Abstract
Mesenchymal stem cells (MSCs) have the capacity for multilineage differentiation and are being explored as a source for stem cell-based therapies. Previous studies have shown that adhesion to extracellular matrix plays a critical role in guiding MSC differentiation to distinct lineages. Here we conducted a focused screen of microRNAs to reveal one microRNA, miR-125b, whose expression changes as a function of cell adhesion. miR-125b expression was upregulated by limiting cell-matrix adhesion using micropatterned substrates, knocking down beta5 integrin, or placing cells in suspension culture. Interestingly, we noted that suspending hMSCs did not induce substantial apoptosis (anoikis) as is typically observed in adherent cells. Although miR-125b appeared to have some effects of on hMSC differentiation, we demonstrated a striking role for miR-125b in protecting hMSCs from anoikis. Knockdown of miR-125b increased anoikis while expressing a mimic protected cells. Mechanistic studies demonstrated that miR-125b protected against anoikis by increasing ERK phosphorylation and by suppressing p53. Lastly, we found that miR-125b expression is quite limited in endothelial cells and MEFs; the rapid anoikis normally observed in these cells is antagonized by expressing a miR-125b mimic; and induced pluripotent stem (iPS) cells generated from the MEFs led to upregulated miR-125b expression. Together, these observations demonstrate a novel link between cell-matrix adhesion, miR-125b expression, and a stem-cell specific survival program triggered in adhesion-limited contexts such as might occur in early development and wound healing.
Introduction
Mesenchymal stem cells (MSCs) are multipotent cells that can differentiate into a variety of tissues, including bone, cartilage, fat and muscle 1. This multipotential differentiation capacity makes these cells an attractive target for cell-based regenerative therapies, such as cardiac repair following myocardial infarction 2. Differentiation is guided by numerous cues, most notably the presence of soluble factors (growth factors, hormones, small molecules) as well as insoluble cues that emanate from interactions of cells with the extracellular matrix. The importance of these insoluble cues is highlighted by several studies showing that cell spreading 3, cell shape 3,4,5,6, matrix stiffness 5,7, and integrin engagement and clustering 8 regulate which lineages can be induced by differentiation medium. Although organization of the actin cytoskeleton and the ability of cells to transduce mechanical forces is a common denominator in these studies, the molecular mechanisms by which these adhesive cues control differentiation remain incompletely understood.
Most mechanistic studies of how cell-matrix adhesion regulates MSC differentiation have focused on well-documented signal transduction pathways such as MAPK signaling 4,9, or secondary messenger systems, such as cAMP 10. Although these pathways are indubitably important, there is increasing evidence that MSC differentiation is guided by the activity of microRNAs. First, the expression of several microRNAs has been shown to correlate with specific lineages including miR-140 for chondrocytes 11, miR-138 for osteoblasts 12, and miR-30, -33a, and -17-92 cluster, -143, -103, for adipocytes 13,14,15,16. Moreover, antagonizing the expression of miR-29b and miR 17-92 blocked MSC differentiation to the osteogenesis and adipogenic lineages, respectively 17,15. Given the emerging importance of microRNAs in regulating cellular differentiation, we hypothesized that cell adhesion might regulate MSC lineage specification through differential expression of microRNAs.
In this study, we performed a focused screen of microRNAs reportedly expressed in hMSCs or its differentiated lineages 18,15,16,11 to determine whether cell adhesion could regulate microRNA expression. We identified one microRNA, miR-125b, that was specifically induced under conditions of low or absent cell-matrix adhesion. Previous studies have shown that miR-125b is expressed in hematopoietic and epidermal stem cells 19, but this microRNA has not been well characterized in hMSCs. Interesting, we found that miR-125b did not promote cellular differentiation, but rather had an unexpected function in promoting cell survival in response to withdrawal of cell-matrix adhesion signals, a process that normally triggers anoikis. This anoikis-resistance phenotype is mediated by the ability of miR-125b to upregulate MEK/ERK signaling while suppressing p53 expression. Moreover, the ability to upregulate miR-125b in response to loss of cell-matrix adhesion appears to be a stem-cell specific phenomenon..
Materials and Methods
Cell culture and reagents
Human mesenchymal stem cells (Lonza) were maintained in DMEM containing 10% fetal bovine serum (Hyclone), 0.3 mg/ml glutamine, 100 mg/ml streptomycin, 100 units/ml penicillin. Experiments were conducted on cells at passage 6 or earlier. Human umbilical vein endothelial cells (HUVEC) were cultured in EGM-2 media (Lonza). For cell suspension studies, cells were plated in F-127 pluronics-treated polystrene dishes. Long-term suspension cultures (24 hours or longer) also included 2% methylcellulose to prevent the cells from settling. For hMSC differentiation studies, osteogenic media (R&D Systems) and adipogenic media (Lonza) were used. Media were changed every 3 days. Cells were then harvested and then assayed for alkaline phosphatase using Fast-Blue, and lipid droplets using Red-oil-O staining as previously described (REF).
Generation of iPS cells
Mouse embryonic fibroblasts (MEF, OCT4-GFP), a gift from Penn iPSC core facility, were maintained in high glucose DMEM containing 10% fetal bovine serum, and 0.3 mg/ml glutamine. iPS reprogramming was performed using a lentivirus containing mouse Oct4, Klf4, and Sox2,(Penn iPSC core facility). Following infection, MEFs were cultured on top of matrigel at seeding density of (3×105/cm2) in DMEM media containing KOSR (Invitrogen), beta-Mercaptoethanol (Sigma), and LIF (Millipore) for 10 days. GFP-positive foci were picked for miRNA analysis.
p53 knock down
p53 knockdown lentivirus were made from pLVTH-sip53 plasmid (Addgene plasmid 12239 20)
3′UTR assay
3′ UTR seed sequences of p53 (sense, AATTCAAGACTTGTTTTATGCTCAGGGTCAACTGCA, anti-sense, GTTGACCCTGAGCATAAAACAAGTCTTG) and LIN28 (sense, AATTC GGTACATGAGCAATCTCAGGGATAGCCTGCA, antisense, GGCTATCCCTGAGATTGCTCATGTACCG) were cloned into pGL3-BS contruct (gift from Dr. Mitchell Weiss)
Caspase-3 activity
Caspase-3 activity was determined by EnzChek Caspase-3 Assay Kit (Invitrogen). The activity was normalized to total DNA content as determined by CyQUANT (Invitrogen) fluorescence.
Live/Dead Viability Assays
Suspended cells were washed PBS 3X to remove excess methylcellulose before seeding on coverslips. Cells were incubated with the live/dead viability solution (calcein AM and ethidium homodimer-1) (Invitrogen) for 30 minutes, as per the manufacturer’s guidelines.
Real time RT-PCR analysis
Total RNA was isolated using Trizol Kit as specified by the manufacturer (Invitrogen). 300ng of total RNA was reverse transcribed by using Multiscribe reverse transcriptase (Applied biosystems) and 5X Taqman microRNA RT primer and 5X Taqman RNU6B RT primer (Applied biosystems). Real-time PCR was performed using Taqman 2X universal PCR master mix and 20X Taqman microRNA Real-time primers or 20X Taqman RNU6B Real-time primer, and monitored using an ABI 7300 system (Applied Biosystems). Data analysis was performed using the ABI Prism 7300 Sequence Detection Systems v1.0 software (Applied Biosystems,). The RT primer and real-time primers for hsa-miR-19a, hsa-miR-24, hsa-miR-103, hsa-miR-107, hsa-miR-140, hsa-miR-125b, hsa-miR-143, hsa-miR-320 are all from Applied Biosystems.
Transfection of siRNA or mimics
Small interfering RNA targeting miR-125b or a miR-125b mimetic were purchased from Thermo Scientific Dharmacon RNAi Technologies (#IH30059505,#C30059503). The siRNA was transfected into hMSC using Lipofectamine RNAiMAX® reagent (Invitrogen).
Results
Expression of miR-125b is regulated by cell-matrix adhesion
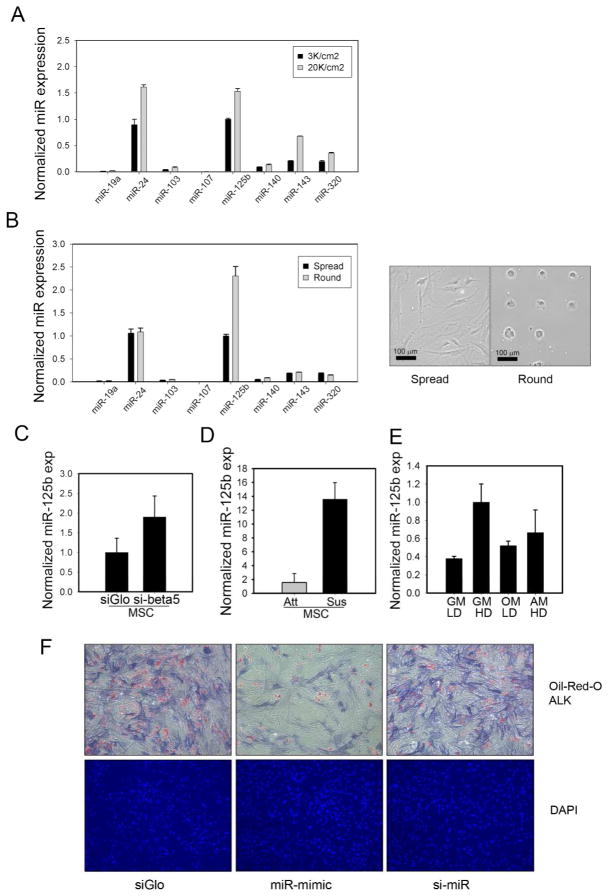
We first tested whether changes in the amount of cell-matrix adhesion could affect the expression of a panel of eight microRNAs reportedly expressed in human mesenchymal stem cells (hMSCs) or differentiated MSCs 18,15,16,11. When plated at high cell seeding densities, hMSCs become crowded and have previously been shown to decrease adhesion against the underlying matrix, which in turn can influence differentiation3. We observed four miRNAs are induced at least 1.5-fold at high seeding densities (Figure 1A). Although changes in cell density can impact cell-matrix adhesion, it also impacts paracrine signaling and cell-cell adhesion. To isolate whether the changes in miRNA levels were a direct consequence of changes in cell-matrix adhesion, we assayed miRNA expression in hMSCs cultured on either continuous fibronectin which promote maximal cell adhesion and spreading (spread cells) or small micropatterns of fibronectin (1024 μm2) which restrict cell spreading (round cells) (Figure 1B). We observed that one miRNA, miR-125b, was elevated two-fold in round compared to spread cells (Figure 1B). To further confirm the specific role of cell-matrix adhesion, we examined whether miR-125b could be induced by specifically antagonizing integrin receptors. Indeed, siRNA-mediated knockdown of beta5 integrin, a major component of the fibronectin receptor, induced miR-125b by two-fold (Figure 1C).
Figure 1.
miR-125b is an adhesion-regulated microRNA whose expression can suppress differentiation. (A) a panel of miRNAs were assayed by qPCR for expression at low (3K/cm2) or high (20K/cm2) seeding densities (B) miRNAs expression as a function of cell spreading; spread (continuous fibronectin) or round (32×32 um2 micropatterns). (C) miR-125b expression was induced by knockdown of beta5 integrin (D) miR-125b expression was strongly induced by cell suspension, suspended (Sus) versus attached (Att) cells (E) miR-125b levels as a function of differentiation, GM:growth media, OM: osteogenic media, AM: Adipogenic media, LD: low density, 3K/cm2, HD: high density, 20K/cm2. (F) miR-125b mimetic (miR mimic) suppressed adipogenic and osteogenic differentiation of hMSCs compared to a transfection control (siGLO) or to miR-125b knockdown cells (si-miR). Oil-red-O staining used to visualize lipid droplets, and Fast Blue staining to visualize alkaline phosphatase (ALP) activity following 1 week of differentiation.
The inverse relationship between levels of cell-matrix adhesion and miR-125b expression suggested that cell-matrix adhesion and/or cell spreading suppresses miR-125b. To test whether cell adhesion per-se suppresses miR-125b, we assayed miR-125b levels in hMSCs suspended in 2% methylcellulose. We found that miR-125b was potently induced by the absence of cell-matrix adhesion (Figure 1D). In fact, the eight-fold induction caused by complete loss of adhesion was greater than the effects of cell density, micropatterning, or integrin knockdown.
miR-125b is not essential for adipogenic or osteogenic differentiation
To investigate a potential role for miR-125b in the differentiation process of hMSCs previously shown to be impacted by high or low seeding densities, we measured miR-125b under conditions that promote either osteogenesis or adipogenesis. miR-125b expression was slightly elevated in either differentiation condition, relative to basal medium; however, miR-125b levels remained below those observed in low cell adhesion contexts (Figure 1E). To investigate whether changes in miR-125b levels could drive differentiation, hMSCs were transiently transfected with a miR-125b antagonist or mimetic prior to differentiation in a bipotential medium that can promote either osteogenic or adipogenic fates 3. Interestingly, miR-125b mimetic strongly inhibited the formation of either adipocytes or osteoblasts (Figure 1F). In contrast, miR-125b knockdown did not appear to perturb hMSC differentiation. Given that miR-125b was not induced by high cell seeding density in bipotential differentiation, and that elevated miR-125b interfered with differentiation, we conclude that the regulation of miR-125b by cell adhesion does not contribute to the process of hMSC differentiation.
miR-125b protects against anoikis
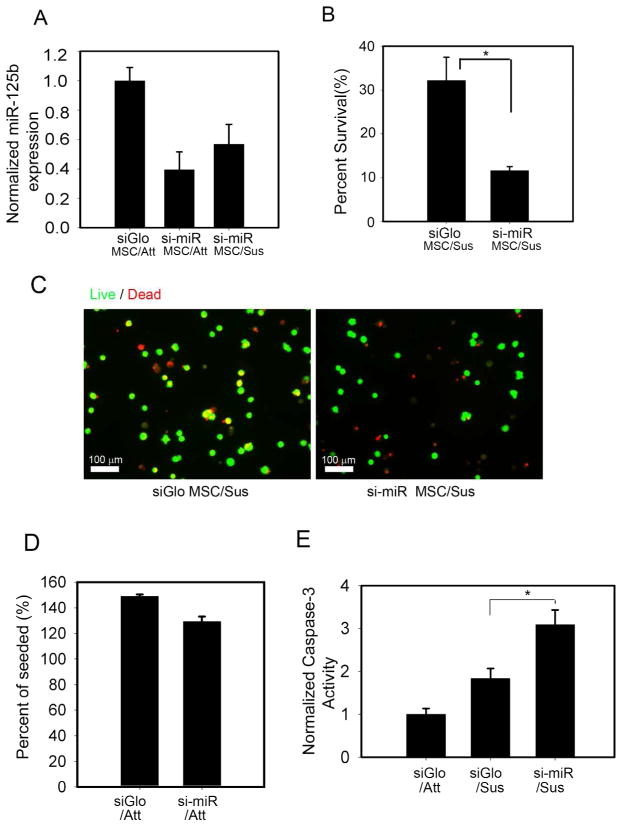
The absence of a positive effect of miR-125b on hMSC differentiation left unanswered the functional significance of miR-125b upregulation in response to limited adhesion. Since we observed the most dramatic induction of miR-125b when hMSC were suspended in methylcellulose, we focused on whether miR-125b might modulate a cellular response to suspension conditions. In most adherent cell types, loss of adhesion triggers cell death in a process known as anoikis 21. We therefore assayed for survival of hMSCs following suspension for either 24 hours, or 7 days. Remarkably, hMSCs were unexpectedly resilient to suspension conditions. While suspension triggers complete cell death in many cells, a sizeable fraction (~ 32%) of hMSCs remained viable in the absence of adhesion for one week (Figure 2B).
Figure 2.
miR-125b protects hMSCs against anoikis. (A) miR-125b expression levels in adherent (Att) or suspended (Sus) hMSCs transfected with miR-125b antagonist (si-miR) (B) Knockdown of miR-125b decreased viability of hMSCs held in suspension. Viability assayed by trypan blue exclusion and reported as percent of cells versus total number seeded (C) Imaging hMSC survival by live (Green cytosolic) versus dead (punctate red nuclear) staining. Note that red fluorescence of siGLO causes some viable cells to be double labeled (yellow). These are easily distinguished from dead cells that exhibit punctate nuclear staining. (D) Knockdown of miR-125b had no effect on cell survival under adherent culture conditions. Percent of seeded exceeds 100% due to cell proliferation. (E) Knockdown of miR-125b promoted apoptosis (caspase 3 activity) in suspended hMSCs. * denotes p<0.05 for Student’s t-test.
To test whether miR-125b played a role in this hMSC survival in suspension culture, we assayed the effects of knockdown of miR-125b on cell survival. Transiently transfecting a miR-125b hairpin inhibitors as an antagonist, we could prevent the induction of miR-125b in response to cell suspension (Figure 2A). Importantly, we observed that knockdown of miR-125b dramatically reduced hMSC survival in suspension (Figure 2B,C). Moreover, this effect was specific to suspended hMSCs since knockdown of miR-125b had no effect on the viability of adherent hMSCs (Figure 2D). To confirm that this change in viability was associated with changes in the levels of apoptosis, we measured the activity level of caspase 3, a terminal effector of apoptosis 22. Consistent with a requirement for upregulation of miR-125b to promote survival in suspended hMSCs, we observed 1.5-fold higher levels of caspase 3 activity in miR-125b knockdown cells compared to controls (Figure 2E). Together, these data show that a subpopulation of hMSCs are refractory to the anoikis pathway triggered by cell suspension, and this resistance to anoikis appears to be largely miR-125b dependent.
miR-125b targets p53 in hMSCs
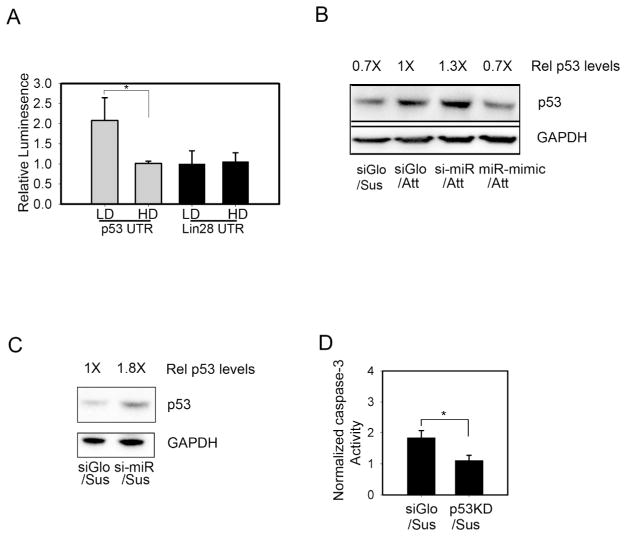
microRNAs bind to complementary sequences in the 3′UTR of mRNAs, and thereby downregulate gene expression by either attenuated translational efficiency or degradation of the targeted mRNA. The repertoire of genes targeted by mir-125b in hMSCs is not yet known; however, p53 and lin28 have been identified as miR-125b targets in several cell types 23,24. To begin to examine whether miR-125b protected cells from anoikis through its reported effects on these genes, we cloned the 3′UTR sequences of these genes into a luciferase plasmid to generate putative reporters for miR-125b activity. We then characterized these reporters under different conditions of cell-matrix adhesion that we had previously observed to trigger elevated miR-125b expression. In this assay, the p53 3′UTR luciferase reporter showed decreased expression at high seeding density, whereas no change occurred in the lin28 reporter (Figure 3A). These results suggested that p53 may be a target for miR-125b regulation in hMSCs.
Figure 3.
miR-125b targets p53 to regulate hMSCs survival. (A) miR-125b targets p53 and LIN28 were analyzed for density-dependent expression (HD, high density; LD, low density) in a 3′UTR luciferase reporter assay in hMSCs (B) p53 levels in adherent hMSCs transfected with transfection control (siGLO), miR-125b antagonist (si-miR), or mimetic (miR mimic). (C) rescue of p53 expression in suspended hMSCs by miR-125b antagonist (D) p53-knockdown (p53 KD) protected hMSCs from anoikis. * denotes p<0.05 for Student’s t-test.
To investigate whether endogenous p53 is specifically targeted by miR-125b, we measured levels of p53 expression in response to direct manipulations of miR-125b levels. In adherent cells, transfection of a miR-125b antagonist upregulated p53 expression, while a miR-125b mimetic suppressed p53 (Figure 3B). Moreover, suspending cells, which upregulates miR-125b, decreased p53 levels. In contrast, miR-125b-knockdown cells showed persistent p53 expression in suspension culture (Figure 3C). Together these results show that p53 is a target of miR-125b in hMSCs.
p53 plays a critical role in mediating apoptosis during anoikis 25. While p53 is reported to increase the sensitivity of cells to anoikis, it is not absolutely required for the anoikis response26. To address whether inhibition of p53 expression by miR-125b may be part of the mechanism by which miR-125b prevents anoikis, we investigated the effects of p53 knockdown in suspended hMSCs. Using a lentivirally encoded p53 shRNA 20, we observed that reduction of p53 was associated with an 80% decrease in caspase 3 activity in suspended hMSCs, (Figure 3D). This decreased level of apoptosis in p53 knockdown cells suggests that p53 suppression by miR-125b may be an effective mechanism to protect hMSCs against anoikis.
miR-125b regulates levels of phospho-ERK in suspended hMSCs
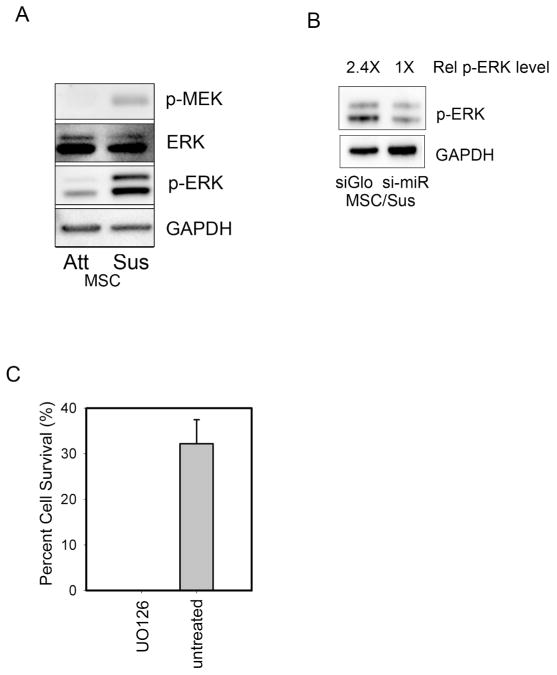
To further characterize how miR-125b protects against anoikis, we examined how signal transduction pathways associated with cell survival changed in suspended hMSCs. As expected, hMSCs showed a dramatic downregulation of phospho-FAK in response to loss of cell adhesion (data not shown). In contrast, suspended hMSCs showed unexpectedly elevated levels of phospho-MEK and phospho-ERK (Figure 4A). Since ERK signaling has been implicated in p53-mediated survival 27 and hMSC viability 28, we examined more closely whether miR-125b was involved with the observed changes in ERK phosphorylation.
Figure 4.
miR-125b expression and hMSC survival are associated with elevated phospho-ERK. (A) suspended hMSCs showed elevated levels of active MEK and phospho-ERK (B) miR-125b knockdown reduced levels of phospho-ERK in suspended hMSCs (C) ERK signaling was absolutely required for survival of hMSCs in suspension, UO126 (MEK inhibitor).
We observed that knockdown of mir-125b strongly attenuated phospho-ERK levels in suspended hMSCs (Figure 4B). To confirm that activation of ERK is functionally relevant to an anoikis protection mechanism in hMSCs, we treated suspended cells with a pharmacological inhibitor of MEK. In the presence of the inhibitor UO126, no cells were able to survive suspension conditions revealing an absolute requirement for MEK-ERK signaling in promoting hMSC survival in suspension (Figure 4C).
Expression of miR-125b is cell-type restricted
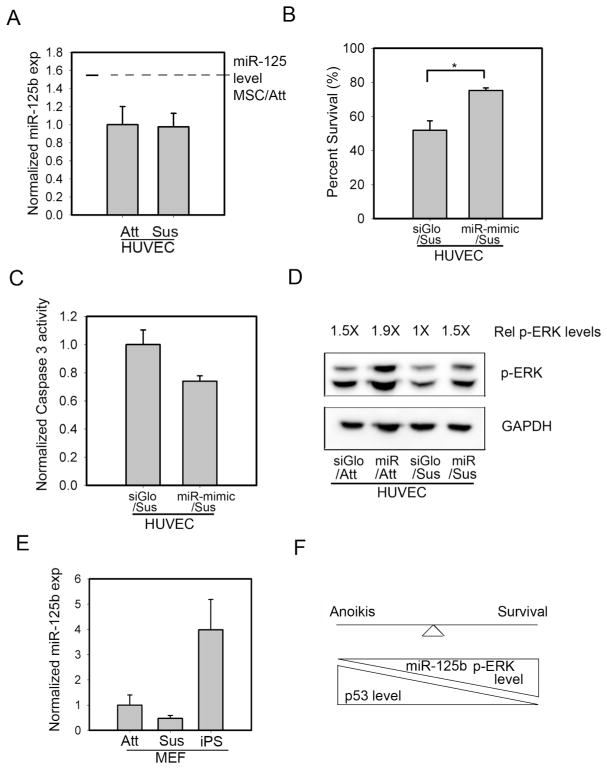
We were interested in exploring whether the upregulation of miR-125b in low adhesion contexts could be observed in other cell types, particularly ones that are known to be susceptible to anoikis. To begin to address this question, we measured miR-125b levels in adherent versus suspended human umbilical vein endothelial cells (HUVECs), a cell type known to be highly dependent on adhesion for survival. We observed that miR-125b is poorly expressed in HUVECs compared to hMSCs; moreover, these cells were unable to upregulate miR-125b in response to suspension (Figure 5A). Despite the absence of miR-125b upregulation in suspended HUVECs, we hypothesized that the ability of miR-125b to protect against anoikis may be conserved across cell types. Indeed, HUVECs transfected with miR-125b mimetic inhibited suspension induced death by nearly half (Figure 5B). In accordance with this improved level of survival, miR-125b mimetic reduced caspase 3 activity in suspended HUVECs (Figure 5C). Since we had previously observed a positive correlation between miR-125b, cell survival, and ERK signaling in hMSCs, we tested whether miR-125b mimetic could regulate ERK phosphorylation in HUVECs. Indeed, we observed that miR-125b mimetic was able to upregulate levels of phospho-ERK in both adherent, and more importantly, in suspended HUVECs (Figure 5D).
Figure 5.
Upregulation of miR-125b in response to suspension is a stem-cell specific phenomenon. (A) miR-125b was expressed at low levels in HUVECs and could not be induced by cell suspension. Dotted line shows basal level of miR-125b expression in attached hMSCs (B) Exogenous miR-125b improved HUVEC survival in suspension conditions (75.2% viability compared to 51.9% of control cells following 3 hrs of suspension and replating) (C) Exogenous miR-125b decreased caspase 3-activity in suspended HUVECs (D) HUVECs did not sustain ERK activation in response to cell suspension; however, exogenous miR-125b elevated phospho-ERK levels (E) miR-125b expression was upregulated upon reprogramming of MEFs to an iPS-like state, but not by cell suspension (F) Qualitative model of how miR-125b regulates stem cell survival in low adhesion contexts. * denotes p<0.05 for Student’s t-test
Given that HUVECs could not upregulate miR-125b in response to loss of cell adhesion, we wondered whether the regulation of miR-125b by changes in cell matrix adhesion is a stem cell specific response. To investigate this possibility, we measured miR-125b levels in mouse embryo fibroblasts (MEF), in adherent or suspension culture, as well as following a reprogramming protocol to generate induced pluripotent cells. While MEFs failed to induce mir-125b in response to suspension conditions (Figure 5E), miR-125b was enriched in MEFs following induction to pluripotency by about 3-fold compared to the parental fibroblasts. These results suggest that robust expression of miR-125b is preferentially associated with stem cells.
Discussion
In vivo, MSCs are resident in a number of different tissues 29, but also transiently circulate in the bloodstream in inflammatory settings perhaps to home to sites of injury 30. Here, we provide the first demonstration that MSCs can resist anoikis, perhaps explaining how MSCs are able to transit through the circulatory system to populate distant sites without undergoing apoptosis. We also show that this response is mediated by upregulation of miR-125b.
Interestingly, miR-125b is reportedly enriched in several stem cells, most notably hematopoietic stem cells (HSCs), as well as skin stem cells 31. miR-125b appears to influence multiple aspects of HSC biology 19, and one study has shown that miR-125b affects proliferation of these cells 32. In skin stem cells, miR-125b has been shown to control the balance between stemness and differentiation 31. One of the hallmarks of HSCs and skin stem cells is that they exist in a specialized niche with minimal cell-matrix interactions33,34. Here, we show in hMSCs that changing cell adhesion itself is a key regulator in upregulating miR-125b. Although the environment of circulating MSC is quite distinct from these stem cell niches, the coincidence of low or absent cell-matrix adhesion with high miR-125b expression is all these setting is quite striking. And this strong association of miR-125b expression, stem cell specificity, and low adhesion is further strengthened by our observation that miR-125b is upregulated in iPS cells, which grow as aggregates and are relatively deficient in cell-matrix contact. In contrast, adhesion-dependent cell types, such as HUVECs and MEFs, show low miR-125b expression that is unresponsive to changes in cell matrix adhesion. Taken together, these observations suggest that elevated miR-125b may be a general mechanism employed specifically by stem cells to enable their survival, and perhaps maintain their stemness, in the context of low or no signaling from cell-matrix adhesion.
How does elevated miR-125b promote cell survival in the absence of adhesion? Upon cell suspension, cells lose integrin-dependent signals. Typically, this loss of integrin signaling causes marked reduction of phospho-Akt and phospho-ERK, and hence a reduction in pro-survival signaling35,36. Surprisingly, while losing integrin-dependent signaling such as FAK phosphorylation, hMSCs show robust ERK phosphorylation and ultimately increased cell survival. Moreover, pharmacological inhibition of MEK completely ablates the ability of hMSCs to survive in suspension. These observations suggest that miR-125b may regulate survival through MEK/ERK signaling. Although the mechanistic link between miR-125b and MEK/ERK signaling remains to be elucidated, it is possible that miR-125b can inhibit the expression of suppressors of this pathway. Indeed, miR-125b is predicted to target SMEK1, a MEK1 phosphatase (targetscan.org). In addition, it is interesting to note that ERK signaling can be downregulated by dual specificity phosphatases in response to p53 signaling27. Thus, suppression of p53 by miR-125b may contribute to sustained ERK signaling. The ability of miR-125b to target p53 likely plays a major role in promoting resistance to anoikis. In fact, direct knockdown of p53 was also observed to protect hMSCs against anoikis. These results are consistent with several studies showing that loss of p53 confers resistance to anoikis37,38. Interestingly, loss of p53 in response to suspension has been attributed to MDM2-based degradation in MEFs and several cancer cell lines 39. The observation that miR-125b can also target p53 suggests an additional level of regulation that may enable long-term suppression of p53 in the absence of cell adhesion. Whether and how miR-125b mediated suppression and MDM-2 based p53 degradation cooperated to regulate p53 levels in normal and cancer cells is an interesting question for future studies.
Because p53 is implicated in a diverse set of cellular processes including cell cycle control, apoptosis and regulating genomic stability, it may seem somewhat surprising that stem cells might adopt a survival mechanism contingent on reducing the levels of this critical protein. Indeed, loss of p53 function can have deleterious effects on MSC functionality. For example, mutations in p53 have been implicated in age-related transformation of MSCs 40, and the proliferation of MSCs is particularly sensitive to p53 levels41. Moreover, p53 appears to have a role in MSC differentiation, in particular during osteogenic commitment 42 Nonetheless, it is important to point out that MSCs are non-proliferative in cell suspension conditions, and these cells may only exist in this state during quiescences or when transiently circulating. Importantly, upon attachment to a surface (an in vitro analog of engraftment into a tissue), we observed that miR-125b levels were reduced to normal levels in hMSCs (data not shown)
In conclusion, this study demonstrates that miR-125b is an adhesion-regulated microRNA that functions to protect stem cells against anoikis. miR-125b appears to confer resistance to anoikis by upregulating phospho-ERK while suppressing p53 levels. These findings have important implications for how stem cells survive in the unique developmental, wounding, and circulatory contexts of reduced or absent cell-matrix adhesion signaling.
Acknowledgments
We thank Jennifer Leight for technical assistance with the caspase activity assays and Michael Yang and Duc Nguyen for HUVEC cells. We also thank Lin Gao, Wenli Yang, Frederick Anokye-Danso and Ed Morrisey for help with iPS cells and Jeroen Eyckmans and Mark Breckenridge for reading the manuscript.
Funded by:
National Institutes of Health (GM74048)
Penn Center for Engineering Cells and Regeneration
American Heart Association (10POST4220014)
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Author’s contribution: X.Y., D.M.C. conception and design, collection of data, data analysis and interpretation, manuscript writing; C.S.C. conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
References
- 1.Pittenger MF, Mosca JD, McIntosh KR. Human mesenchymal stem cells: progenitor cells for cartilage, bone, fat and stroma. Curr Top Microbiol Immunol. 2000;251:3–11. doi: 10.1007/978-3-642-57276-0_1. [DOI] [PubMed] [Google Scholar]
- 2.Boyle AJ, McNiece IK, Hare JM. Mesenchymal stem cell therapy for cardiac repair. Methods Mol Biol. 660:65–84. doi: 10.1007/978-1-60761-705-1_5. [DOI] [PubMed] [Google Scholar]
- 3.McBeath R, Pirone DM, Nelson CM, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 4.Kilian KA, Bugarija B, Lahn BT, et al. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu J, Wang YK, Yang MT, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao L, McBeath R, Chen CS. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 28:564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Huebsch N, Arany PR, Mao AS, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyers VE, Zayzafoon M, Gonda SR, et al. Modeled microgravity disrupts collagen I/integrin signaling during osteoblastic differentiation of human mesenchymal stem cells. J Cell Biochem. 2004;93:697–707. doi: 10.1002/jcb.20229. [DOI] [PubMed] [Google Scholar]
- 10.Siddappa R, Martens A, Doorn J, et al. cAMP/PKA pathway activation in human mesenchymal stem cells in vitro results in robust bone formation in vivo. Proc Natl Acad Sci U S A. 2008;105:7281–7286. doi: 10.1073/pnas.0711190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyaki S, Nakasa T, Otsuki S, et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskildsen T, Taipaleenmaki H, Stenvang J, et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A. 108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaragosi LE, Wdziekonski B, Brigand KL, et al. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 12:R64. doi: 10.1186/gb-2011-12-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Li YC, Wang J, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapinas K, Kessler C, Ricks T, et al. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 285:25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goff LA, Boucher S, Ricupero CL, et al. Differentiating human multipotent mesenchymal stromal cells regulate microRNAs: prediction of microRNA regulation by PDGF during osteogenesis. Exp Hematol. 2008;36:1354–1369. doi: 10.1016/j.exphem.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connell RM, Chaudhuri AA, Rao DS, et al. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvesen GS. Caspases: opening the boxes and interpreting the arrows. Cell Death Differ. 2002;9:3–5. doi: 10.1038/sj.cdd.4400963. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le MT, Teh C, Shyh-Chang N, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grossmann J. Molecular mechanisms of “detachment-induced apoptosis--Anoikis”. Apoptosis. 2002;7:247–260. doi: 10.1023/a:1015312119693. [DOI] [PubMed] [Google Scholar]
- 26.McGill G, Shimamura A, Bates RC, et al. Loss of matrix adhesion triggers rapid transformation-selective apoptosis in fibroblasts. J Cell Biol. 1997;138:901–911. doi: 10.1083/jcb.138.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Y, Liu YX, Jin YJ, et al. PAC1 phosphatase is a transcription target of p53 in signalling apoptosis and growth suppression. Nature. 2003;422:527–531. doi: 10.1038/nature01519. [DOI] [PubMed] [Google Scholar]
- 28.Pricola KL, Kuhn NZ, Haleem-Smith H, et al. Interleukin-6 maintains bone marrow-derived mesenchymal stem cell stemness by an ERK1/2-dependent mechanism. J Cell Biochem. 2009;108:577–588. doi: 10.1002/jcb.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Karp JM, Leng Teo GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4:206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Stokes N, Polak L, et al. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell. 8:294–308. doi: 10.1016/j.stem.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bousquet M, Harris MH, Zhou B, et al. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A. 107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianco P. Minireview: The stem cell next door: skeletal and hematopoietic stem cell “niches” in bone. Endocrinology. 152:2957–2962. doi: 10.1210/en.2011-0217. [DOI] [PubMed] [Google Scholar]
- 34.Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khwaja A, Rodriguez-Viciana P, Wennstrom S, et al. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. Embo J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlaepfer DD, Hanks SK, Hunter T, et al. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994;372:786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 37.Vitale M, Di Matola T, Bifulco M, et al. Apoptosis induced by denied adhesion to extracellular matrix (anoikis) in thyroid epithelial cells is p53 dependent but fails to correlate with modulation of p53 expression. FEBS Lett. 1999;462:57–60. doi: 10.1016/s0014-5793(99)01512-4. [DOI] [PubMed] [Google Scholar]
- 38.Nikiforov MA, Hagen K, Ossovskaya VS, et al. p53 modulation of anchorage independent growth and experimental metastasis. Oncogene. 1996;13:1709–1719. [PubMed] [Google Scholar]
- 39.Lewis JM, Truong TN, Schwartz MA. Integrins regulate the apoptotic response to DNA damage through modulation of p53. Proc Natl Acad Sci U S A. 2002;99:3627–3632. doi: 10.1073/pnas.062698499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Fan X, Kovi RC, et al. Spontaneous expression of embryonic factors and p53 point mutations in aged mesenchymal stem cells: a model of age-related tumorigenesis in mice. Cancer Res. 2007;67:10889–10898. doi: 10.1158/0008-5472.CAN-07-2665. [DOI] [PubMed] [Google Scholar]
- 41.Armesilla-Diaz A, Elvira G, Silva A. p53 regulates the proliferation, differentiation and spontaneous transformation of mesenchymal stem cells. Exp Cell Res. 2009;315:3598–3610. doi: 10.1016/j.yexcr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Molchadsky A, Shats I, Goldfinger N, et al. p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One. 2008;3:e3707. doi: 10.1371/journal.pone.0003707. [DOI] [PMC free article] [PubMed] [Google Scholar]