Abstract
The emergence of whole genome sequencing (WGS) technologies as primary research tools has allowed for the detection of genetic diversity in Mycobacterium tuberculosis (Mtb) with unprecedented resolution. WGS has been used to address a broad range of topics, including the dynamics of evolution, transmission and treatment. Here, we have analyzed 55 publically available genomes to reconstruct the phylogeny of Mtb, and we have addressed complications that arise during the analysis of publically available WGS data. Additionally, we have reviewed the application of WGS to the study of Mtb and discuss those areas still to be addressed, moving from global (phylogeography), to local (transmission chains and circulating strain diversity), to the single patient (clonal heterogeneity) and to the bacterium itself (evolutionary studies). Finally, we discuss the current WGS approaches, their strengths and limitations.
Keywords: Whole genome sequencing, evolution, heterogeneity, Mycobacterium tuberculosis
1) Intro
With the rapid development of whole genome sequencing (WGS) technologies, we have unprecedented capacity to detect genetic diversity in Mycobacterium tuberculosis (Mtb). WGS is particularly powerful when applied to Mtb, which is characterized by a relatively low amount of genetic diversity. WGS data has allowed us to reconstruct the phylogeny of Mtb, and in the process learn a great deal about it's geographic distribution1,2. More recently, studies have investigated the dynamics of evolution, transmission and treatment across shorter time scales3–7 By sequencing strains from small outbreaks and single infections, groups have sought to understand the unique evolutionary dynamics inherent in the spread of tuberculosis5. Using WGS, we can address a broad range of topics - from questions on the transmission and fitness of clinical strains to how Mtb evolves over long and short time scales. Here we have reviewed the insights gained from the use of WGS and discuss those areas still to be addressed, moving from global (phylogeography), to local (transmission chains and circulating strain diversity), to the single patient (clonal heterogeneity), to the bacterium itself (evolutionary studies), and finally discussing the platform of WGS, its strengths and current limitations.
2) Global Diversity
WGS has been proposed as a sort of “gold standard” for strain typing in Mtb. As such, it clarifies previous strain typing approaches used for phylogenetic and epidemiologic studies. The standard genotyping methods are based on repetitive elements that provide limited functional information and are highly prone to convergent evolution, limiting their application to phylogenic reconstructions. The discriminative power of these methods varies, the results of different methods do not always agree, and the diversity of the markers can complicate the analysis1,8. For example, Niemann et al sequenced two isolates of the rapidly spreading Mtb Beijing genotype clone from a high incidence region (Karakalpakstan, Uzbekistan)9. The isolates possessed the same genotype by IS6110 restriction fragment length polymorphism (RFLP) and mycobacterial interspersed repetitive unit – number tandem repeat (MIRU-VNTR) patterns; however, WGS demonstrated that they differed at 131 separate sites, including one large deletion. Thus, typing methods can miss substantial amounts of genetic diversity, and where the overall diversity of circulating clones is limited, standard typing measures are insufficient to discriminate between strains.
As the cost of WGS drops, it is more feasible to consider WGS as a primary tool for the typing of Mtb strains that is not subject to the limitations of standard methodologies. However, use of WGS sequence data is subject to its own set of errors. To demonstrate the power and the pitfalls of this approach, we undertook a phylogenetic analysis of Mtb strains using all of the publically available Mtb genomes. We utilized 55 whole (or nearly whole) genomes downloaded and assembled from GenBank, the Broad Institute, or obtained through personal communication from the authors of published papers by January 15 of 2011 (Table 1). While each genome included more than 4 million bases, these genomes were not fully assembled, nor were they annotated in the same manner. To enable an inclusive comparison, we blasted these genomes against annotated genes of the reference strain F11 to form multiple strain alignments for each gene, including all available strains. Variable sequence positions or SNPs, where at least 1 of 55 sequences differed from other sequences, were then extracted and concatenated, in the order they appear in the reference F11 genome, to create an abbreviated multiple alignment of SNP positions from all 55 sequences. A phylogenetic tree was built from these sequences by molecular parsimony using PAUP10 (Figure 1A). Genes or regions of the genes that were misaligned, where one or several sequences had a high density of SNPs in close proximity, indicating possible either sequencing or alignment problems, were excluded from the analysis using a computational clean-up algorithm; the resulting trees are provided in Figure 1. The 17740 aligned positions were in 3324 genes and included a mutation in at least one of 55 strains that differed from the other sequences.
Table 1.
Summary of 55 strains used in the analysis
| Strain | Genotype | GB ID | Source | Place of isolation | Year | DR Status | Clustered SNPs | Comment |
|---|---|---|---|---|---|---|---|---|
| F11 | F11 | 148719718 | Broad CDB | Western Cape SA | DS | 0 | ||
| V2475 | KZN | 297718568 | GB, Ioerger2009 | Durban, KZN, SA | 1994 | MDR | 3 | Resistant to RIF INH STR |
| 1435 | KZN | 253318418 | Broad CDB | Durban, KZN, SA | 1994 | MDR | 28 | Resistant to RIF INH STR |
| R506 | KZN | 297718569 | GB, Ioerger2009 | Durban, KZN, SA | 2006 | XDR | 3 | Resistant to INH RIF STR OFL KAN |
| 605 | KZN | 289552250 | Broad CDB | Tugela Ferry, KZN, SA | 2005 | XDR | 28 | First full XDR genome, resistant to INH RIF STR OFL KAN ETH |
| 4207 | KZN | 295687135 | Broad | Durban, KZN, SA | 1995 | DS | 87 | |
| GM1503 | 193506183 | Broad DDB | Gambia | 2002 | 1501 * | |||
| 98R604 ** | 220031339 | Broad CDB | MDR | 1403 * | genomesonline.org notes the patient from KZN, SA | |||
| SUMu001..012 | multiple | Broad NMR | Canada | DS | 28..281 | 12 endemic Canadian strains | ||
| H37Ra | H37 | 148503909 | GB | 1905 | DS | 38 | Avirulent strain derived from virulent H37 | |
| H37Rv | H37 | 57116681 | GB | 1905 | DS | 43 | Virulent strain derived from original virulent H37 | |
| Haarlem | Haarlem | 115299839 | Broad CDB | Netherlands | MDR | 590 | Has an accelerated transmission rate in crowded conditions | |
| CDC1551 | 50952454 | GB | KY/TN, USA | ~1994 | DS | 387 | 1994–1996 outbreak in a rural community; highly contagious | |
| C | 81248475 | Broad CDB | New York, USA | 1997 | DS | 569 | Caused a large proportion of new tuberculosis cases in New York | |
| HN878 | Beijing | 315064806 | GB | Houston, TX, USA | 1990s | DS | 27 | Hyper-virulent; part of TB outbreak in the 1990s |
| 210 | Beijing | 261746034 | GB | TX, US | ~1993 | DS | 382 | 1993–1995 outbreak. One of the most virulent M.tb strains; |
| T85 | 189490718 | Broad DDB | San Francisco, CA | 1998 | 617 | Patient born in China | ||
| R1390 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | Mono | 27 | Resistant to INH | |
| X189 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | XDR | 27 | Resistant to INH RIF AMI CAP OFL KAN | |
| R1441 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | Mono | 27 | Resistant to INH | |
| R1505 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | MDR | 27 | Resistant to INH RIF | |
| X122 | Beijing | 312987796 | GB, Ioerger2010 | Western Cape, SA | preXDR | 27 | Resistant to INH RIF OFL | |
| R1909 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | MDR | 27 | Resistant to INH RIF EMB | |
| R1842 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | Mono | 27 | Resistant to INH | |
| X29 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | preXDR | 27 | Resistant to INH RIF AMI STR KAN | |
| X132 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | preXDR | 9 | Resistant to INH RIF AMI CAP STR KAN | |
| R1207 | Beijing | 312984523 | GB, Ioerger2010 | Western Cape, SA | MDR | 9 | Resistant to INH RIF | |
| X28 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | XDR | 9 | Resistant to INH RIF AMI CAP OFL STR KAN | |
| X156 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | preXDR | 9 | Resistant to INH RIF AMI CAP STR KAN | |
| X85 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | XDR | 9 | Resistant to INH RIF AMI OFL KAN ETH | |
| R1746 | Beijing | Not in GB | Ioerger2010 | Western Cape, SA | MDR | 9 | Resistant to INH RIF | |
| W148 | Beijing | 326590567 | Broad CDB | MDR | 324 | Highly multi-drug resistant | ||
| 021987 | 189215797 | Broad DDB | San Francisco, CA | 2002 | 1876 * | Patient born in South Korea | ||
| 94M4241A | 189212844 | Broad DDB | San Francisco, CA | 1994 | 1665 * | Patient born in China | ||
| EAS054 | 189213750 | Broad DDB | San Francisco, CA | 1993 | 1043 * | Patient born in India | ||
| T17 | 192338437 | Broad DDB | San Francisco, CA | 1995 | 1808 * | Patient born in The Philippines | ||
| T46 | 225935560 | Broad DDB | San Francisco, CA | 1996 | 1514 * | Patient born in The Philippines | ||
| T92 | 189213624 | Broad DDB | San Francisco, CA | 1999 | 3200 * | Patient born in The Philippines | ||
| CPHLA | 225935638 | Broad DDB | California | 638 | Patient born in South Africa | |||
| K85 | M.africanum | 225935793 | Broad DDB | Netherlands | 896 | |||
| GM041182 | M.africanum | Not in GB | TBDB | West Africa | 820 | |||
| AF2122|97 | M.bovis | 31742509 | GB | United Kingdom | 1997 | 869 | Isolated from a cow with necrotic lesions in lung | |
| BCGTokyo172 | M.bovis | 224771496 | GB | 603 | BCG vaccine substrain | |||
| BCGPasteur ** | M.bovis | 121491530 | GB | 731 | Was used to produce BCG vaccine |
Abbreviations: GB – GenBank; Broad – Broad Institute M.tb databases; CDB – Comparative Database; DDB – Diversity Database; NMR – Natural Mutation Rate Project; TBDB – genome.tbdb.org; Ioerger2010 – Ioerger et al, 2010 (PMID: 21110864); Ioerger2009 – Ioerger et al, 2009 (PMID: 19890396)
Strains that are highly enriched for clusters of potentially problematic bases
98R604 and BCGPasteur strains have longer names: 98R604|INH|RIF|EM and BCGPasteur1173P2
We utilized 55 whole (or nearly whole) genomes downloaded and assembled from GenBank, the Broad Institute, or obtained through personal communication from the authors of published papers by January of 2011. This table provides the number of clustered SNPs for each sequence. For several strains high numbers of clustered SNPs were unusual (8 strains have greater than 1000 regional clusters of SNPS), and indicated these sequences tended to be more problematic than the others. While such clustered SNP regions were excluded from our phylogenetic analysis, the not-clustered, stand-alone SNPs in those same strains were included in our collection of SNPs for phylogenetic analysis. Whenever available for the near full-length genomes, we recorded isolate's sample history: the year and geographic location of isolate collection, and patient history including place of birth and drug resistance status. The total number of bases where at least one of 55 strains differed from other sequences was 17740, in 3324 genes.
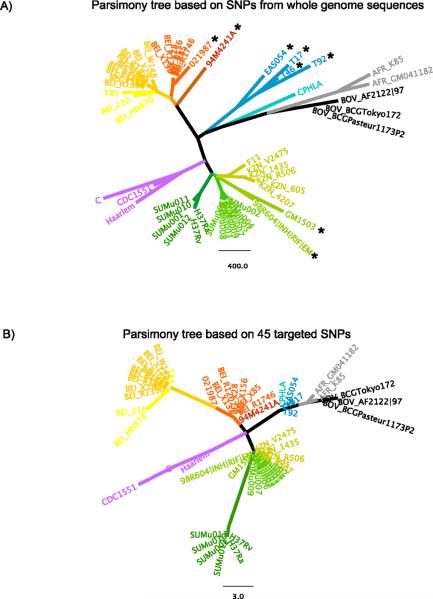
Figure 1. Comparison of phylogenies based on whole genome and targeted SNPs from the available WGS data (Table 1).
The phylogenies were reconstructed using molecular parsimony in PAUP software10. Different lineages are colored in distinct colors, with sub-lineages colored in different shades of the same color. The coloring scheme is preserved between A and B figures.
A) Parsimony tree based on 17740 SNP positions taken from 3324 genes, where at least 1 of 55 sequences differed from other sequences. SNPs were extracted from multiple sequence alignments created for each gene using the following algorithm. Each annotated gene, from a total of 3941 genes for the reference strain F11 from GenBank, was used to perform pair-wise BLASTN search against the other 54 whole genomes. If multiple partial sequences in a genome were found to be homologous to the same gene in F11, we selected the version with the longest coverage, requiring minimum similarity score of 75% and minimum length of coverage of 75%. If the resulting partial sequence was shorter than the gene in F11, it was augmented by adjacent chromosome sequence to the length of the gene of F11. The homologous genes from 55 strains were then assembled into files so that each gene (except for three genes which were uniquely found in F11) had a corresponding file. After assembling homologous sequences, the sequences were aligned using MUSCLE (version 3.8.31) downloaded from www.drive5.com to generate nucleotide multiple sequence alignments for homologous gene files (total 3596) that has at least one non-identical sequence. Since in some strains N- and/or C-terminal gene ends were augmented compared to original BLASTN hit to reflect the full length of the gene in F11, they could be in fact not homologous to F11. In this case these ends did not align within F11 gene and were excluded from the alignment according to F11 gene start and end positions. The resulting gene alignments were then used to generate artificial SNP sequences, which contained a concatenated version of all positions where a SNP was identified (where at least 1 of 55 sequences differed from other sequences). To minimize inclusion of sequencing errors in these artificial SNP sequences, we excluded SNPs that appeared to be clustered in a local region, specifically where three SNPs were found in a 10 base pair window when compared to genes in F11. Furthermore we manually checked the files that have are enriched for clusters of mutations and excluded 70 problematic files. Sequences that are highly enriched for clusters of potentially problematic bases (Table 1) are indicated with the stars. The strains that have the highest number of clustered SNPS (Table 1) also have unusually long terminal branches, a further indication of sequencing artifacts in these strains.
B) Parsimony tree based on 45 SNP positions from Filliol et al1 taken from the same set of 55 genomes.
The relatively high number of SNPs we identified is in part the result of natural variation, as we have included all genes from 55 nearly complete globally diverse strains, and it is in part a consequence of the technical error present in some sequences. In particular, several strains out of the 55 available were highly enriched in regional clusters of SNPs, suggesting potentially problematic base calls (Table 1). The number of clustered SNPs was unusually high in several strains (8 strains have greater than 1000 regional clusters of SNPS), which indicates these sequences tended to be noisier than the others. While such clustered SNP regions were excluded from our phylogenetic analysis, the not-clustered, stand-alone SNPs in those same strains were included in our collection of SNPs for phylogenetic analysis. It is difficult to algorithmically resolve whether any given mutation is of biological origin or is a sequencing artifact. Not surprisingly, the strains that have the highest number of clustered SNPs (Table 1) also have unusually long terminal branches (Figure 1A), a further indication of a higher frequency of sequencing error in these strains. As our intent was to review the whole genome data currently publically available, however, we included all 55 sequences here. While the long terminal branch lengths are likely to be contributed to by sequencing error, the approximate location of these sequences in the tree is of interest, as the phylogenetically informative SNPs that determine the branching order are likely to be valid. Also, all 55 genomes may offer useful sequence information in particular genes of interest. However, potential problems in these strains should be considered for any subsequent analysis. In particular, polymorphisms relating to drug-resistance, and further studies based on these whole genomes should include manual inspection of the alignments.
The reconstructed phylogeny of 55 whole genomes (Figure 1A) was compared to a phylogeny based on a minimal set 45 SNP positions determined by Filliol et al1 to be sufficient to resolve and differentiate Mtb and M.bovis isolates (Figure 1B). In general, all main lineages were preserved and there is a high level of correspondence between the two trees. Two of the three Beijing strain sub-lineages present in the whole genome tree converged into 1 sub-lineage in the 45-SNP tree. Additionally, F11, the KZN strains, and the SUMu strains from Canada were related but clearly distinguishable on the whole genome tree, but they were collapsed on the 45-SNP trees. In contrast, one sub-lineage of the Beijing genotype formed its own lineage on the 45 SNP trees (orange cluster, Figure 1A, 1B). Similarly, H37 and close SUMu strains from Canada appeared further from F11, KZN and other SUMu strains from Canada on the 45-SNP tree than they appeared on a whole genome tree. Taken together, this confirms that the 45 positions determined by Filliol et al can successfully resolve the same main lineages that appear in the whole genome analysis and thus can be used for initial characterization of isolates, but WGS provides added resolution which may be of value to applications that require finer resolution data.
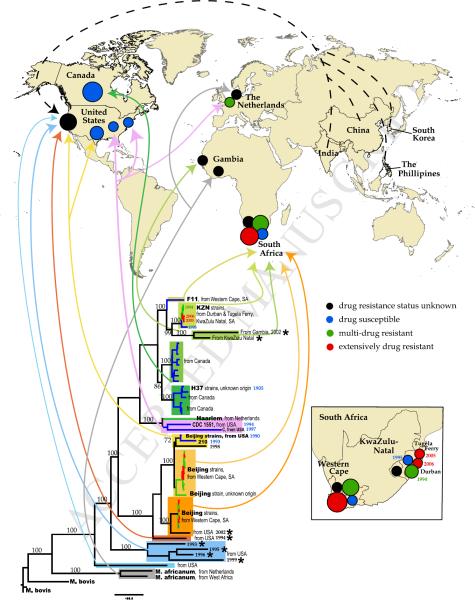
Whenever available for the near full-length genomes, we recorded isolate's sample history: the year and geographic location of isolate collection, and patient history including place of birth and drug resistance status (Table 1). This allowed us to relate the isolates phylogeny with their geographic location and the date of sampling (Figure 2). In agreement with Filliol et al, the 55 whole genomes fall into 7 major lineages. We found 4 distinctive sub-lineages of the Beijing strain: 2 sub-lineages formed from strains isolated in USA in 1990–1998 and 2 sub-lineages formed from strains isolated in Western Cape, South Africa. Perhaps not surprisingly, one of the South African Beijing lineages also includes a strain isolated in San Francisco in 2002 (Table 1, Figure 2).
Figure 2. Phylogenetic lineages and geographic mapping.
Whenever available for the near full-length genomes, we recorded isolate's sample history: the year and geographic location of isolate collection, and patient history including place of birth and drug resistance status (Table 1). This allowed us to relate the isolates phylogeny with their geographic location and the date of sampling. Lower part of the figure: same phylogenetic tree that as shown in Figure 1A. The numbers on the brunches represent bootstrap values obtained using 200 bootstrap replicates. Sequence names are removed except several reference strains, but the most relevant information (genotypes, isolation place, year of sampling) is noted on the tree. As in Figure 1A, sequences that are highly enriched for clusters of potentially problematic bases (Table 1) are indicated with the stars. Each lineage is shown with a distinct background box color, which corresponds to the colors of Figure 1. The sub-lineages are shown with the shades of the same background box color. For example, the Beijing strain-related lineage is shown in shades of yellow and orange. Additionally, drug resistance status is indicated by branch color. 4 colors for branches are used: black – drug resistant status of the strain is unknown; blue -- drug susceptible (DS); green – mono-resistant to INH or multi-drug resistant (MDR); red – extensively drug resistant (XDR), or pre-XDR (resistant to either fluoroquinolones or aminoglycosides). Arrows that are the same color as the background boxes from which they originate show the geographic places of the sequence isolation on the world map at the upper portion of the figure. The black, blue, green and red circles on the map correspond to the drug status of the isolates, the same coloring scheme as in the tree. The relative size of the circles corresponds to the number of whole genomes isolated in the particular location. The dashed black arrows pointing to the black circle in California represent isolates that were sequenced in San Francisco from patients that were born in India, China, South Korea and The Phillipines. The more detail geographic distribution of whole genome isolates from South Africa is shown on insert at the right lower corner of the figure.
In agreement with previous studies1,2,9, the tree revealed large genetic distance between isolates that were designated related to the Beijing strain. The distances between different strains within the Beijing sub-lineages appear to be roughly comparable to the distances between different F11 and KwaZulu Natal (KZN) lineages, hence the latter are marked by the same light green background color on Figure 2. The distances between the F11 / KZN sub-cluster and Canadian SUMu sequences and H37Rv and H37Ra (colored in distinct shades of green on Figure 2) are comparable to the distances between Beijing sub-clusters. Thus the genetic distances in the hierarchy of relationships in TB lineages are not always consistently represented by commonly used nomenclature conventions and reference strains.
2) Local Diversity
WGS derived phylogenies build a framework upon which questions about evolution, transmission, and drug resistance can be asked. The last of these is of particular interest as drug resistant strains have stymied the treatment of tuberculosis, leading to the need for novel therapeutics and carefully designed treatment regimens. When different levels of drug resistance are observed among highly related strains, such as the KZN strains and the Beijing strains from the Western Cape of South Africa, it provides an opportunity for improved clarity and resolution in mapping the acquisition and spread of drug resistance.
Using such comparisons, it has become clear that highly drug resistant strains are emerging independently in the same geographic locale to a greater degree than previously appreciated. For example, in the Western Cape region of South Africa, the Beijing XDR strains are closely intermingled with MDR strains (Figure 2). Standard genotyping methods suggested that the Beijing XDR strains emerged once but were undergoing clonal expansion and transmission in the region. However, WGS-based phylogenetic analysis revealed the independent appearance of distinct XDR resistance mutations within different MDR sub-lineages of the Beijing genotype4. Similarly, in KwaZulu Natal, South Africa, the XDR KZN strains, isolated in 2005 and 2006, have a slightly longer phylogenetic distance from their most recent common ancestor of the KZN lineage than the MDR KZN strains, isolated in 1994, suggesting at first glance that the XDR KZN strains evolved stepwise from the MDR strains. However, upon further inspection of the WGS data, the XDR strains and MDR strains have different rifampicin and pyrazinamide resistance mutations, indicating that these strains emerged independently from monoresistant isolates3.
The high resolution of WGS based phylogenetic analysis has been informative in other public health settings, most notably in the context of outbreak tracing. A recent outbreak of Mtb was detected in Vancouver, British Columbia, and by standard typing methods, it was defined as a single clonal outbreak5. The authors initially applied MIRU/VNTR to determine a source case and transmission chain; however, the MIRU-VNTR pattern was identical across all isolates and the addition of contact tracing did not reveal a source case. The authors sequenced the genomes of 36 isolates, 32 from the outbreak and 4 historical isolates from the region with an identical MIRU-VNTR pattern. Concatenation of polymorphic loci and subsequent phylogenetic analysis revealed a dendrogram with two primary branches, indicating two distinct transmission chains. Overlaying the phylogenetic analysis with a social network analysis, Gardy et al identified the transmission chain through which the infection spread. This study serves as the prototype for the use of WGS in outbreak tracing for Mtb, demonstrating it's effectiveness particularly in low-endemic countries where even high resolution (24-loci) MIRU-VNTR is likely to be uninformative.
3) Individual diversity
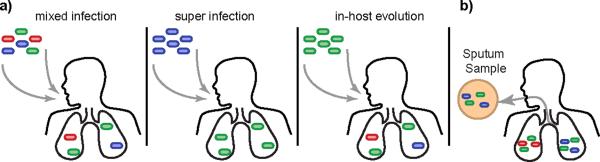
The resolution provided by WGS also allows researchers to investigate long-standing assumptions about the nature of tuberculosis. While Mtb infection is typically thought to be clonally homogeneous, recent studies have challenged this idea suggesting there is more heterogeneity within the infecting bacterial population than expected11,12. There are several potential mechanisms by which bacterial population heterogeneity might exist within a host – (a) an individual may be simultaneously infected by multiple strains, (b) an individual may be super-infected, or reinfected by a new strain, or (c) genetic diversity may arise in the bacterial population spontaneously during the course of infection (Figure 3). While the level of heterogeneity created by each of these mechanisms varies, WGS provides both the depth of coverage and sensitivity necessary to accurately assess population heterogeneity and begin to probe its functional consequences.
Figure 3.
A) In-host heterogeneity complicates the study of the genetic diversity and makes treatment and diagnosis of tuberculosis more difficult. There are several potential mechanisms by which heterogeneity might exist within the host – (a) a host may be simultaneously infected by multiple strains, (b) a host may be super-infected, or reinfected by a new strain, or (c) heterogeneity may arise spontaneously during the course of infection. B) Experimental results suggest that lesions are both genetically independent and genetically distinct, such that bacteria sampled from one lesion may not represent the true diversity present within the patient. Thus sputa samples are likely to under-estimate the amount of diversity present within a single patient, and serial sputa samples may not originate from the same lesion within the host.
Recurrent Mtb infection is a common clinical problem. Many studies have used a variety of genotyping techniques to identify differences between the original strain and the recurrent one. Where these strains are discordant, patients are typically assumed to have been infected with a second strain13–18. Indeed, mixed infections have been reported to occur in up to 54% of patients sampled19. Clinically, infection with multiple strains can lead to apparent differences in drug susceptibility20–22, which may only be one aspect of the differences between strains16 making diagnosis and treatment significantly more complicated. It is likely that with additional investigation and better sampling methods, mixed infections will be increasingly recognized, and the application of WGS will provide greater resolution in these studies, indentifying genetically distinct though highly related strains.
Within-host evolution of strains is another important source of genetic heterogeneity, and the principle source of de novo drug resistances. Saunders et al use deep resequencing of serial sputum isolates to characterize genetic diversity in a single patient infected with drug susceptible Mtb and in whom drug resistance evolved7. Serial isolates were obtained over a 12-month period from presentation with an initial drug susceptible infection through stepwise development of multiple drug resistance. Interestingly, only mutations conferring drug resistance were identified, with no additional mutations identified in non-repetitive regions. These results emphasize the strong bottleneck created by antibiotic exposure, and suggest that neither the host immune system nor exposure to antibiotics generates a hypermutable state in Mtb. However, limits in sampling and tracking in vivo populations of Mtb make it difficult to fully understand the evolutionary dynamics of patient isolates, and such isolates can only be obtained from patients with active disease making it difficult to assess the evolution of the bacterium throughout all stages of infection.
Indeed, until recently, it was assumed that Mtb has little capacity to acquire new mutations in the host because the bacterium is presumed to be quiescent through much of that time. However, by applying WGS to Mtb isolates from the cynomolgus macaque model, Ford et al. have recently shown that the mutation rate (mutations/bp/day) in active and latent disease is roughly equivalent, suggesting that bacteria continue to acquire genetic diversity, even during latency6. Additionally, their results suggest that lesions are both genetically independent and genetically distinct, such that bacteria sampled from one lesion may not represent the true diversity present within the patient (Figure 3b). Thus, if extrapulmonary dissemination occurs from any one lesion, the extrapulmonary sites would be genetically related to that lesion but distinct from others. These results have particular relevance when taken in the light of our primary sample source - sputa. Only bacteria present in cavitary lesions open to the airway will be present in sputa samples, and the cavitary lesions producing sputa may change dynamically during the course of infection as old lesions resolve and new lesions form. Future studies investigating in-host bacterial diversity may provide additional insight into these issues.
The overall picture of bacterial diversity is clearly a complex one – with diversity existing between patients, within a patient, and within a lesion. Mixed Mtb infection (whether by mixed inoculum, super-infection, or in-host evolution) raises a set of important questions. Could apparent phenotypic classifications (such as drug resistance) based on culture sometimes be incorrect due to mixed populations, leading to misdiagnosis in clinical settings? Can we look at key SNPs from pooled sweeps of colonies to estimate their frequencies in mixed cultures and rapidly detect low frequencies of drug resistance mutations in an individual? Finally, is Mtb superinfection enhanced in HIV high prevalent populations? For HIV-infected individuals, rapid HIV-1 depletion of Mtb-specific CD4 T cells can aggravate Mtb infections23, raising the possibility that immune dysfunction in HIV-infected people may make them more susceptible to serial Mtb infection, including superinfection with drug resistant strains. WGS will allow us to implement carefully crafted studies to address these important questions.
4) Bacterial diversity
The accumulation of WGS data allows us to assess the genetic diversity across the genome, seeking signatures of selective pressure. Selection can be quantified by relating the ratio of nonsynonymous genetic changes to synonymous changes (dN/dS), where a dN/dS greater than one is thought to reflect positive selection for increased diversity. Recently, Comas and colleagues used WGS to assess selection in a panel of 21 clinical strains24. Not surprisingly, essential genes had a lower dN/dS (0.53) than non-essential genes (0.66), indicating that while the genome overall is under purifying selection, essential genes are under greater purifying selective pressure. The authors then examined an experimentally defined set of T-cell epitopes25, hypothesizing that these might represent regions of increased functional diversity as a mechanism of immune evasion. Intriguingly, the T-cell epitopes analyzed appear to show the greatest amount of sequence conservation, with a dN/dS less than that of essential genes (0.25 for the epitope-coding region of the ORF). This may suggest that Mtb growth and transmission requires T-cell recognition, and therefore that the bacterium actually benefits from the host T-cell response. While further work is needed to clarify the dynamics of host-pathogen co-evolution in Mtb, these early results suggest that Mtb may depart from classic paradigms.
5) Challenges in WGS
While the capacity to perform low cost, high quality whole genome sequencing has transformed phylogenetic and population analyses in Mtb, there are some limitations with the current sequencing methodologies that can significantly skew our interpretation of the data. Most of the analyses described above hinge on the power of WGS to identify SNPs. Deletion or insertion of entire genes or large regions can be detected relatively easily (by absence of expected reads, or presence of novel contigs relative to a reference genome), and gene loss in particular has been frequently noted as a source of variability among mycobacteria26–28. However, polymorphisms in repetitive regions, gene duplications, chromosomal rearrangements, and copy-number changes of tandem repeats, are more challenging to detect by next-generation sequencing methods, such as Illumina, and can have significant biologic consequences. Paired-end read technology is quickly becoming the standard in generating WGS data, and offers some solutions to the problem of resolving these otherwise inaccessible regions.
Repetitive Regions
The limitations in sequencing repetitive regions apply to several genes and repeat elements scattered throughout the Mtb genome. These include some lipid biosynthetic genes as well as insertion elements, including IS6110, MIRUs and the clustered regularly interspaced short palindromic repeats (CRISPR) elements, which have been exploited extensively for strain typing. One example of the pitfalls associated with the sequencing of these regions has emerged from the debate over whether there is indeed purifying selection on T cell epitopes as suggested by Comas et al24. Uplekar et al. recognized that the genes encoding many of the ESX proteins, a family of secreted proteins that are represent strong CD4 and CD8 T cell antigens, were extremely homologous and thus poorly assessed by Illumina technology. Thus, the authors used Sanger sequencing to resequence these genes from a panel of clinical isolates and found, contrary to the previously mentioned report, that some of these genes are highly polymorphic, in part because of high levels of recombination29. When this diversity is taken into account, these antigens appear to be under diversifying selection. Similarly other important antigens including the PE and PPE genes are simply not accessible to short read sequencing technology and thus may obscure significant antigenic variation.
Genomic Duplications
Large-scale genomic duplications have been observed among mycobacterial strains, and in some cases, are postulated to have an influence on phenotype. For example, some members of the Beijing strain family have recently been found to have a large-scale duplication of ~350kb in the region of Rv3128c to Rv3427c30, including DosR, the transcriptional regulator of the hypoxic response31. This type of polymorphism is difficult to detect with short reads because there are multiple alternative ways to build contigs, producing ambiguity in assembly.
Current advances in data analysis, including de Bruijn graph methods32 and methods of statistical analysis33 are designed to detect signatures of large duplications, often using variations in depth of coverage to detect when large regions have been copied.
Genomic rearrangements
Genome rearrangements are difficult to detect with short reads because of the localized nature of the lesion. Genomic sequence on either side of the cross-over point might be well-covered by reads, but detection of the rearrangement point itself requires longer reads (e.g. from Roche 454) that span the discontinuity, or paired-end/mate-pair data as evidence of the connectivity. However, genome rearrangements have not been reported among M. tuberculosis strains (although there is a chromosomal inversion between M. tuberculosis and M. leprae34. The wild-type (drug-sensitive) KZN 4207 strain isolate from the KwaZulu-Natal region of South Africa was initially reported to have an inversion (http://www.broadinstitute.org/annotation/genome/mycobacterium_tuberculosis_spp/), although sequencing of the same strain in another lab did not find evidence for this inversion3. In practice, the large-scale genomic stability of Mtb justifies the use of a comparative assembly approach35 in which sequencing data for new M. tuberculosis strains are aligned against H37Rv, F11, or the genome sequence of another representative strain for comparative analysis.
6) Conclusion
Because of the paucity of genetic diversity in Mtb, WGS is a uniquely powerful tool, providing both the sensitivity to detect rare genetic events, and the broad applicability to detect multiple forms of genetic change. Already, we have learned a great deal about the bacterium and the nature of the disease. Early reports show surprising amounts of heterogeneity in bacterial populations, even between strains with identical MIRU/VNTR, RFLP patterns, or spoligotypes. Many questions remain, however. Perhaps chief amongst these is the true nature of in-host diversity: its clinical consequences and the mechanisms behind it. WGS has the capacity to address these and other questions with minimal bias and unprecedented sensitivity. However, it will be important to remember that current WGS methodologies do not cover all regions of the genome and there is likely to be important biology hidden in these uncharted areas.
BOX : Questions to be addressed by WGS
-
1)
Could apparent phenotypic classifications (such as drug resistance) based on culture sometimes be incorrect due to mixed populations, leading to misdiagnosis in clinical settings?
-
2)
Can we look at key SNPs from pooled sweeps of colonies to estimate their frequencies in mixed cultures and rapidly detect low frequencies of DR mutations in an individual?
-
3)
What is the true extent of Mtb in-host heterogeneity, and is it enhanced in HIV positive populations?
-
4)
Are certain hosts or host microenvironments more conducive to the development of genetic diversity?
-
5)
How can sequencing technology be modified and enhanced to allow for the complete de-novo assembly of the Mtb genome, including GC-rich repetitive regions?
-
6)
Given the relatively limited sampling of XDR Mtb strains (Figure 2), the global dissemination of XDR Mtb (provide WHO web link?), and the fact that drug resistance can evolve independently in individuals under treatment, will novel mechanisms of drug resistance be identified through more extensive sampling and contrasting drug resistance profiles using WGS?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Filliol I, Motiwala AS, Cavatore M, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188(2):759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershberg R, Lipatov M, Small PM, et al. High Functional Diversity in Mycobacterium tuberculosis Driven by Genetic Drift and Human Demography. Plos Biol. 2008;6(12):e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioerger TR, Koo S, No E-G, et al. Genome analysis of multi- and extensively-drug-resistant tuberculosis from KwaZulu-Natal, South Africa. PLoS ONE. 2009;4(11):e7778. doi: 10.1371/journal.pone.0007778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ioerger TR, Feng Y, Chen X, et al. The non-clonality of drug resistance in Beijing-genotype isolates of Mycobacterium tuberculosis from the Western Cape of South Africa. BMC Genomics. 2010;11(1):670. doi: 10.1186/1471-2164-11-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardy JL, Johnston JC, Sui SJH, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364(8):730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 6.Ford CB, Lin PL, Chase MR, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43(5):482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunders NJ, Trivedi UH, Thomson ML, et al. Deep resequencing of serial sputum isolates of Mycobacterium tuberculosis during therapeutic failure due to poor compliance reveals stepwise mutation of key resistance genes on an otherwise stable genetic background. J. Infect. 2011;62(3):212–217. doi: 10.1016/j.jinf.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Rozo-Anaya JC, Ribón W. Molecular tools for Mycobacterium tuberculosis genotyping. Rev Salud Publica (Bogota) 2010;12(3):510–521. [PubMed] [Google Scholar]
- 9.Niemann S, Köser CU, Gagneux S, et al. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS ONE. 2009;4(10):e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0604s00. Chapter 6:Unit 6.4. [DOI] [PubMed] [Google Scholar]
- 11.García de Viedma D, Marín M, Ruiz MJ, Bouza E. Analysis of clonal composition of Mycobacterium tuberculosis isolates in primary infections in children. Journal of Clinical Microbiology. 2004;42(8):3415–3418. doi: 10.1128/JCM.42.8.3415-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín A, Herránz M, Serrano M, Bouza E, de Viedma D. Rapid clonal analysis of recurrent tuberculosis by direct MIRU-VNTR typing on stored isolates. BMC Microbiol. 2007;7(1):73. doi: 10.1186/1471-2180-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandera A, Gori A, Catozzi L, et al. Molecular epidemiology study of exogenous reinfection in an area with a low incidence of tuberculosis. Journal of Clinical Microbiology. 2001;39(6):2213–2218. doi: 10.1128/JCM.39.6.2213-2218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caminero JA, Pena MJ, Campos-Herrero MI, et al. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med. 2001;164(7):1165–1170. doi: 10.1164/ajrccm.164.7.2101031. [DOI] [PubMed] [Google Scholar]
- 15.Chaves F, Dronda F, Alonso-Sanz M, Noriega AR. Evidence of exogenous reinfection and mixed infection with more than one strain of Mycobacterium tuberculosis among Spanish HIV-infected inmates. AIDS. 1999;13(5):615–620. doi: 10.1097/00002030-199904010-00011. [DOI] [PubMed] [Google Scholar]
- 16.García de Viedma D, Marín M, Ruiz Serrano MJ, Alcalá L, Bouza E. Polyclonal and compartmentalized infection by Mycobacterium tuberculosis in patients with both respiratory and extrarespiratory involvement. J INFECT DIS. 2003;187(4):695–699. doi: 10.1086/368368. [DOI] [PubMed] [Google Scholar]
- 17.Nardell E, McInnis B, Thomas B, Weidhaas S. Exogenous reinfection with tuberculosis in a shelter for the homeless. N Engl J Med. 1986;315(25):1570–1575. doi: 10.1056/NEJM198612183152502. [DOI] [PubMed] [Google Scholar]
- 18.Small PM, Shafer RW, Hopewell PC, et al. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N Engl J Med. 1993;328(16):1137–1144. doi: 10.1056/NEJM199304223281601. [DOI] [PubMed] [Google Scholar]
- 19.Stavrum R, Mphahlele M, Ovreås K, et al. High diversity of Mycobacterium tuberculosis genotypes in South Africa and preponderance of mixed infections among ST53 isolates. Journal of Clinical Microbiology. 2009;47(6):1848–1856. doi: 10.1128/JCM.02167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan G, Post FA, Moreira AL, et al. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect Immun. 2003;71(12):7099–7108. doi: 10.1128/IAI.71.12.7099-7108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turett GS, Fazal BA, Justman JE, et al. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis. Clin Infect Dis. 1997;24(3):513–514. doi: 10.1093/clinids/24.3.513. [DOI] [PubMed] [Google Scholar]
- 22.Horn DL, Hewlett D, Haas WH, et al. Superinfection with rifampin-isoniazid-streptomycin-ethambutol (RISE)-resistant tuberculosis in three patients with AIDS: confirmation by polymerase chain reaction fingerprinting. Ann. Intern. Med. 1994;121(2):115–116. doi: 10.7326/0003-4819-121-2-199407150-00007. [DOI] [PubMed] [Google Scholar]
- 23.Geldmacher C, Ngwenyama N, Schuetz A, et al. Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J. Exp. Med. 2010;207(13):2869–2881. doi: 10.1084/jem.20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comas I, Chakravartti J, Small PM, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42(6):498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst JD, Lewinsohn DM, Behar S, et al. Tuberculosis (Edinburgh, Scotland) Vol 88. 2008. Meeting Report: NIH Workshop on the Tuberculosis Immune Epitope Database; pp. 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato-Maeda M, Bifani PJ, Kreiswirth BN, Small PM. The nature and consequence of genetic variability within Mycobacterium tuberculosis. J. Clin. Invest. 2001;107(5):533–537. doi: 10.1172/JCI11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosch R, Gordon SV, Marmiesse M, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99(6):3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsolaki AG, Hirsh AE, Deriemer K, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci USA. 2004;101(14):4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uplekar S, Heym B, Friocourt V, Rougemont J, Cole ST. Comparative genomics of esx genes from clinical isolates of Mycobacterium tuberculosis provides evidence for gene conversion and epitope variation. Infect Immun. 2011;79(10):4042–4049. doi: 10.1128/IAI.05344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domenech P, Kolly GS, Leon-Solis L, Fallow A, Reed MB. Massive Gene Duplication Event among Clinical Isolates of the Mycobacterium tuberculosis W/Beijing Family. J Bacteriol. 2010;192(18):4562–4570. doi: 10.1128/JB.00536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts DM, Liao RP, Wisedchaisri G, Hol WGJ, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. Journal of Biological Chemistry. 2004;279(22):23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci USA. 2001;98(17):9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiang DY, Getz G, Jaffe DB, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Meth. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisen JA, Heidelberg JF, White O, Salzberg SL. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1(6) doi: 10.1186/gb-2000-1-6-research0011. RESEARCH0011.1–0011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pop M, Phillippy A, Delcher AL, Salzberg SL. Comparative genome assembly. Brief. Bioinformatics. 2004;5(3):237–248. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]