Summary
The flagellum of Campylobacter jejuni provides motility essential for commensal colonization of the intestinal tract of avian species and infection of humans resulting in diarrheal disease. Additionally, the flagellar type III secretion system has been reported to secrete proteins such as CiaI that influence invasion of human intestinal cells and possibly pathogenesis. The flagellar regulatory system ultimately influences σ28 activity required for expression of the FlaA major flagellin and other flagellar filament proteins. In this work, we discovered that transcription of ciaI and four genes we propose annotating as feds (for flagellar co-expressed determinants) is dependent upon σ28, but these genes are not required for motility. Instead, the Feds and CiaI are involved in commensal colonization of chicks, with FedA additionally involved in promoting invasion of human intestinal cells. We also discovered that the major flagellin influences production, stability or secretion of σ28-dependent proteins. Specific transcriptional and translational mechanisms affecting CiaI were identified and domains of CiaI were analyzed for importance in commensalism or invasion. Our work broadens the genes controlled by the flagellar regulatory system and implicates this system in coordinating production of colonization and virulence determinants with flagella, which together are required for optimal interactions with diverse hosts.
Keywords: Campylobacter jejuni, σ 28, flagellar motility, commensalism, colonization
Introduction
Campylobacter jejuni colonizes both animal and human hosts to result in different outcomes of infection. C. jejuni is a natural commensal organism of the intestinal tract of many wild and domesticated animals, especially avian species. In chickens, C. jejuni promotes a prolonged asymptomatic colonization of the ceca and large intestines, which leads to contamination of poultry meats for human consumption (Beery et al., 1988; Friedman et al., 2004). During infection of humans, C. jejuni invasion of intestinal and colonic epithelial cells is thought to contribute to inflammation and destruction of the epithelium (van Spreeuwel et al., 1985). Both invasion and the host inflammatory response likely contribute to pathogenesis of disease, which ranges from mild, watery diarrhea to a severe, bloody diarrheal syndrome.
To understand factors required by C. jejuni to colonize hosts, we previously used a 1-day old chick model of commensalism to identify 29 C. jejuni genes necessary for wild-type levels of colonization of the chick ceca (Hendrixson & DiRita, 2004). Other studies revealed that C. jejuni requires specific transport systems, metabolic pathways, cytochrome c peroxidases, protein glycosylation systems, capsular polysaccharide production and fibronectin-binding proteins to promote commensalism (Jones et al., 2004; Karlyshev et al., 2004; Velayudhan et al., 2004; Bingham-Ramos & Hendrixson, 2008; Davis et al., 2009; Flanagan et al., 2009; Ribardo & Hendrixson, 2011). An ideal in vivo virulence model that mimics C. jejuni diarrheal disease in humans remains elusive. Instead, cell culture models of infection are available to assess the ability of C. jejuni to invade and survive within human small intestinal or colonic epithelial cells. One transposon mutant screen revealed the importance of oxidative stress resistance and fumarate metabolism in promoting C. jejuni entry into or survival within colonic epithelial cells (Novik et al., 2010).
Flagellar motility is one factor of C. jejuni required for both commensal colonization of poultry and infection of human volunteers (Black et al., 1988; Nachamkin et al., 1993; Wassenaar et al., 1993; Hendrixson & DiRita, 2004; Wosten et al., 2004). Furthermore, flagella and flagellar motility are required for interactions with and invasion of human intestinal epithelial cells (Wassenaar et al., 1991; Grant et al., 1993; Yao et al., 1994). In addition to secreting proteins for flagellar biosynthesis, the C. jejuni flagellar type III secretion system (T3SS) has been implicated in secretion of proteins required for interactions with eukaryotic cells (Konkel et al., 1999; Konkel et al., 2004; Song et al., 2004; Poly et al., 2007). Some of these proteins have been annotated as the Campylobacter invasion antigens (Cias) (Konkel et al., 1999). Production of Cia proteins has been reported to be induced by bile salts, with flagellum-dependent secretion of these proteins requiring either a factor produced by intestinal cells or components of serum (Rivera-Amill et al., 2001; Konkel et al., 2004; Malik-Kale et al., 2008). C. jejuni mutants lacking different Cia proteins (CiaB, CiaC, and CiaI) are reduced in their ability to invade or survive within eukaryotic cells (Konkel et al., 1999; Christensen et al., 2009; Buelow et al., 2011). However, conflicting reports exist on the universal requirement of Cia proteins for cell invasion among different C. jejuni strains (Goon et al., 2006; Novik et al., 2010). Other proteins of C. jejuni, such as FlaC and the FspA proteins, are secreted by the flagellum but are not dependent on bile salts for expression or serum for secretion. FlaC, a flagellin-like protein involved in motility, is required for full level of invasion of eukaryotic cells (Song et al., 2004). Two related FspA proteins are produced by different C. jejuni strains (Poly et al., 2007). Of these proteins, FspA2, but not FspA1 promotes apoptosis of eukaryotic cells.
As in many motile bacteria, regulation of flagellar gene expression and biosynthesis is a complex process in C. jejuni (Lertsethtakarn et al., 2011). Two alternative σ factors in C. jejuni control expression of flagellar genes encoding extracytoplasmic components of the flagellar organelle (Hendrixson et al., 2001). σ54 is required for expression of most flagellar rod and hook genes, whereas σ28 is required for expression of flaA, encoding the major flagellin, and other filament genes (Hendrixson & DiRita, 2003; Carrillo et al., 2004; Wosten et al., 2004; Wosten et al., 2010). The flagellar T3SS components (including FlhA, FlhB, FliP, and FliR), the FlgSR two-component regulatory system, and the FlhF GTPase form a regulatory system required to initiate transcription by σ54-RNA polymerase holoenzyme (Hendrixson & DiRita, 2003; Joslin & Hendrixson, 2008; Balaban et al., 2009; Joslin & Hendrixson, 2009; Boll & Hendrixson, 2011). A current model proposes that the FlgS histidine kinase may sense the formation or activity of the flagellar T3SS to result in activation of the FlgR response regulator for σ54-dependent expression of flagellar rod and hook genes (Joslin & Hendrixson, 2008, 2009). The FlhF GTPase is also essential for σ54-dependent flagellar gene expression, but specific details regarding its requirement are not yet known (Balaban et al., 2009). Like in other motile bacteria, activity of σ28 in C. jejuni is repressed by the FlgM anti-σ factor until the flagellar rod and hook are synthesized (Wosten et al., 2004; Wosten et al., 2010). After rod and hook formation, FlgM is secreted from the cytoplasm to relieve σ28 from repression, which results in flaA expression necessary for filament synthesis. Additional mechanisms involving FlhF and the putative FlhG ATPase spatially and numerically regulate flagellar biosynthesis so that only one flagellum is produced per bacterial pole (Balaban et al., 2009; Balaban & Hendrixson, 2011). These two proteins, along with the flagellar MS ring and switch complex also influence a mechanism to spatially control cell division (Balaban & Hendrixson, 2011).
A previously reported microarray analysis provided information regarding potential members of the σ28 regulon (Carrillo et al., 2004). This analysis not only identified genes encoding proteins known to be involved in motility, but also genes with functions other than in flagellar motility that potentially may be dependent on σ28 for expression (Carrillo et al., 2004; Goon et al., 2006). Genes encoding the FspA proteins described above are dependent on σ28 for expression (Poly et al., 2007). Cj0977 (encoded by Cjj81176_0996 in the C. jejuni 81-176 genome; for the remainder of this work, this gene will be referred to as 0996 to be consistent with the annotation of the 81-176 genome) is another member of the σ28 regulon initially not thought to be required for motility (Goon et al., 2006). However, further analysis revealed that 0996 is required for flagellar motility in liquid media, but not in semi-solid motility agar (Novik et al., 2010). This motility defect may explain the reduced invasion and pathogenicity observed with a C. jejuni 0996 mutant (Goon et al., 2006).
In this work, we provide an extensive analysis that establishes the σ28 regulon of C. jejuni and identifies a new class of C. jejuni virulence and colonization factors. We identified five σ28-dependent genes (including ciaI) that are not required for motility, but are required for wild-type levels of commensal colonization of poultry. In addition to CiaI, we found another σ28-dependent factor involved in invasion of epithelial cells. Further exploration of CiaI analyzed potential domains for a role in commensal colonization or invasion and revealed an influence of bile salts on translation of a specific ciaI mRNA. Due to their co-expression with flagellar genes and their requirement for wild-type levels of colonization, and in some cases virulence, we propose annotating four of these previously uncharacterized proteins as Feds (for flagellar co-expressed determinants). Furthermore, this work establishes the flagellar system of C. jejuni as a regulatory system required for expression of genes not only required for motility, but also for genes with broader functions than previously realized that include commensalism and virulence.
Results
fliA expression is partially dependent on the σ54 regulatory pathway
A previous analysis of flagellated and aflagellated C. jejuni NCTC11168 strains revealed that expression of fliA, encoding σ28, was partially reduced in the aflagellated mutant (Carrillo et al., 2004). In addition, a potential σ54-binding site was identified within flhG, which is two genes upstream of fliA. Therefore, we hypothesized that fliA may be a member of the σ54 regulon and examined if factors belonging to the σ54 regulatory pathway influence fliA expression.
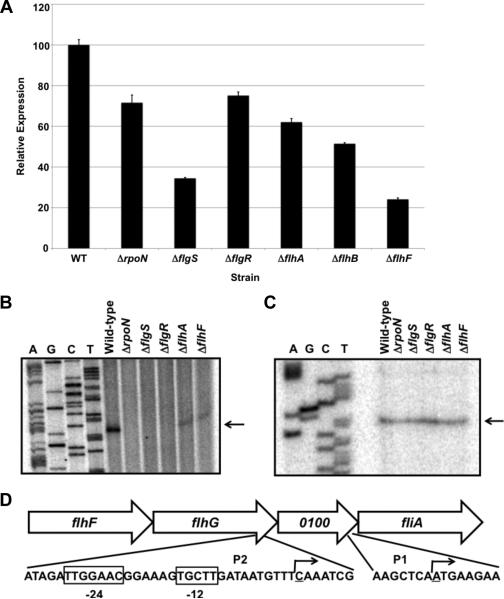
For all analyses in this work, we examined C. jejuni 81-176, a strain capable of infecting human volunteers and promoting commensal colonization of the chick cecum (Black et al., 1988; Hendrixson & DiRita, 2004). We compared expression of the fliA::astA transcriptional reporter in wild-type C. jejuni 81-176 SmR ΔastA and isogenic mutants lacking σ54 (ΔrpoN), the FlgSR two-component system (ΔflgS or ΔflgR), flagellar T3SS components (ΔflhA or ΔflhB), or the FlhF GTPase (ΔflhF). Expression of fliA::astA was reduced approximately ~25-80% in these mutants relative to wild-type C. jejuni (Figure 1A), suggesting that fliA is partially dependent on the σ54 regulatory pathway for expression. We identified two transcriptional start sites for fliA by primer extension analysis (Figures 1B and 1C). The transcriptional start site for the σ54-independent promoter (P1) was located 27 nucleotides upstream of the fliA start codon (Figure 1C and 1D). The transcriptional start site for a σ54-dependent promoter (P2) was located 382 nucleotides upstream of fliA and within the 3’ end of flhG (Figure 1B and 1D). Consistent with this observation, a highly conserved σ54-binding site was found directly upstream of the P2 transcriptional start site (Figure 1D).
Figure 1. Analysis of fliA expression and transcriptional start sites in wild-type and mutant C. jejuni strains.
(A) Arylsulfatase assays measuring expression of a fliA::astA transcriptional fusion in wild-type C. jejuni 81-176 SmR and isogenic mutant strains lacking a component of the σ54 regulatory pathway. The amount of fliA::astA expression in each strain is relative to wild-type C. jejuni, which was set to 100 units. Error bars indicate standard error of the average arylsulfatase activity analyzed from three samples. The reporter activity in each mutant was significantly different from the activity in the wild-type strain (P-value < 0.05). (B and C) Primer extension analyses to identify transcriptional start sites for fliA. Two different primers were used to identify transcriptional start sites dependent (B) and independent (C) of the σ54 regulatory pathway. Primer extension reactions were performed with RNA from C. jejuni 81-176 SmR or isogenic mutant strains lacking a component of the σ54 regulatory pathway. Reactions were run alongside and to the right of a sequencing ladder generated with the same primer used in primer extension reactions. Arrows indicate transcriptional start sites. (D) Location of the transcriptional start sites for fliA. The transcriptional start site generated from the σ54-dependent promoter (P2) is located within the 3’ end of flhG. Boxed nucleotides indicate conserved -24 and -12 binding sites for σ54. The transcriptional start site from the σ54-independent promoter (P1) is located immediately upstream of fliA and within Cjj81176_0100. The underlined nucleotides and bent arrows indicate transcriptional start sites.
Establishment of the σ28 regulon and identification of flagellar co-expressed determinants (Feds)
Initial data from a previous microarray analysis suggested involvement of σ28 and FlhA, a flagellar T3SS protein and component of the σ54 regulatory pathway, in the expression of approximately 30 genes, with some genes encoding proteins not previously associated with a role in flagellar motility in other motile bacteria (Carrillo et al., 2004). Therefore, we analyzed expression of these genes to determine if they indeed compose part of the C. jejuni σ28 regulon.
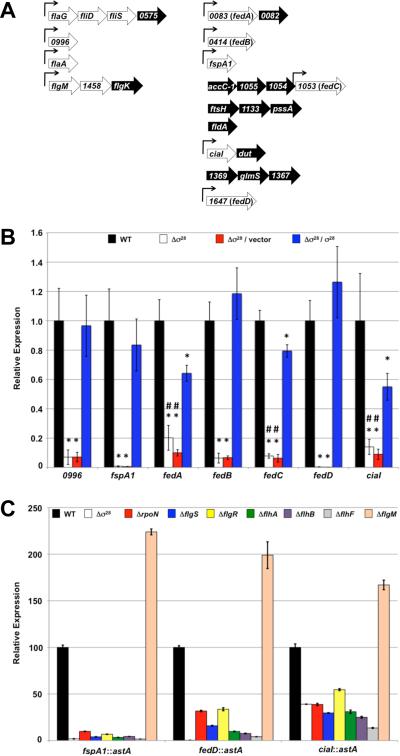
Expression of potential σ28-dependent genes in wild-type C. jejuni 81-176 SmR and an isogenic σfliA mutant (for the remainder of this report, fliA will be referred to as σ28 for clarity and to avoid confusion with flaA encoding the major flagellin) was compared by semi-quantitative real-time RT-PCR (qRT-PCR) (Figure 2A). Of 27 genes examined, expression of 12 genes was reduced 4- to over 100-fold in the Δσ28 mutant (Figure 2A and Table S1). Transcription of these genes increased upon complementation with a plasmid expressing Δ28 in trans. In addition, expression of fliS, encoding a putative flagellin chaperone, was reduced 25% in the Δσ28 mutant and complementation with σ28 in trans resulted in two-fold overexpression of the gene (Table S1). Transcription of the remaining 14 genes was not affected by deletion of σ28, and these genes were not further analyzed.
Figure 2. Identification and analysis of expression of the C. jejuni σ28 regulon.
(A) Individual genes and operons examined for potential dependence on σ28 for expression. Genes and operons were selected for investigation based on a previous microarray analysis that suggested at least partial dependence on σ28 and FlhA, a flagellar T3SS protein and component of the σ54 regulatory pathway, for expression (Carrillo et al., 2004). Numerical annotation of each gene and location of genes in potential operons are based on the C. jejuni 81-176 genome (Fouts et al., 2005). Genes in the left column are homologous to known flagellar genes or have been shown to be involved in flagellar motility of C. jejuni. Genes to the right are not homologous to any known flagellar genes. Genes in white were verified in this work to be at least partially dependent on σ28 for expression in C. jejuni 81-176 SmR. Genes in black were not found to depend on σ28 for wild-type levels of expression. Bent arrows indicate location of consensus σ28-binding sites in the promoter for each gene or operon. (B) Semi-quantitative real time RT-PCR (qRT-PCR) analysis of expression of 0996, fspA1, the fed genes, and ciaI in wild-type C. jejuni 81-176 SmR (black) and an isogenic Δσ28 mutant (white). Also included for analysis were the Δσ28 mutant strains complemented with vector alone (pECO102; red) or pECO102 expressing σ28 in trans (blue). Expression of each gene in each mutant is relative to the expression in wild-type C. jejuni, which was set to 1. All strains were examined in triplicate and the error bars indicate the standard error. Statistically-significant differences in gene expression between wild-type C. jejuni and Δσ28 mutant strains with or without σ28 in trans are indicated (* P-value < 0.05). Statistically-significant differences in gene expression between C. jejuni Δσ28 with σ28 expressed in trans and the Δσ28 mutants alone or with empty vector are indicated (# P-value < 0.05). (C) Arylsulfatase assays measuring expression of fspA1::astA, fedD::astA, and ciaI::astA transcriptional fusions in wild-type C. jejuni or mutant strains lacking σ28, FlgM, or a component of the σ54 regulatory pathway. Strains used in analysis include: wild-type C. jejuni 81-176 SmR (black) and isogenic mutants lacking σ28 (white), rpoN (encoding σ54; red), flgS (blue), flgR (yellow), flhA (green), flhB (purple), flhF (grey), and flgM (peach). The amount of expression of the transcriptional reporter in each mutant is relative to wild-type C. jejuni, which was set to 100 units. Error bars indicate the standard error of the average arylsulfatase activity analyzed from three samples. The reporter activities in each mutant were significantly different from the activity in the wild-type strain (P-value < 0.05).
Of the 13 σ28-dependent genes, seven encode proteins that either have been verified for a role in flagellar motility or are predicted to be involved in motility (Figure 2A). These genes include flaA (encoding the major flagellin; (Wassenaar et al., 1991; Grant et al., 1993; Nachamkin et al., 1993)), flaG (encoding a filament length control protein; (Kalmokoff et al., 2006)), fliD (encoding the putative filament cap; (Golden & Acheson, 2002; Konkel et al., 2004)), fliS (encoding the putative flagellin chaperone; (Golden & Acheson, 2002)), flgM (encoding the anti-σ factor that regulates σ28 activity; (Hendrixson & DiRita, 2003; Wosten et al., 2004; Wosten et al., 2010)), Cjj81176_1458 (encoding a possible chaperone for flagellar hook-associated proteins), and 0996 (encoding a protein required for motility in low viscosity media; (Novik et al., 2010)). However, six genes encode proteins with no homology to any known flagellar proteins. These genes and our proposed annotation based on additional findings described below include: fspA1 (Poly et al., 2007), ciaI (Buelow et al., 2011), Cjj81176_0083 (fedA), Cjj81176_0414 (fedB), Cjj81176_1053 (fedC), and Cjj81176_1647 (fedD) (Figure 2A). Similar to expression of fspA1, expression of the fed genes and ciaI was reduced 5- to 100-fold in the Δσ28 mutant compared to wild-type C. jejuni (Figure 2B and Table S1; (Poly et al., 2007)). Transcription of these genes was largely restored by complementation with σ28 in trans, with no statistically-significant differences in transcription of 0996, fspA1, fedB, or fedD between wild-type and complemented strains. Although expression of fedA, fedC, and ciaI did not reach wild-type levels upon complementation with σ28, transcription of these genes in the complemented strain was significantly higher than in the Δσ28 mutant with or without the empty vector (Figure 2B).
Considering that the feds and ciaI were dependent on σ28 for expression, we predicted that expression of these genes would decrease in σ54-regulatory pathway mutants. These mutants have reduced expression of σ28 and lack rod and hook biogenesis, which can inhibit residual σ28 activity due to cytoplasmic retention of the FlgM anti-σ factor (Figure 1A; (Hendrixson & DiRita, 2003; Wosten et al., 2004; Joslin & Hendrixson, 2008; Balaban et al., 2009; Joslin & Hendrixson, 2009; Wosten et al., 2010)). In addition, we predicted that transcription of these genes would increase in a ΔflgM mutant, due to derepression of σ28 activity (Wosten et al., 2004; Wosten et al., 2010). As expected, expression of fspA1::astA and fedD::astA transcriptional fusions was reduced approximately 3- to 62-fold (Figure 2C). Expression of ciaI::astA was more modestly reduced about 2- to 4-fold in σ54-regulatory pathway mutants and the Δσ28 mutant (Figure 2C). In addition, expression of these transcriptional fusions increased 67 to 123% in a ΔflgM mutant, which possesses augmented σ28-dependent activity due to lack of FlgM-mediated repression (Figure 2C).
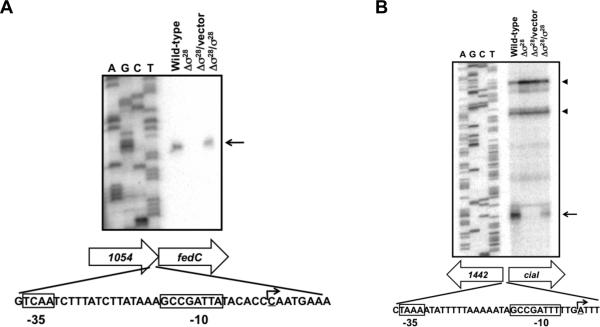
We characterized the promoter of fedC since a previous bioinformatic analysis did not identify a potential σ28-binding site upstream of this gene (Carrillo et al., 2004). In addition, we characterized the ciaI promoter because expression of this gene was not entirely dependent upon σ28. By primer extension analysis, we identified a single transcriptional start site 61 nucleotides upstream of the fedC start codon that was absent in the Δσ28 mutant and restored upon complementation with σ28 in trans (Figure 3A). Immediately upstream of the σ28-dependent fedC transcriptional start site and within the 3’ end of the upstream gene Cjj81176_1054, a potential σ28 consensus-binding site was identified (Figure 3A). Similarly, we identified a σ28-dependent transcriptional start site for ciaI 28 nucleotides upstream of the start codon of the gene (Figure 3B). As with the fedC promoter, a potential σ28-binding site upstream of this transcriptional start site was identified (Figure 3B; (Carrillo et al., 2004)). We also identified one or two other potential transcriptional start sites for ciaI further upstream and into the coding sequence of Cjj81176_1442 that were not dependent on σ28 (Figure 3B). These alternative transcriptional start sites may contribute to residual expression of ciaI in the Δσ28 mutant (Figure 2B and 2C and Table S1). These data are strong evidence that the fed genes and ciaI are members of the σ28 regulon and dependent on the flagellar regulatory system for expression.
Figure 3. Identification of transcriptional start sites for fedC and ciaI.
Primer extension analyses identified transcriptional start sites for fedC (A) and ciaI (B). Strains analyzed include wild-type C. jejuni 81-176 SmR and an isogenic Δσ28 mutant. Additional strains included in analysis were the C. jejuni 81-176 Δσ28 mutant complemented with vector alone (pECO102) or pECO102 expressing σ28Δ in trans. Primer extension reactions were performed using RNA from wild-type C. jejuni and mutant strains. Reactions were run alongside and to the right of a sequencing ladder generated with the same primer used in primer extension reactions. Arrows indicate transcriptional start sites generated from σ28-dependent promoters. Arrowheads include one or two possible transcriptional start sites generated from σ28-independent promoters for ciaI. Shown below each gel are the locations of σ28-dependent transcriptional start sites for fedC and ciaI. Boxed nucleotides indicate conserved -35 and -10 binding sites for σ28. The underlined nucleotides and bent arrows indicate transcriptional start sites.
The Fed proteins and CiaI are required for commensal colonization but not for flagellar motility
Although dependent on σ28 for expression, the fed genes and ciaI do not encode proteins with homology to any known motility proteins. FedA is most homologous to single-domain hemerythrins, which are diiron- and oxygen-binding proteins primarily found in anaerobic and microaerobic bacteria and some invertebrates (French et al., 2008). FedC contains a putative DnaJ-domain, suggesting the protein may be part of the Hsp70 chaperone machinery and involved in protein folding or degradation (Genevaux et al., 2007). FedB and FedD are not homologous to any proteins with known functions.
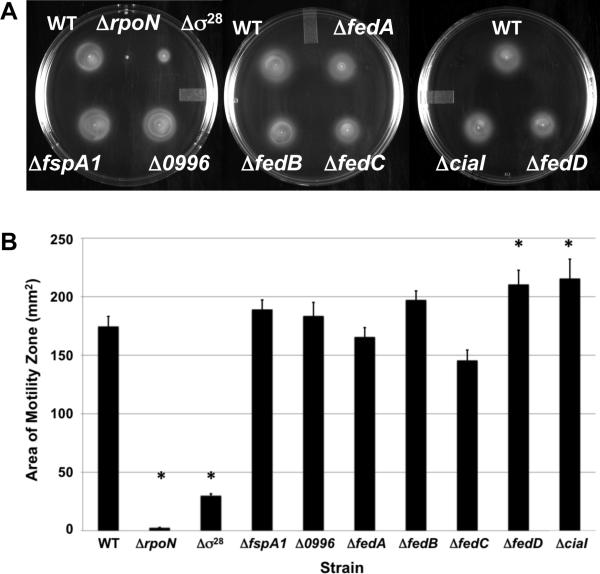
To characterize a role for the Fed proteins, CiaI, and other σ28-dependent proteins in the biology of C. jejuni, we constructed in-frame chromosomal deletions of each gene in C. jejuni strain 81-176 SmR. All strains grew similarly to wild-type C. jejuni in Mueller-Hinton (MH) broth at 37 °C in microaerobic conditions (Figure S1). We also examined the C. jejuni Δσ28 mutant as a control. We noted that the Δσ28 mutant demonstrated a growth defect in standing broth cultures, which may be due to the reduced motility of the mutant (Figure S1 and Figure 4A and 4B). As observed in semi-solid motility agar and in liquid broth by dark-field microscopy, no significant motility defects were observed in any of the Δfed mutants or the ΔciaI mutant (Figure 4A and 4B; data not shown). In fact, deletion of fedD and ciaI appeared to cause a slight, but statistically-significant, increase in motility (Figure 4B). As controls, we examined motility in the ΔrpoN mutant, which lacks σ54 and expression of many flagellar rod and hook genes, and the Δσ28 mutant, which expresses reduced levels of flaA (Table S1). Both mutants were defective for motility in motility agar (Figure 4A and 4B). Similar to previous observations (Novik et al., 2010), the C. jejuni Δ0996 mutant was non-motile in liquid broth, but motile in semi-solid media (Figure 4A and 4B; data not shown).
Figure 4. Motility phenotype of mutants lacking σ28-dependent genes.
Assays were performed by stabbing cultures of wild-type C. jejuni 81-176 SmR and isogenic mutants at similar optical densities in MH motility medium containing 0.4% agar. Plates were incubated in microaerobic conditions at 37 °C for 30 hours. (A) Motility of wild-type C. jejuni and each mutant strain in motility agar. (B) Area of motility zone for each wild-type and mutant C. jejuni strains as determined by averaging six assays. Error bars indicate the standard error of the average area. Statistically-significant differences in motility between wild-type C. jejuni and mutant strains are indicated (* P-value < 0.05).
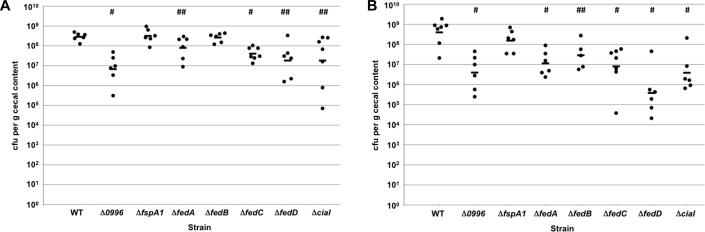
We previously found that a C. jejuni 81-176 SmR Δσ28 mutant was defective for commensal colonization of chicks (Hendrixson & DiRita, 2004). The colonization defect of this mutant was thought to be due solely to its greatly reduced motility phenotype. However, we considered if the fed genes or ciaI, which require σ28 for expression, may be necessary for colonization and contribute to the reduced colonization capacity of the Δσ28 mutant. For these experiments, we infected 1-day old chicks orally with either 104 or 102 cfu of wild-type C. jejuni or mutants lacking one of the feds or ciaI. At seven days post-infection, the levels of wild-type C. jejuni in the ceca of chicks given an oral inoculum of 104 cfu averaged 2.8 × 108 cfu per gram of cecal content (Figure 5A). In contrast, the ΔfedA, ΔfedC, ΔfedD, and ΔciaI mutants colonized at 4- to 16-fold lower levels than the wild-type strain in the ceca of chicks, which were statistically-significant differences (Figure 5A). However, the ΔfspA1 and the ΔfedB mutants did not show a colonization defect when administered at this inoculum. As expected, the Δ0996 mutant was reduced 42-fold for colonization compared to wild-type C. jejuni, which is likely due to its non-motile phenotype under certain conditions. When the inoculum was lowered to 102 cfu, all mutants except for the ΔfspA1 mutant showed statistically-significant colonization defects relative to wild-type C. jejuni. These colonization defects ranged from 14-fold lower for the C. jejuni ΔfedB mutant to over 1,000-fold lower for the ΔfedD mutant (Figure 5B). These data indicate that many of the σ28-dependent genes not required for motility are required for optimal commensal colonization of chicks.
Figure 5. Commensal colonization capacity of wild-type C. jejuni and mutant strains.
One-day old chicks were orally inoculated with approximately 104 (A) or 102 (B) cfu of wild-type C. jejuni 81-176 SmR or isogenic mutant strains. Each dot represents the amount of C. jejuni recovered from the ceca of each chick seven days post-infection. The geometrical mean for each group is depicted by the horizontal bar. Statistical analysis was performed using the Mann-Whitney U test (## P < 0.05; # P < 0.01).
FedA is involved in invasion of epithelial cells
Because a ciaI mutation in another C. jejuni strain was previously shown to possess a two-fold invasion defect for human intestinal epithelial cells (Buelow et al., 2011), we examined the fed mutants for defects in invasion of T84 colonic epithelial cells at six hours post-infection by using a standard gentamicin-protection assay. In these assays, approximately 2.2% of the wild-type C. jejuni inoculum was found intracellularly at the end of the assay (Table 1). Consistent with previous reports, the ΔciaI and Δ0996 mutants demonstrated 2- and 10-fold reductions in invasion, respectively (Goon et al., 2006; Buelow et al., 2011). Upon examination of each fed mutant, only the ΔfedA mutant showed a significantly reduced invasion capacity, which was approximately 10-fold lower than the wild-type strain. This invasion defect was similar to the levels of the Δσ28 mutant, which is minimally motile and expresses reduced levels of fedA (Figure 2B and Figure 4A and 4B). Thus, in addition to being a determinant for commensal colonization of chicks, FedA is also a virulence determinant required for invasion of human colonic epithelial cells.
Table 1.
Invasion capacity of wild-type C. jejuni 81-176 SmR and mutant strains for T84 colonic cells.
| Strain | Invasion of T84 cells (% inoculum)a |
|---|---|
| Wild-type | 2.23 ± 0.28 |
| Δ σ 28 | 0.16 ± 0.06* |
| Δ 0996 | 0.20 ± 0.05* |
| Δ fspA1 | 2.59 ± 0.15 |
| Δ fedA | 0.22 ± 0.05* |
| Δ fedB | 1.80 ± 0.34 |
| Δ fedC | 2.17 ± 0.38 |
| Δ fedD | 1.61 ± 0.30 |
| Δ ciaI | 1.11 ± 0.35* |
Percent invasion was determined by comparing the number of intracellular bacteria surviving a 2 h gentamicin treatment of infected T84 cells compared to the number of bacteria in the infecting inoculum (approximately 3.0 × 106 cfu). Each assay was performed in triplicate, and at least three biological replicates were performed. The average percent invasion +/- standard error for each strain is presented. Statistically-significant differences in invasion between wild-type C. jejuni and mutant strains are indicated
P-value < 0.05.
A ciaI transcript is dependent on DOC for translation
Previous analysis in C. jejuni strain F38011 suggested that transcription of ciaI and production of Cia proteins are augmented in the presence of the bile salt sodium deoxycholate (DOC) (Malik-Kale et al., 2008). However, our results presented above indicated that approximately 60 - 85% of ciaI transcription originates from the σ28-dependent promoter in C. jejuni 81-176 (Figure 2B and Table S1). Therefore, we analyzed if expression of ciaI from σ28-independent and -dependent promoters in C. jejuni 81-176 strains was increased in the presence of 0.1% DOC, a concentration previously shown to be required for CiaI production (Buelow et al., 2011). When wild-type and Δσ28 mutant strains were grown in the presence of DOC, we actually observed a 13 to 30% decrease in ciaI::astA expression, respectively (Figure S2). Growth in higher concentrations of DOC resulted in similar decreased levels of ciaI::astA expression (data not shown). Therefore, contrary to a previous report analyzing CiaI production in a different strain of C. jejuni, we were unable to link DOC to a mechanism regulating transcription of ciaI in C. jejuni 81-176 (Rivera-Amill et al., 2001).
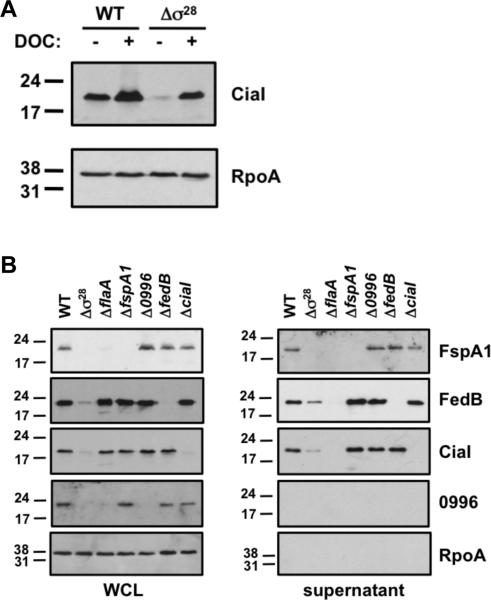
To determine if production of CiaI was influenced by DOC, proteins from whole-cell lysates of wild-type and Δσ28 mutant strains grown on media with and without 0.1% DOC were examined by immunoblot analysis. We observed an increase in CiaI levels in wild-type C. jejuni 81-176 SmR grown in the presence of DOC (Figure 6A). Furthermore, we discovered almost complete dependence on DOC for CiaI production in the Δσ28 mutant. Levels of RpoA, the α component of RNA polymerase, or other σ28-dependent proteins such as FspA1, FedB, and 0996 did not increase with growth in the presence of DOC (Figure 6A and data not shown), suggesting that the effect of DOC was specific for translation of CiaI. These findings suggest a DOC-dependent post-transcriptional mechanism that influences production of CiaI specifically from a σ28-independent transcript.
Figure 6. Analysis of production and secretion of CiaI and other σ28-dependent proteins.
(A) Production of CiaI in wild-type C. jejuni 81-176 SmR and an isogenic Δσ28 mutant. Strains were grown on MH agar in the absence (-) or presence (+) of 0.1% sodium deoxycholate (DOC). Equal amount of proteins in whole-cell lysates (WCL) were examined by immunoblot analysis using antiserum against CiaI or RpoA, the α subunit of RNA polymerase. Molecular weight markers are indicated in kDa. (B) Production of σ28-dependent proteins and CiaI in bacterial strains and supernatants after growth in MH broth. Wild-type C. jejuni 81-176 SmR and isogenic mutants lacking σ28, flaA (encoding the major flagellin), fspA1, 0996, fedB, and ciaI were grown in MH broth for 4 h at 37 °C in microaerobic conditions. WCL and supernatant proteins were recovered and analyzed by immunoblots using antiserum specific for each protein or RpoA. Molecular weight markers are indicated in kDa.
FedB and CiaI are secreted by the flagellum in the absence of serum
Secretion of CiaI and other Cias from C. jejuni strain F38011 was previously found to be dependent on both flagella and serum (Rivera-Amill et al., 2001; Buelow et al., 2011). In contrast, a previous investigation found that the FspA proteins, which are other σ28-dependent proteins, are secreted by the flagellum of C. jejuni 81-176 in the absence of serum (Poly et al., 2007). To analyze secretion of FspA1, FedB, CiaI and 0996, we generated or obtained antisera specific for these proteins. We were unable to generate antisera to FedA, FedC, and FedD as these proteins were refractory to purification or antisera generation. In addition, these proteins could not be detected with antiserum specific for a 6XHis tag as addition of this epitope to the N- or C-terminus of these proteins made the proteins unstable in C. jejuni (data not shown).
After growth of wild-type and mutant C. jejuni strains in MH broth alone, proteins from whole bacteria and supernatants were analyzed. We also analyzed an 81-176 SmR ΔflaA mutant, which lacks the major flagellin. Secretion of Cia proteins has been shown to be reduced in a C. jejuni F38011 flaA mutant (Konkel et al., 2004). As a negative control, we analyzed a Δσ28 mutant, which lacks these proteins or produces the proteins at greatly reduced levels (Figure 6B). As previously reported, FspA1 was secreted from wild-type C. jejuni 81-176 SmR in MH broth without addition of serum, but not from the ΔflaA mutant (Figure 6B; (Poly et al., 2007)). RpoA was only observed in the whole-cell lysates of these strains, suggesting that our procedures were adequate for recovering secreted proteins. In addition, the 0996 protein remained associated with bacteria as previously reported (Goon et al., 2006). We also noted that FspA1 and 0996 were absent in whole-cell lysate of the ΔflaA mutant (Figure 6B).
Contrary to a previous report, we observed that CiaI was secreted abundantly in MH broth alone without serum addition (Figure 6B; (Buelow et al., 2011)). Secretion of CiaI was dependent on flagella as this protein was absent from supernatants of the ΔflaA mutant. In addition, we found FedB to be secreted in MH broth alone in a flagellum-dependent manner (Figure 6B). In the absence of FlaA and secretion, both CiaI and FedB were stable in C. jejuni (Figure 6B). In addition, deletion of any σ28-dependent gene analyzed with the exception of flaA did not impair secretion or stability of other σ28-dependent proteins. These results identified FedB as a new flagellum-dependent secreted protein and suggest that serum-dependent secretion of CiaI is not a universal feature among C. jejuni strains. Furthermore, our results suggest that FlaA directly or indirectly plays a role in the production, stability or secretion of σ28-dependent proteins.
The potential nucleotide-binding domain of CiaI, but not the dileucine motif, mildly influences colonization and invasion
A previous study indicated that CiaI contains a dileucine motif that may be important for C. jejuni to promote invasion of eukaryotic cells (Buelow et al., 2011). This domain has been suggested to function as an endosomal-targeting motif. Analysis of CiaI-GFP ectopically produced in HeLa cells revealed a punctate distribution, but a GFP fusion to CiaI with a mutated dileucine motif appeared diffuse (Buelow et al., 2011). Together, these data suggested that the dileucine motif of CiaI may be important in localizing the protein to endosomal vesicles and influencing the biology or survival of intracellular C. jejuni. However, the effect of CiaI with a mutation of the dileucine motif on invasion when produced from C. jejuni was not examined (Buelow et al., 2011). In addition, bioinformatic analysis indicates that CiaI may also contain a putative ATP- or GTP-binding motif. We tested if either of these domains are required for CiaI to function as a commensal colonization factor or a virulence determinant for C. jejuni. Therefore, we replaced wild-type ciaI on the chromosome of C. jejuni 81-176 SmR with ciaIK42A, which is predicted to disrupt the nucleotide-binding motif, or ciaILL153-154AA, which disrupts the dileucine motif. We attempted to analyze potential ATP- or GTP-binding activity or hydrolysis with purified wild-type CiaI, but the recombinant protein did not show either activity in vitro (data not shown).
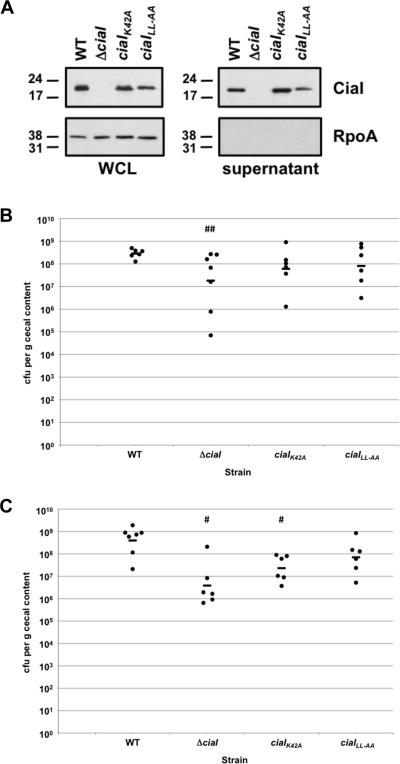
Immunoblot analysis revealed that the mutant proteins were produced and secreted from C. jejuni, with perhaps a slight decrease in production or stability of the CiaILL153-154AA protein (Figure 7A). Both mutants demonstrated a slight 4- to 5-fold decreased colonization capacity for chicks relative to wild-type C. jejuni 81-176 SmR when administered at an inoculum of 104 cfu (Figure 7B). However, these decreases were not significant compared to the level of colonization promoted by the wild-type strain. When administered at an inoculum of 102 cfu, the ciaIK42A mutant demonstrated a 17-fold decrease in commensal colonization, which was a statistically-significant difference from wild-type C. jejuni (Figure 7C). The ciaILL153-154AA mutant only showed a 6-fold decrease in colonization when administered at an inoculum of 102 cfu, which was not statistically significant. Neither mutant was as defective for commensal colonization as the ΔciaI mutant at either inoculum.
Figure 7. Analysis of the commensal colonization capacity of C. jejuni ciaI mutants.
(A) Production of CiaI proteins in bacteria and supernatants after growth in MH broth. Wild-type C. jejuni 81-176 SmR and isogenic mutants lacking ciaI, or containing ciaIK42A or ciaILL153-153AA (ciaILL-AA) were inoculated in MH broth and grown for 4 h at 37 °C in microaerobic conditions. Proteins from whole-cell lysates (WCL) and supernatants were recovered and examined by immunoblot analysis using antiserum specific CiaI or RpoA. For both WCL and supernatants, equal amount of proteins were loaded across strains. Molecular weight markers are indicated in kDa. (B and C) Commensal colonization capacity of wild-type C. jejuni 81-176 SmR and ciaI mutants. One-day old chicks were orally inoculated with 104 cfu (B) or 102 cfu (C) of C. jejuni strains. Each dot represents the amount of C. jejuni recovered from the ceca of each chick seven days post-infection. The geometrical mean for each group is depicted by the horizontal bar. Statistical analysis was performed using the Mann-Whitney U test (## P < 0.05; # P < 0.01). For wild-type C. jejuni and the ΔciaI mutant, the same data is shown as in Figure 5A and 5B.
We also tested the C. jejuni ciaI mutants for invasion of T84 cells. Whereas the ΔciaI mutant was reduced approximately two-fold for invasion, the invasion capacity of the ciaIK42A mutant was reduced only about 30%, which was not statistically significant (Table 2). Despite a previous report attributing the CiaI dileucine motif as being important for vesicular localization and possibly invasion, we did not detect a significant invasion defect of the C. jejuni ciaILL153-154AA mutant in vitro. These results suggest that the dileucine motif is likely not important for CiaI to promote invasion of C. jejuni. Instead, our results suggest that the putative nucleotide-binding domain of CiaI may have a limited role in commensal colonization or invasion by C. jejuni.
Table 2.
Invasion capacity of wild-type C. jejuni 81-176 SmR and ciaI mutant strains for T84 colonic cells.
| Strain | Invasion of T84 cells (% inoculum)a |
|---|---|
| Wild-type | 2.08 ± 0.22 |
| Δ ciaI | 1.28 ± 0.13* |
| ciaI K42A | 1.44 ± 0.27 |
| ciaI LL153-154AA | 1.74 ± 0.18 |
Percent invasion was determined by comparing the number of intracellular bacteria surviving a 2 h gentamicin treatment of infected T84 cells compared to the number of bacteria in the infecting inoculum (approximately 3.0 × 106 cfu). Each assay was performed in triplicate, and at least three biological replicates were performed. The average percent invasion +/- standard error for each strain is presented. Statistically-significant differences in invasion between wild-type C. jejuni and mutant strains are indicated
P-value < 0.05.
Discussion
Flagella and flagellar motility are well-established virulence and colonization factors of C. jejuni required for infection of humans to promote diarrheal disease and natural commensal colonization of avian species (Black et al., 1988; Nachamkin et al., 1993; Wassenaar et al., 1993; Hendrixson & DiRita, 2004; Wosten et al., 2004). Flagella not only provide chemotactic motility required for bacterial migration to proper replicative niches in hosts, but also likely facilitate initial contact of C. jejuni with human intestinal and colonic epithelial cells for subsequent invasion (Wassenaar et al., 1991; Grant et al., 1993; Yao et al., 1994). Furthermore, the flagellum and its secretory system have been implicated in secretion of some proteins that are not required for motility, including the Cia proteins, FspA proteins, and FlaC, which may modulate either interactions with or the biology of eukaryotic cells (Konkel et al., 2004; Song et al., 2004; Poly et al., 2007).
The intricate regulatory pathway that governs σ54-dependent gene expression in C. jejuni involves the flagellar T3SS, the FlgSR two-component system, and the FlhF GTPase (Hendrixson & DiRita, 2003; Wosten et al., 2004; Joslin & Hendrixson, 2008; Balaban et al., 2009; Joslin & Hendrixson, 2009). Because the σ54 regulon largely includes flagellar rod and hook genes (Boll & Hendrixson, 2011), disruption of the σ54 regulatory pathway results in an aflagellated and non-motile phenotype, which has consequences for the ability of C. jejuni to promote commensalism or disease in multiple hosts. In this work, we established a more direct role for the flagellar regulatory system in commensalism and pathogenesis of disease. We found that fliA, encoding σ28, is a member of the σ54 regulon of C. jejuni 81-176. Furthermore, in addition to σ28 being required for expression of flagellin and other filament genes, we discovered that σ28 is directly involved in expression of ciaI and four genes we annotated as feds. We found that CiaI and each Fed protein are required for wild-type levels of commensal colonization of poultry, and that CiaI and FedA are involved in invasion of colonic epithelial cells. Because expression of the feds and ciaI is largely dependent on σ28 and the σ54 regulatory pathway, our results broaden the number of genes whose expression is influenced by the flagellar regulatory system to include those involved in motility and also genes directly involved in commensalism and invasion.
Our findings indicate that a substantial proportion of genes within the C. jejuni σ28 regulon have functions other than in flagellar motility. Previous analysis revealed that C. jejuni fspA1 is a σ28-dependent gene, but a mutant lacking fspA1 did not demonstrate a noticeable in vitro motility defect (Poly et al., 2007). Similarly, we did not detect any in vitro motility defects in mutants lacking CiaI or any Fed proteins. Instead, we found these proteins are required for wild-type levels of commensal colonization of the chick ceca. Mutants lacking a single Fed protein or CiaI demonstrated 4- to over a 1000-fold commensal colonization defects. Furthermore, we confirmed the previously noted slight invasion defect of the ΔciaI mutant and found that the ΔfedA mutant possessed a 10-fold defect in invasion of T84 colonic cells (Buelow et al., 2011). The only σ28-dependent factor not found to be involved in motility, commensalism, or invasion was FspA1.
Through our analysis, we propose that the Feds and CiaI are a new collection of colonization and virulence factors co-expressed with flagella. Currently, we do not know how CiaI or each Fed functions for C. jejuni in vivo to promote optimal levels of commensal colonization of chicks. FedA, which we found to be involved in both commensalism and invasion, shares most homology to hemerythrins. These proteins are found mainly in anaerobic and microaerobic bacteria and some invertebrates, but the biological function of many bacterial hemerythrins is unknown (French et al., 2008). FedA contains a conserved domain that has been shown in a few characterized bacterial hemerythrins to bind iron and oxygen (Karlsen et al., 2005; Kao et al., 2008). As such, hemerythrins of anaerobes and microaerobes are predicted to function in biological processes involving iron or oxygen. Thus, we hypothesize that FedA may be involved in one or more iron- or oxygen-dependent activities for C. jejuni during commensalism or invasion. FedC contains a C-terminal region with homology to a DnaJ domain. Proteins with DnaJ domains often interact with DnaK or similar proteins to serve as a cochaperone in the Hsp70 chaperone machine, which assists in protein folding or degradation in bacteria (Genevaux et al., 2007). Mutants lacking the DnaJ-like cochaperone component of an Hsp70 chaperone system are often sensitive to thermal or oxidative stress. Due to the increased body temperature of avian species compared to humans (42 °C versus 37 °C), one possibility is that FedC may be required for folding or maintaining stability of one or more specific proteins essential for colonization of poultry. FedB and FedD do not share homology with any proteins of known function. Although we have ruled out in vitro motility defects of mutants lacking the Fed proteins, it is possible that the mutants possess motility defects in certain in vivo settings.
Our work uncovered some significant findings for CiaI that differed from prior investigations regarding its expression, production, and importance in C. jejuni biology. Previous work suggested that transcription of ciaI and production of the encoded protein are induced by DOC in C. jejuni strain F38011 (Malik-Kale et al., 2008; Buelow et al., 2011). Furthermore, secretion of CiaI was previously determined to be both flagellum- and serum-dependent (Buelow et al., 2011). A defect in invasion of human intestinal epithelial cells of a ciaI mutant was hypothesized to be due to CiaI localizing to and influencing development of C. jejuni-containing vacuoles for intracellular survival (Buelow et al., 2011). Ectopic production of CiaI in HeLa cells suggested that a dileucine motif in CiaI is essential for the protein to localize to vesicles and perhaps C. jejuni-containing vacuoles.
In this study, we discovered specific details regarding the regulated transcription and production of CiaI. First, we found that σ28 and the flagellar regulatory system are responsible for approximately 60 - 85% of the transcription of ciaI in C. jejuni 81-176. Consistent with this finding, we identified a σ28-dependent transcriptional start site and at least one possible σ28-independent transcriptional start site. Expression from neither promoter was induced upon growth in the presence of DOC, which counters a previous study that indicated ciaI transcription was increased when C. jejuni strain F38011 was grown with DOC (Rivera-Amill et al., 2001). Instead, we discovered that translation of CiaI was induced by DOC specifically from the σ28-independent transcript. These results suggest that a majority of CiaI is dependent on σ28 and the flagellar regulatory system for production, but not DOC. In addition, residual CiaI is produced by a DOC-dependent mechanism that influences translation of a ciaI mRNA that originates independently of σ28 and the flagellar regulatory system. It is interesting to speculate that DOC-dependent production of CiaI may be important for the bacterium in vivo when the flagellar regulatory cascade may be inactive and not promote expression of the σ28 regulon. Whether translation, rather than transcription of other Cia proteins, such as CiaB and CiaC, are induced by DOC remains to be determined. In additional analysis, we did not observe DOC to be required for production of any other σ28-dependent protein. Consistent with previous analysis of secretion of FspA1, we found that CiaI and FedB also did not require serum to be secreted in a flagellum-dependent manner in C. jejuni 81-176 (Poly et al., 2007). These results suggest that serum is not universally required for flagellum-dependent secretion among C. jejuni strains.
Our work also provides new insights into the role of CiaI in the biology of C. jejuni. We found that CiaI is a commensal colonization determinant as the ΔciaI mutant displayed 16- to over a 100-fold defects in cecal colonization of chicks. We were able to verify a mild, two-fold defect at an early step in invsion previously reported for the ΔciaI mutant in a different strain of C. jejuni (Buelow et al., 2011). However, the ciaILL153-154AA mutant did not demonstrate an invasion defect at 6 h post-infection, which suggest that the dileucine motif of CiaI previously determined to be required for vesicular-localization of the protein may not influence invasion. In addition, alteration of the dileucine motif did not influence the ability of C. jejuni to promote commensal colonization of chicks. We did note a mild invasion and colonization defect when the putative nucleotide-binding motif of CiaI was altered, but these defects of the ciaIK42A mutant were not as severe as the ΔciaI mutant. As a note, we were unable to verify an in vitro nucleotide-binding or -hydrolysis activity of recombinant CiaI. Even though CiaI appears to be involved in invasion of human intestinal cells, we believe that CiaI is likely required for a different function in commensalism. During colonization of chicks, C. jejuni primarily localizes to the mucosa layer atop the cecal and intestinal epithelium, with little invasion of epithelial cells evident (Beery et al., 1988). As a commensal in the intestinal tract of the natural avian host, we do not expect CiaI to be influencing an invasion mechanism of invasion for C. jejuni. The role of CiaI in colonization of poultry remains to be fully elucidated.
We also noticed a curious requirement for FlaA, the major flagellin of the flagellar filament of C. jejuni, in the production, stability or secretion of multiple proteins encoded by σ28-dependent genes. We found that both CiaI and FedB are dependent on FlaA for secretion. In the absence of FlaA, both proteins remain stably associated with the bacterium. We also found that FspA1 requires FlaA for either production or stability. As a result, FspA1 is not found in the whole-cell lysate or in the secreted protein fraction in the absence of FlaA. Lastly, the 0996 protein, which we and others did not find to be secreted (Goon et al., 2006), requires FlaA for either production or stability in C. jejuni. These results suggest an additional function of FlaA outside of its role as a flagellin composing the filament for flagellar motility. Currently, it is unknown if the effect of FlaA on these proteins is direct or indirect. For instance, it is possible that FlaA may need to be secreted first to alter the flagellar secretory system for secretion of other σ28-dependent proteins. Alternatively, these proteins may directly complex with FlaA for increased stability or secretion or FlaA may be involved in a mechanism influencing translation of the proteins. It is intriguing to speculate that in addition to being a secreted flagellin, FlaA may have some chaperone activity for other σ28-dependent proteins. This latter possibility may provide a reason why these proteins are part of the σ28 regulon and co-expressed with FlaA.
Previous work from our group identified a requirement of flagellar components for proper spatial regulation of division (Balaban & Hendrixson, 2011). In this work, we continued to expand our understanding of the requirements of flagella and the flagellar regulatory system in the biology of C. jejuni. This study demonstrates that the flagellar regulatory system of C. jejuni is directly required for expression of genes essential for broad biological functions, such as motility, commensalism and virulence. Furthermore, we established the Fed proteins and CiaI as a new class of colonization factors co-expressed with flagella. Due to the dependence of many of these proteins on FlaA for stability or secretion, our findings suggest a possible new function for the major flagellin in C. jejuni. Continued exploration will undoubtedly further contribute to our understanding of the global requirement of flagella for many diverse aspects of C. jejuni biology.
Materials and Methods
Bacterial strains and plasmids
All strains and plasmids used in this study are listed in Tables S2 and S3 in Supporting Information. C. jejuni strain 81-176 was originally isolated from a patient with gastroenteritis (Korlath et al., 1985). Subsequent studies verified the capacity of this strain to infect human volunteers and promote commensal colonization of chicks (Black et al., 1988; Hendrixson & DiRita, 2004). C. jejuni was typically grown in microaerobic conditions (85% N2, 10% CO2, 5% O) on Mueller-Hinton (MH) agar or in MH broth at 37 °C. As required, antibiotics were added to MH media at the following concentrations: 10 μg/ml trimethoprim (TMP), 20 μg/ml chloramphenicol, 50 μg/ml kanamycin, 30 μg/ml cefoperazone or 0.5, 1, 2, or 5 mg/ml streptomycin. All C. jejuni strains were stored at -80 °C in a 85% MH broth and 15% glycerol solution. For routine growth to perform most experiments, C. jejuni strains were grown from frozen stocks for 48 h in microaerobic conditions at 37 °C, then streaked on MH agar and grown for additional 16 h in identical conditions. Escherichia coli DH5α, XL1-Blue and BL21 strains were grown on Luria-Bertani (LB) agar or in LB broth containing 100 μg/ml ampicillin, 50 μg/ml kanamycin or 15 μg/ml chloramphenicol as appropriate. All E. coli strains were stored at -80 °C in a 80% LB broth and 20% glycerol solution.
Construction of mutants
C. jejuni mutants were constructed by electroporation following previously described methods (Hendrixson et al., 2001). For cloning of each gene to be deleted from the C. jejuni 81-176 SmR (DRH212) chromosome, DNA fragments containing approximately 750 bases upstream and downstream of each gene were amplified by PCR using primers containing 5’ BamHI restriction sites. Each fragment was then cloned into the BamHI site of pUC19 to create the following plasmids: pDRH3022 (pUC19::fspA1), pABT253 (pUC19::Cjj81176_0996), pABT115 (pUC19::ciaI), pABT249 (pUC19::fedA), pABT113 (pUC19::fedB), pABT328 (pUC19::fedC), and pABT111 (pUC19::fedD) (Table S3). To create restriction sites within the coding sequence of some genes, point mutations were introduced into plasmids by PCR-mediated mutagenesis (Makarova et al., 2000). These reactions created an EcoRV site in fspA1 (pDRH3025), a PmeI site in fedA (pABT322), and StuI sites in Cjj81176_0996 (pABT329), fedC (pABT355), and fedD (pABT152). A SmaI-digested cat-rpsL cassette was obtained from pDRH265 and ligated into plasmids in the appropriate restriction sites to interrupt each gene (Table S3; (Hendrixson et al., 2001)).
Each plasmid generated above was electroporated into C. jejuni 81-176 SmR (DRH212) to interrupt each respective gene on the chromosome with the cat-rpsL cassette. Transformants were recovered on MH agar containing chloramphenicol. Mutations were verified by colony PCR and the following isogenic mutants of 81-176 SmR were obtained: ABT103 (fspA1::cat-rpsL), ABT366 (Cjj81176_0996::cat-rpsL), ABT214 (ciaI::cat-rpsL), ABT353 (fedA::cat-rpsL), ABT261 (fedB::cat-rpsL), ABT370 (fedC::cat-rpsL), and ABT233 (fedD::cat-rpsL) (Table S3).
PCR-mediated mutagenesis was employed to construct in-frame deletions of genes that were originally cloned into pUC19 (see above and Table S3; (Makarova et al., 2000)). After sequencing to verify correct construction of in-frame deletions, the following plasmids were obtained: pABT205 (pUC19::ΔfspA1), pABT325 (pUC19::ΔCjj81176_0996), pABT173 (pUC19::ΔciaI), pABT280 (pUC19::ΔfedA), pABT357 (pUC19::ΔfedB), pABT356 (pUC19::ΔfedC), and pABT164 (pUC19::ΔfedD) (Table S3). These plasmids were electroporated into strains containing cat-rpsL interruptions of the respective genes on the chromosome. Transformants were recovered on MH agar with 0.5, 1, 2 or 5 mg/ml of streptomycin and then screened for chloramphenicol sensitivity. Deletion of each gene was verified by colony PCR, which resulted in creation of the following 81-176 SmR mutant strains: ABT361 (ΔfspA1), ABT501 (ΔCjj81176_0996), ABT279 (ΔciaI), ABT477 (ΔfedA), ABT473 (ΔfedB), ABT472 (ΔfedC), and ABT278 (ΔfedD).
Creation of 81-176 SmR ΔastA ΔflgM was accomplished by electroporation of 81-176 SmR ΔastA (DRH461) with pDRH552 to interrupt flgM on the chromosome with a cat-rpsL cassette (Hendrixson & DiRita, 2003). Transformants were recovered on MH agar containing chloramphenicol. Colony PCR verified interruption of flgM with the cat-rpsL cassette, resulting in 81-176 SmR ΔastA flgM::cat-rpsL (DRH557). This strain was then electroporated with pDRH565 to replace flgM::cat-rpsL with the in-frame ΔflgM mutation (Hendrixson & DiRita, 2003). Transformants were recovered on MH agar containing 0.5, 1, 2 or 5 mg/ml of streptomycin and then screened for chloramphenicol sensitivity. Deletion of flgM on the chromosome was verified by colony PCR to result in creation of 81-176 SmR ΔastA ΔflgM (DRH604).
To create plasmids containing astA transcriptional fusions to genes of interest, a SmaI-digested astA-kan cassette from pDRH580 was ligated into the EcoRV site of fspA1 in pDRH3025, the EcoRV site of ciaI in pABT115, the HpaI site of fliA in pDRH263, and the StuI site of fedD in pABT152 (Hendrixson & DiRita, 2003). As a result, astA transcriptional fusions were created in the following plasmids: pABT405 (fliA::astA-kan), pDRH3027 (fspA1::astA-kan), pABT119 (ciaI::astA-kan), and pABT236 (fedD::astA-kan). These plasmids were then electroporated into 81-176 SmR ΔastA (DRH461) and isogenic strains lacking σ28, FlgM, or components of the σ54 regulatory pathway. Transformants were recovered on MH agar containing kanamycin and acquisition of the astA transcriptional reporter at the native locus on the chromosome of each gene was verified by colony PCR (Table S2).
PCR-mediated mutagenesis was used to introduce point mutations in the coding sequence of ciaI in pABT115 to result in pABT673 (pUC19::ciaIK42A) and pABT674 (ciaILL153-154AA) (Makarova et al., 2000). These plasmids were electroporated into ABT214 to replace ciaI::cat-rpsL on the chromosome with genes encoding the mutant proteins. Transformants were recovered on MH agar containing 0.5, 1, 2 or 5 mg/ml of streptomycin and screened for chloramphenicol sensitivity. Putative transformants were verified by colony PCR and sequencing to result in 81-176 SmR ciaIK42A (ABT704) and 81-176 SmR ciaILL153-154AA (ABT706).
Gene expression analysis
Arylsulfatase assays were employed to measure the level of expression of astA transcriptional fusions located on the chromosome of C. jejuni strains as previously described (Henderson & Milazzo, 1979; Yao & Guerry, 1996; Hendrixson & DiRita, 2003). Each strain was analyzed in triplicate and each assay was performed three times. The level of expression of each transcriptional fusion in mutant strains was calculated relative to the expression in wild-type C. jejuni 81-176 SmR ΔastA, which was set to 100 units.
Semi-quantitative real time RT-PCR (qRT-PCR) was performed by extracting total RNA from wild-type and mutant C. jejuni 81-176 SmR strains with Trizol (Invitrogen) according to manufacturer's instructions. RNA was treated with DNaseI (GenHunter) and diluted to a final concentration of 30 - 50 ng/μl. For qRT-PCR analysis, 2.5 μg RNA was mixed with 0.2 μM forward and reverse primers and 0.1 μl Multiscribe reverse transcriptase along with Sybr green PCR mix (Applied Biosystems). A control sample was prepared by omitting the reverse transcriptase. A 7500 real-time PCR system (Applied Biosystems) was used to perform the reactions with the following conditions: 48 °C for 30 min and 95 °C for 10 min, then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Detection of gyrA or secD served as endogenous controls for normalization of results. Relative expression of each gene was calculated using the ΔΔCT method and reported as the level of expression compared to wild-type C. jejuni 81-176 SmR, which was set to 1. Each assay was performed in triplicate.
Primer extension analyses
RNA was isolated from wild-type C. jejuni 81-176 SmR and isogenic mutants. To identify transcriptional start sites for fliA, two primers were used: the primers (5’-TGTAAGAAAGCACAAGCTCA-3’) and (5’-AAAGGCTTCATCTATACTAA-3’) bound 94 bases upstream and 223 bases downstream of the start codon, respectively. For identification of the ciaI transcriptional start site, a primer (5’-CATCAAGATCATTTTGTGTG-3’) that bound 119 bases downstream from the start codon was used. The primer (5’-AAGTCTTTAAAATACTGAAA-3’) was used for analysis of the transcriptional start site of fedC, which bound 74 nucleotides downstream of the start codon. The primers were end-labeled with γ32[P]-ATP using polynuclotide kinase from the Excel Cycle-Sequencing kit (Epicentre Tech). The end-labeled primer was then mixed with RNA and Superscript II reverse transcriptase (Invitrogen) to generate labeled cDNA. The cDNA was analyzed on a 6% acrylamide gel by running alongside a sequencing ladder generated using the same end-labeled primer and a plasmid containing each gene. The gel was dried and analyzed using a Storm 820 Phosphorimager according to manufacturer's instructions (Amersham Biosciences).
Motility Assays
C. jejuni strains were suspended from plates in MH broth and diluted to an OD600 0.8. Each bacterial strain was stabbed into semisolid MH motility media containing 0.4% agar using an inoculation needle. The motility of each strain was tested six times. The plates were incubated for 30 h at 37 °C in microaerobic conditions and the area of the motility zone for each strain was calculated and averaged. For darkfield microscopy, the cultures were further diluted 1:10 in MH broth. Strains were immediately analyzed for motility by applying 3 μl of culture between a glass slide and glass coverslip.
Analysis of in vitro growth
Wild-type and mutant C. jejuni 81-176 SmR strains were suspended from plates in MH broth and diluted to an OD600 0.7 to 1.0. Fifty milliliters of MH broth with TMP were inoculated with 3 ml of each diluted bacterial culture and placed in 500 mL flasks. Flasks were then incubated at 37 °C in microaerobic conditions without shaking for 48 h. OD600 readings were taken at time 0, 4, 8, 24, and 48 h. The experiment was repeated three times and the OD600 readings at each time point were averaged.
Chick colonization assays
The ability of wild-type or mutant C. jejuni 81-176 SmR strains to colonize the ceca of chicks after oral inoculation was determined as previously described (Hendrixson & DiRita, 2004). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 d at 37.8 °C with appropriate humidity and rotation in a Sportsman II model 1502 incubator (Georgia Quail Farms Manufacturing Company). Approximately 12 to 36 h after hatching, chicks were orally infected with 100 μl of PBS or MH broth containing approximately 102 or 104 cfu of a single wild-type C. jejuni or mutant strain. To prepare strains for infection, C. jejuni strains were suspended from plates after growth at 37 °C in microaerobic conditions and diluted in PBS or MH broth to obtain the appropriate inoculum for oral gavage of chicks. Dilutions of the inocula were spread on MH agar to determine the number of bacteria in each inoculum. Seven days post-infection, chicks were sacrificed and the cecal contents were recovered and suspended in PBS or MH broth. Serial dilutions were spread on MH agar containing TMP and cefoperazone. Bacteria were grown for 72 h at 37 °C in microaerobic conditions and then counted to determine the cfu per gram of cecal contents.
In vitro invasion assays
Internalization of C. jejuni into T84 colonic epithelial cells was assessed using a gentamicin-protection assay. Semi-confluent monolayers of T84 cells (2.5 × 105 cells/ml) were seeded in 24-well tissue culture plates in DME/F12 (HyClone) with 5% FBS 24 h before infection. Wild-type and mutant C. jejuni 81-176 SmR strains were suspended from plates in MH broth to an OD600 0.4 and then diluted 1:10 in MH broth. Prior to infection, media was removed from the T84 cells and 300 μl of tissue culture media were added back to the cells. Monolayers were then infected with 15 μl of each diluted bacterial culture (~3 × 106 cfu per monolayer). Each inoculum was diluted and plated on MH agar to verify the actual number of bacteria used to infect each monolayer. Tissue culture plates were then centrifuged for 5 min at 960 rpm at room temperature to enhance contact between C. jejuni and colonic epithelial cells. The plates were then incubated for 4 h at 37 °C in a 5% CO2 incubator. T84 cells were washed three times with PBS and fresh tissue culture media containing 250 μg/ml of gentamicin was added to the monolayer. After a 2 h incubation at 37 °C in 5% CO2, cells were rinsed three times with PBS. Monolayers were released from the plates with 0.25% trypsin and the cells were disrupted by repeated pipetting. Serial dilutions were then spread on MH agar. After incubation for 72 h at 37 °C in microaerobic conditions, the number of internalized bacteria were determined. Percent invasion was determined by dividing the number of internalized bacteria by the number of bacteria in the inoculum.
Generation of antisera
Specific antiserum to C. jejuni proteins was generated from purified recombinant proteins with N- or C-terminal 6XHis- or maltose-binding protein (MBP)-fusions. For generation of a N-terminal 6XHis-tag to FspA1, primers containing 5’ in-frame BamHI restriction sites to codon 2 and the stop codon were used to amplify fspA1 from the C. jejuni 81-176 genome. The BamHI-digested PCR product was then ligated into pQE30, generating pABT363. This plasmid was then transformed into E. coli XL-1 Blue for protein induction and purification from the soluble fraction with Ni-NTA agarose according to manufacturer's instructions (QIAGEN).
Cjj81176_0996 was amplified from the C. jejuni 81-176 genome by PCR with a primer containing a 5’ in-frame NdeI restriction site fused to codon 2. The other primer contained a 5’ BamHI restriction site and an in-frame 6XHis-tag fused to the last codon of the gene. This PCR fragment was then digested with NdeI and BamHI and ligated into NdeI- and BamHI-digested pT7-7 to create pABT522. This plasmid was then transformed into E. coli BL21 (DE3) for protein production and purification from the soluble fraction with Ni-NTA agarose according to manufacturer's instructions (QIAGEN).
For purification of recombinant CiaI, primers containing 5’ in-frame BamHI restriction sites were used. In addition, one of the primers contained an in-frame 6XHis-tag to the last codon of ciaI. The BamHI-digested PCR product was cloned into pMal-C2x to create pABT613, which encoded a MBP-CiaI-6XHis tag protein. The plasmid was transformed into E. coli BL21 (DE3) cells for protein production and purification from the soluble fraction with amylose resin according to manufacturer's instructions (New England Biolabs). Each protein was then used to immunize five mice for production of polyclonal antisera (Cocalico Biologicals, Inc).
Analysis of production and secretion of σ28-dependent proteins
C. jejuni strains were suspended from MH agar plates into MH broth to an OD600 of 0.6. For each strain, 20 ml of diluted culture were incubated at 37 °C in microaerobic conditions without shaking for 4 h. At the end of the incubation period, final OD600 measurements were obtained. For preparation of proteins from whole-cell lysates (WCL), 1 ml of culture for each strain was pelleted in a microcentrifuge at full speed for 3 min, washed once with PBS and resuspended in 25 μl of PBS and 25 μl of 2X SDS-PAGE loading buffer for a total volume of 50 μl. For recovery of supernatant proteins, the remaining 19 ml of culture were centrifuged for 30 min at 13,000 rpm. The supernatants were recovered and the centrifugation step was repeated to ensure removal of all bacteria. Proteins present in the supernatant were precipitated by combining 18 ml of supernatant with 2 ml of trichloroacetic acid (TCA; 10% final concentration) followed by a 30 min incubation on ice. Precipitated proteins were recovered by centrifugation for 15 min at 10,000 rpm. The protein pellets were rinsed with 0.5 ml of cold acetone and dried. Precipitated proteins were resuspended in 20 μl of 1M Tris, pH 8.0 and 20 μl of 2X SDS-PAGE loading buffer.
All protein samples were boiled for 5 min prior to loading on 12.5% SDS-PAGE gels. For wild-type C. jejuni WCL, 10 μl of WCL (representing 200 μl of culture) were analyzed for detection of all proteins except FspA1 and FedB. For these latter proteins, 15 μl (for analysis of FspA1) and 2.5 μl (for analysis of FedB) of WCL were loaded onto 12.5% SDS-PAGE gels. The volumes of WCL of mutant strains loaded onto gels were normalized based on the final OD600 readings of wild-type and mutant strains to ensure analysis of equal amounts of proteins between strains. For supernatant samples, 10 μl of precipitated proteins were separated by 12.5% SDS-PAGE. Duplicate 12.5% SDS-PAGE gels were analyzed by silver staining to verify equal loading of proteins. For immunoblot analysis, primary murine antisera was used at the following concentrations to detect proteins: α-FspA1 M140, 1:1500 and 1:2000 for WCL and supernatant proteins, respectively; α-Cjj81176_0996 M151, 1:2000; α-CiaI M154, 1:2000; and α-RpoA M59, 1:2500 (Sommerlad & Hendrixson, 2007). For detection of FedB, polyclonal rabbit antiserum donated by Dr. Patricia Guerry (Naval Medical Research Center) was used at a dilution of 1:10,000 or 1:7500 for WCL and supernatant proteins, respectively. A 1:10,000 dilution of a horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit antiserum (Bio-Rad) was used as the secondary antibody. Immunoblots were developed by using the Western Lightning Plus ECL kit (Perkin-Elmer).
Statistical analysis
Tests for statistical significance in gene expression, motility and invasion assays were conducted by using the Student's t test (two-tailed distribution with two-sample, equal variance calculations). As indicated in figures or figure legends, statistically-significant differences between relevant strains possessed P-values < 0.05. For chick colonization assays, statistical analyses were performed by the Mann-Whitney U test, with statistically-significant differences between wild-type and mutant strains indicated with P-values < 0.01 or 0.05.
Supplementary Material
Acknowledgements
We thank Dr. Deborah Ribardo for assistance in chick colonization experiments. This work was supported by NIH grant R01AI065539 and associated research supplements 3R01AI065539-05S1 and 3R01AI065539-06S1.
References
- Balaban M, Hendrixson DR. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog. 2011;7:e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban M, Joslin SN, Hendrixson DR. FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J Bacteriol. 2009;191:6602–6611. doi: 10.1128/JB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beery JT, Hugdahl MB, Doyle MP. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl Environ Microbiol. 1988;54:2365–2370. doi: 10.1128/aem.54.10.2365-2370.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham-Ramos LK, Hendrixson DR. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect Immun. 2008;76:1105–1114. doi: 10.1128/IAI.01430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- Boll JM, Hendrixson DR. A specificity determinant for phosphorylation in a response regulator prevents in vivo cross-talk and modification by acetyl phosphate. Proc Natl Acad Sci U S A. 2011;108:20160–20165. doi: 10.1073/pnas.1113013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelow DR, Christensen JE, Neal-McKinney JM, Konkel ME. Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol Microbiol. 2011;80:1296–1312. doi: 10.1111/j.1365-2958.2011.07645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo CD, Taboada E, Nash JH, Lanthier P, Kelly J, Lau PC, Verhulp R, Mykytczuk O, Sy J, Findlay WA, Amoako K, Gomis S, Willson P, Austin JW, Potter A, Babiuk L, Allan B, Szymanski CM. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem. 2004;279:20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
- Christensen JE, Pacheco SA, Konkel ME. Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol Microbiol. 2009;73:650–662. doi: 10.1111/j.1365-2958.2009.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LM, Kakuda T, DiRita VJ. A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J Bacteriol. 2009;191:1631–1640. doi: 10.1128/JB.01394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan RC, Neal-McKinney JM, Dhillon AS, Miller WG, Konkel ME. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect Immun. 2009;77:2399–2407. doi: 10.1128/IAI.01266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CE, Bell JM, Ward FB. Diversity and distribution of hemerythrin-like proteins in prokaryotes. FEMS Microbiol Lett. 2008;279:131–145. doi: 10.1111/j.1574-6968.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Reddy S, Ahuja SD, Helfrick DL, Hardnett F, Carter M, Anderson B, Tauxe RV. Risk factors for sporadic Campylobacter infection in the United States: A case-control study in FoodNet sites. Clin Infect Dis. 2004;38(Suppl 3):S285–296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- Golden NJ, Acheson DW. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect Immun. 2002;70:1761–1771. doi: 10.1128/IAI.70.4.1761-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon S, Ewing CP, Lorenzo M, Pattarini D, Majam G, Guerry P. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect Immun. 2006;74:769–772. doi: 10.1128/IAI.74.1.769-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CCR, Konkel ME, Cieplak W, Jr., Tompkins LS. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson MJ, Milazzo FH. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. Journal of Bacteriology. 1979;139:80–87. doi: 10.1128/jb.139.1.80-87.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, Akerley BJ, DiRita VJ. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol. 2001;40:214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol. 2003;50:687–702. doi: 10.1046/j.1365-2958.2003.03731.x. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- Jones MA, Marston KL, Woodall CA, Maskell DJ, Linton D, Karlyshev AV, Dorrell N, Wren BW, Barrow PA. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect Immun. 2004;72:3769–3776. doi: 10.1128/IAI.72.7.3769-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslin SN, Hendrixson DR. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J Bacteriol. 2008;190:2422–2433. doi: 10.1128/JB.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslin SN, Hendrixson DR. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol. 2009;191:2656–2667. doi: 10.1128/JB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, Sanders G, Austin J, Kelly J, Szymanski CM. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J Bacteriol. 2006;188:4312–4320. doi: 10.1128/JB.01975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao WC, Wang VC, Huang YC, Yu SS, Chang TC, Chan SI. Isolation, purification and characterization of hemerythrin from Methylococcus capsulatus (Bath). J Inorg Biochem. 2008;102:1607–1614. doi: 10.1016/j.jinorgbio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Karlsen OA, Ramsevik L, Bruseth LJ, Larsen O, Brenner A, Berven FS, Jensen HB, Lillehaug JR. Characterization of a prokaryotic haemerythrin from the methanotrophic bacterium Methylococcus capsulatus (Bath). Febs J. 2005;272:2428–2440. doi: 10.1111/j.1742-4658.2005.04663.x. [DOI] [PubMed] [Google Scholar]
- Karlyshev AV, Everest P, Linton D, Cawthraw S, Newell DG, Wren BW. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology. 2004;150:1957–1964. doi: 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Bacterial secreted proteins are required for the internaliztion of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques. 2000;29:970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- Malik-Kale P, Parker CT, Konkel ME. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J Bacteriol. 2008;190:2286–2297. doi: 10.1128/JB.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachamkin I, Yang X-H, Stern NJ. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol. 1993;59:1269–1273. doi: 10.1128/aem.59.5.1269-1273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novik V, Hofreuter D, Galan JE. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect Immun. 2010;78:3540–3553. doi: 10.1128/IAI.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poly F, Ewing C, Goon S, Hickey TE, Rockabrand D, Majam G, Lee L, Phan J, Savarino NJ, Guerry P. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect Immun. 2007;75:3859–3867. doi: 10.1128/IAI.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribardo DA, Hendrixson DR. Analysis of the LIV system of Campylobacter jejuni reveals alternative roles for LivJ and LivK in commensalism beyond branched-chain amino acid transport. J Bacteriol. 2011;193:6233–6243. doi: 10.1128/JB.05473-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Amill V, Kim BJ, Seshu J, Konkel ME. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J Infect Dis. 2001;183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- Sommerlad SM, Hendrixson DR. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J Bacteriol. 2007;189:179–186. doi: 10.1128/JB.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YC, Jin S, Louie H, Ng D, Lau R, Zhang Y, Weerasekera R, Al Rashid S, Ward LA, Der SD, Chan VL. FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol Microbiol. 2004;53:541–553. doi: 10.1111/j.1365-2958.2004.04175.x. [DOI] [PubMed] [Google Scholar]
- van Spreeuwel JP, Duursma GC, Meijer CJ, Bax R, Rosekrans PC, Lindeman J. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut. 1985;26:945–951. doi: 10.1136/gut.26.9.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J, Jones MA, Barrow PA, Kelly DJ. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect Immun. 2004;72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, Bleumink-Pluym NMC, van der Zeijst BAM. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. Embo J. 1991;10:2055–2061. doi: 10.1002/j.1460-2075.1991.tb07736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, van der Zeijst BAM, Ayling R, Newell DG. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J Gen Microbiol. 1993;139(Pt 6):1171–1175. doi: 10.1099/00221287-139-6-1171. [DOI] [PubMed] [Google Scholar]
- Wosten MM, van Dijk L, Veenendaal AK, de Zoete MR, Bleumink-Pluijm NM, van Putten JP. Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol Microbiol. 2010;75:1577–1591. doi: 10.1111/j.1365-2958.2010.07079.x. [DOI] [PubMed] [Google Scholar]
- Wosten MMSM, Wagenaar JA, van Putten JPM. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J Biol Chem. 2004;279:16214–16222. doi: 10.1074/jbc.M400357200. [DOI] [PubMed] [Google Scholar]
- Yao R, Burr DH, Doig P, Trust TJ, Niu H, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol. 1994;14:883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]
- Yao R, Guerry P. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J Bacteriol. 1996;178:3335–3338. doi: 10.1128/jb.178.11.3335-3338.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.