation model complements and expands the well-established DAP-PGN pattern recognition IMD activation model by explaining how innate immune responses might be similarly activated in response to infection, inflammation and tissue injury (Fig. 6 and refs. 22 and 23). It points to an interesting and rather simple possibility of the involvement of proteases, virulence factors as well as some other specific “danger/damage” signals for the host cells to recognize pathogenic microbes and mount effective immune responses in addition to pattern recognition.
The new model is attractive because mammalian innate immunity and pattern recognition receptors lack the receptor diversity to match the remarkable diversity of potential pathogens.76–80 By detecting the infection-induced “loss of well-being” signal through the monitoring of structural integrity of a small number of innate immunity receptors (PGRP-LC in the IMD pathway) and ligands (Spz in the TOLL pathway), it can send an unambiguous, irreversible and highly effective “danger/damage” signal to the host cells in response to infection and tissue damage by a diverse array of pathogenic microbes. A locally activated proteolytic cascade initiating from a small number of host innate immune receptors/sensors would allow for selective activation of specific innate defense pathways, and confinement of host defense to specific sites of infection/injury to initiate local tissue repair and fight off infection in a temporally and spatially controlled manner. In addition, downregulation of the innate immune receptors during active infection might provide a regulatory feedback mechanism for host cells to fine tune their innate immune responses and mount an optimal host defense against the invading microorganisms. An irreversible cleavage of the sentinel receptors would also preclude a microorganism's ability to evade innate immune pattern recognition receptors through adaptive mutations.
In addition to these unique features listed above, we believe this working model has two additional merits. One, it helps to explain how inflammatory pathways contribute to tissue regeneration and repair in wound-healing in the absence of pathogen infiltration. Second, the model helps to explain how a small number of sentinel receptors like PGRP-LC can be so effective in activating the host defense system to combat specific pathogenic microbes while remaining unaffected by a diverse array of commensal microbes continually present in the environment. There are trillions of commensal microorganisms (1014 microbes) co-existing peacefully with us in complex ecosystem; in a healthy individual, commensal microbes outnumber host cells by a ratio of up to 20:1.81 Drosophila hosts millions of resident microbes without eliciting TOLL and IMD activation in healthy animals raised in normal laboratory environment while certain commensal bacteria could become pathogenic to flies raised in germ-free environment.82 How Drosophila controls its resident microbiota communities and how Drosophila distinguishes pathogenic microbial infection in the midst of commensal microbes remain an interesting but unanswered question.83,84 Since many exogenous and endogenous immune agonists are known to be cleaved products/shed substances from pathogenic microbes or host cells, it is likely that infection/injury-induced specific digestive enzymes/proteases may play a role in generating the PAMPs as the immune agonists, modulate immune receptors/sensors and in turn play a role in activating the innate immunity pathways. During the continual cell cycles of our symbiotes, commensal bacterial cell wall metabolites/byproducts, many of which are similar to pathogenic PAMPs, are being constantly shed but are also actively monitored and recognized by the host immune cells.85–87 Yet the common microbial associated patterns that are shed by commensal bacteria very rarely elicit any host immune responses in healthy animals (except for animals raised in germ-free environment). And though exactly what enables a host cell to differentiate between a pathogenic vs. a nonpathogenic microbe remains an unanswered question in biology. It is extremely likely that some other mechanisms in addition to the pattern recognition strategy may exist.41,87–90
Our understanding of innate immunity has advanced with the realization that innate immunity receptors/sensors are able to recognize both exogenous and endogenous signals; for example, mammalian TLRs, once thought to recognize only exogenous substances, have recently been shown to also have endogenous TLR agonists.77,80,88,89,91,92 How endogenous and exogenous immune agonists/stimuli are generated during infection is unclear but based on the size and nature of these cleaved products, an infection- and inflammation-dependent enzyme-mediated cleavage event for the production of these substances may be inferred. Since bacterial cell wall components are cleaved by digestive enzymes during infection to generate PAMPs, it is conceivable that host innate immunity receptors/sensors on the cell surface may be similarly exposed to the proteolytic events during pathogen-host warfare.
Protease cascades are commonly activated in host-pathogen interaction, inflammation, tissue damage and remodeling, cell invasion and cancer metastasis.26–29,93 Since protease release is often associated with the presence of pathogenic microbes, in fact, the involvement of serine proteases in mammalian immunological responses has been widely implicated.31,36–38,40 For example, the Drosophila IMD pathway exhibits homology to the mammalian tumor necrosis factor (TNFα) signaling pathway. Although no mammalian TNF superfamily of receptors have been shown to be cleaved or activated by proteolysis, many ligands for the TNF superfamiliy receptors can be released from the cell surface by a limited proteolysis through distinct types of proteases.94,95 Also, cell surface proteolysis and proteinase-activated receptors (PARs) are involved in many important biological processes. Hence, it is conceivable that some host immune receptors/sensors could be cleaved by an infection-induced damage-dependent proteolysis in a manner similar to that described for Spz, PARs and Notch.1,27,96–99
As pathogenicity of bacteria is not necessarily related to production of known exogenous PAMP stimuli of innate defense, mechanisms in addition to pattern recognition of these agonists may exist. It is known that serine proteases play key roles in TOLL pathway activation.11–13,74,100 Thus, there remains a possibility that the infection-induced protease-mediated modulation of innate immunity receptors/ligands/sensors may serve as a generic “loss of well-being” signal in pathogen-host interaction and inflammation/wound healing. As such, we would like to propose that in conjunction with PAMP recognition, the infection-induced protease release (whether pathogen-derived, host-derived or more likely of mixed origins) during pathogen-host antagonism might be an additional determinant in differentiating pathogenic microbes. Although much work is needed to be done to prove this model, we are convinced that additional cues in addition to pattern recognition may be necessary for the host cells to distinguish commensal vs. pathogenic microbes and activate multiple host defense mechanisms.
Acknowledgments
We would like to dedicate this paper to Dr. Paul Leibson, a beloved and well-respected immunologist, who passed away on August 6, 2007. We are grateful to Paul Leibson, Karen Hedin, Hirohito Kita, Larry R. Pease, Henry Chang, Grazia Isaya, Jim Maher, Whyte Owen, Adina Bailey and Nick Zagorski for critical reading of the manuscript. We thank Kathryn Anderson, Tony Ip, Lousia Wu, Julien Royet, Mika P. Rämet, Pascal Manfruelli, Stephen Hou and the Bloomington Drosophila Stock Center for fly stocks and plasmids. We thank Robin Patel for bacterial stocks. We thank Benjamin J. Madden at the Mayo proteomic core facilities for proteomic identification of PGRP-LC cleavage products. R.L.S. is supported by the Mayo Clinic Pobanz Family Predoctoral Research Fellowship. J.L.P is supported in part by National Institutes of Health Grant (AI53733). A.H.T and this work are supported by a startup fund to A.H.T. from the Mayo Foundation and in part by National Institutes of Health Grant (GM 069922).
Abbreviations
- AMP
antimicrobial peptide
- IMD
immune deficiency
- LPS
lipopolysaccharide
- MMP
matrix metalloproteinase
- PGRP
peptidoglycan-recognition protein
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- PAMP
pathogen-associated molecular pattern
- ECM
extracellular matrix
Declaration of Potential Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 3.Royet J, Reichhart JM, Hoffmann JA. Sensing and signaling during infection in Drosophila. Curr Opin Immunol. 2005;17:11–17. doi: 10.1016/j.coi.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, et al. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- 5.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 6.Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297:114–116. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- 7.Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, et al. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 2006;25:5005–5014. doi: 10.1038/sj.emboj.7601363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Grampositive bacteria. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- 10.Leulier F, Lemaitre B. Toll-like receptors--taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 11.Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, et al. A Spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. 2006;10:45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, et al. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 13.Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, et al. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- 15.Hu X, Yagi Y, Tanji T, Zhou S, Ip YT. Multimerization and interaction of Toll and Spätzle in Drosophila. Proc Natl Acad Sci U S A. 2004;101:9369–9374. doi: 10.1073/pnas.0307062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 17.Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 18.Choe KM, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 19.Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci U S A. 2002;99:13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem. 2003;278:26319–26322. doi: 10.1074/jbc.C300184200. [DOI] [PubMed] [Google Scholar]
- 21.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/S1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 24.Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellroth P, Karlsson J, Håkansson J, Schultz N, Goldman WE, Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci U S A. 2005;102:6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ossovskaya VS, Bunnett NW. Protease-activated receptors: contribution to physiology and disease. Physiol Rev. 2004;84:579–621. doi: 10.1152/physrev.00028.2003. [DOI] [PubMed] [Google Scholar]
- 28.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 29.Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. doi: 10.1016/S0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- 30.Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 31.Belaaouaj A, McCarthy R, Baumann M, Gao Z, Ley TJ, Abraham SN, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 32.Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/S0145-305X(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 33.Theopold U, Li D, Fabbri M, Scherfer C, Schmidt O. The coagulation of insect hemolymph. Cell Mol Life Sci. 2002;59:363–372. doi: 10.1007/s00018-002-8428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Söderhäll K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol. 1998;10:23–28. doi: 10.1016/S0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 35.Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 2000;289:1185–1188. doi: 10.1126/science.289.5482.1185. [DOI] [PubMed] [Google Scholar]
- 36.Volanakis JE. Overview of the complement system. In: Volanakis JE, Frank MM, editors. The Human Complement System in Health and Disease. New York: Marcel Dekker, Inc; 1998. pp. 9–32. [Google Scholar]
- 37.Tkalcevic J, Novelli M, Phylactides M, Iredale JP, Segal AW, Roes J. Impaired immunity and enhanced resistance to endotoxin in the absence of neutrophil elastase and cathepsin G. Immunity. 2000;12:201–210. doi: 10.1016/S1074-7613(00)80173-9. [DOI] [PubMed] [Google Scholar]
- 38.Wipke BT, Allen PM. Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol. 2001;167:1601–1608. doi: 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J, Nathan C, Jin W, Sim D, Ashcroft GS, Wahl SM, et al. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111:867–878. doi: 10.1016/S0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 40.Dahlbäck B. Blood coagulation and its regulation by anticoagulant pathways: genetic pathogenesis of bleeding and thrombotic diseases. J Intern Med. 2005;257:209–223. doi: 10.1111/j.1365-2796.2004.01444.x. [DOI] [PubMed] [Google Scholar]
- 41.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 42.Brunn GJ, Bungum MK, Johnson GB, Platt JL. Conditional signaling by Toll-like receptor 4. FASEB J. 2005;19:872–874. doi: 10.1096/fj.04-3211fje. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt RL, Trejo TR, Plummer TB, Platt JL, Tang AH. Infection-induced proteolysis of PGRP-LC controls the IMD activation and melanization cascades in Drosophila. FASEB J. 2008;22:918–929. doi: 10.1096/fj.06-7907com. [DOI] [PubMed] [Google Scholar]
- 44.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/S1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 45.Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3:91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- 46.Asha H, Nagy I, Kovacs G, Stetson D, Ando I, Dearolf CR. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics. 2003;163:203–215. doi: 10.1093/genetics/163.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang AH, Tu C-PD. Pentobarbital-induced changes in Drosophila glutathione S-transferase D21 mRNA stability. J Biol Chem. 1995;270:13819–13825. doi: 10.1074/jbc.270.23.13819. [DOI] [PubMed] [Google Scholar]
- 48.Tang AH, Tu C-PD. Biochemical characterization of Drosophila dlutathione S-transferase D1 and D21. J Biol Chem. 1994;269:27876–27884. [PubMed] [Google Scholar]
- 49.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/S0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- 51.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 53.Johnson GB, Brunn GJ, Kodaira Y, Platt JL. Receptor-mediated monitoring of tissue well-being via detection of soluble heparan sulfate by Toll-like receptor 4. J Immunol. 2002;168:5233–5239. doi: 10.4049/jimmunol.168.10.5233. [DOI] [PubMed] [Google Scholar]
- 54.Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 55.Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 56.Lander AD, Selleck SB. The elusive functions of proteoglycans: in vivo veritas. J Cell Biol. 2000;148:227–232. doi: 10.1083/jcb.148.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wrenshall LE, Cerra FB, Carlson A, Bach FH, Platt JL. Regulation of murine splenocyte responses by heparan sulfate. J Immunol. 1991;147:455–459. [PubMed] [Google Scholar]
- 58.Ihrcke NS, Platt JL. Shedding of heparan sulfate proteoglycan by stimulated endothelial cells: evidence for proteolysis of cell-surface molecules. J Cell Physiol. 1996;168:625–637. doi: 10.1002/(SICI)1097-4652(199609)168:3<625::AID-JCP15>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 59.Dziarski R. Peptidoglycan recognition proteins (PGRPs) Mol Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Chaput C, Boneca IG. Peptidoglycan detection by mammals and flies. Microbes Infect. 2007;9:637–647. doi: 10.1016/j.micinf.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 61.Parks WC, Wilson CL, López-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 62.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Page-McCaw A, Serano J, Santé JM, Rubin GM. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell. 2003;4:95–106. doi: 10.1016/S1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- 64.Llano E, Adam G, Pendás AM, Quesada V, Sánchez LM, Santamariá I, et al. Structural and enzymatic characterization of Drosophila Dm2-MMP, a membrane-bound matrix metalloproteinase with tissue-specific expression. J Biol Chem. 2002;277:23321–23329. doi: 10.1074/jbc. M200121200. [DOI] [PubMed] [Google Scholar]
- 65.Ashida M, Brey PT. Recent advances in research on the insect prophenoloxidase cascade. In: Brey PT, Hultmark D, editors. Molecular Mechanisms of Immune Response in Insects. London: Chapman and Hall; 1998. pp. 135–172. [Google Scholar]
- 66.De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, et al. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell. 2002;3:581–592. doi: 10.1016/S1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 67.Sodeinde OA, Subrahmanyam YV, Stark K, Quan T, Bao Y, Goguen JD. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 68.Lundrigan MD, Webb RM. Prevalence of ompT among Escherichia coli isolates of human origin. FEMS Microbiol Lett. 1992;76:51–56. doi: 10.1111/j.1574-6968.1992.tb05438.x. [DOI] [PubMed] [Google Scholar]
- 69.Stumpe S, Schmid R, Stephens DL, Georgiou G, Bakker EP. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol. 1998;180:4002–4006. doi: 10.1128/jb.180.15.4002-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to α-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/JB.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ulvatne H, Haukland HH, Samuelsen O, Krämer M, Vorland LH. Proteases in Escherichia coli and Staphylococcus aureus confer reduced susceptibility to lactoferricin B. J Antimicrob Chemother. 2002;50:461–467. doi: 10.1093/jac/dkf156. [DOI] [PubMed] [Google Scholar]
- 72.Kramer RA, Brandenburg K, Vandeputte-Rutten L, Werkhoven M, Gros P, Dekker N, et al. Lipopolysaccharide regions involved in the activation of Escherichia coli outer membrane protease OmpT. Eur J Biochem. 2002;269:1746–1752. doi: 10.1046/j.1432-1327.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- 73.McCarter JD, Stephens D, Shoemaker K, Rosenberg S, Kirsch JF, Georgiou G. Substrate specificity of the Escherichia coli outer membrane protease OmpT. J Bacteriol. 2004;186:5919–5925. doi: 10.1128/JB.186.17.5919-5925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chasan R, Anderson KV. The role of easter, an apparent serine protease, in organizing the dorsal-ventral pattern of the Drosophila embryo. Cell. 1989;56:391–400. doi: 10.1016/0092-8674(89)90242-0. [DOI] [PubMed] [Google Scholar]
- 75.Saunders WB, Bayless KJ, Davis GE. MMP-1 activation by serine proteases and MMP-10 induces human capillary tubular network collapse and regression in 3D collagen matrices. J Cell Sci. 2005;118:2325–2340. doi: 10.1242/jcs.02360. [DOI] [PubMed] [Google Scholar]
- 76.Medzhitov R, Janeway CA., Jr. Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 77.Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 78.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 79.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 80.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 81.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 82.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 83.Silverman N, Paquette N. Immunology. The right resident bugs. Science. 2008;319:734–735. doi: 10.1126/science.1154209. [DOI] [PubMed] [Google Scholar]
- 84.Muyskens JB, Guillemin K. Bugs inside Bugs: what the fruit fly can teach us about immune and microbial balance in the gut. Cell Host Microbe. 2008;3:117–118. doi: 10.1016/j.chom.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 85.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 86.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 87.Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- 88.Beg AA. Endogenous ligands of Toll-like receptors: implications for regulating inflammatory and immune responses. Trends Immunol. 2002;23:509–512. doi: 10.1016/S1471-4906(02)02317-7. [DOI] [PubMed] [Google Scholar]
- 89.Johnson GB, Brunn GJ, Tang AH, Platt JL. Evolutionary clues to the functions of the Toll-like family as surveillance receptors. Trends Immunol. 2003;24:19–24. doi: 10.1016/S1471-4906(02)00014-5. [DOI] [PubMed] [Google Scholar]
- 90.Tang AH, Brunn GJ, Cascalho M, Platt JL. Pivotal advance: endogenous pathway to SIRS, sepsis, and related conditions. J Leukoc Biol. 2007;82:282–285. doi: 10.1189/jlb.1206752. [DOI] [PubMed] [Google Scholar]
- 91.Janeway CA., Jr. The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 92.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 93.List K, Szabo R, Molinolo A, Sriuranpong V, Redeye V, Murdock T, et al. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev. 2005;19:1934–1950. doi: 10.1101/gad.1300705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aggarwal BB. Signalling pathways of the TNF super-family: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 95.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/S0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 96.Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 97.Coughlin SR, Camerer E. PARticipation in inflammation. J Clin Invest. 2003;111:25–27. doi: 10.1172/JCI17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 99.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 100.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]