Abstract
Non-coding RNAs (ncRNAs) are now recognized as active participants in controlling many biological processes. Indeed, these products of transcription can even control the process of transcription itself. In the past several years, ncRNAs have been found to regulate transcription of single genes, as well as entire transcriptional programs, affecting the expression of hundreds to thousands of genes in response to developmental or environmental signals. Compared to more classical protein regulators, the list of ncRNAs that regulate mRNA transcription in mammalian cells is still quite small; however, the rate at which new ncRNA transcriptional regulators are being discovered is rapid, suggesting that models for how gene expression is controlled will continue to be redefined as this field develops.
ncRNAs are key regulators of mammalian transcription
The paradigmatic view that the regulation of transcription is mediated solely by protein factors has changed. Indeed, this shift in established thought is occurring throughout biology as ncRNAs are increasingly demonstrated to be active participants in regulating fundamental processes in cells. Moreover, the advent of high-throughput sequencing technologies has revealed that the majority of DNA contained in mammalian genomes is transcribed [1–3]. This finding begs the question of whether all of these cellular transcripts have functions, or are simply the byproducts of transcriptional noise. Although issues such as these remain topics for debate, it is evident that the number of ncRNAs found to have biological function is rapidly increasing and that these functional ncRNAs act in a diversity of capacities, many of which directly control gene expression.
In this review, we discuss recent research on the functions of mammalian ncRNAs that regulate the first step of gene expression: transcription of protein-encoding genes by RNA polymerase II (Pol II). We limit our discussion primarily to ncRNAs that are ~200 nt in length or greater, and refer to these molecules as lncRNAs (long non-coding RNAs). The lncRNA transcriptional regulators can be loosely categorized into two groups: those that mediate changes in chromatin and those that modulate the activity of transcription factors, although the distinction between these categories is not black and white. The examples discussed below are not all-inclusive, but are meant to highlight emerging themes as the field of lncRNA transcriptional regulators continues to expand. In the interest of space, we do not discuss the examples of Xist (X-inactive specific transcript) and the lncRNAs that mediate genomic imprinting; they have been covered in recent reviews [4–6].
ncRNAs that control transcription by mediating changes in chromatin structure
The structure of chromatin (the assembly of DNA and histone proteins that constitute eukaryotic genomes) regulates the accessibility of DNA to Pol II and transcription factors, and therefore is integral to transcriptional control. Chromatin structure can be altered by specific post-translational modifications, such as methylation and acetylation, that are added to or removed from the histone proteins. For example, tri-methylation of histone H3 lysine 4 (H3K4me3) at promoters of genes and H3K36me3 in the transcribed regions are linked to gene activation, whereas H3K9me3, H3K27me3, and H4K20me3 are linked to repression [7].
Although enzymatic complexes responsible for placing the methyl marks on histones are largely defined, in many cases the mechanisms by which these complexes target specific regions of the genome, and the mechanisms by which domains of specifically modified chromatin are established, are not well understood. A growing body of literature suggests that ncRNAs are integral participants in these processes. For example, a functional genomics approach has revealed thousands of intergenic regions with chromatin signatures indicative of active transcription; their transcripts are referred to as lincRNAs (long intergenic non-coding RNAs) [8]. Strikingly, approximately a third of lincRNAs tested in human cells associate with chromatin modifying complexes [9], and many lincRNAs expressed in mouse ES cells control both pluripotency and differentiation, most likely by associating with chromatin modifying complexes [10]. In addition, many intronic and intergenic ncRNAs have been found by subjecting chromatin from human fibroblasts to RNA-seq [11], also suggesting that lncRNAs might be integral participants in controlling chromatin structure.
The examples below describe lncRNAs that regulate chromatin structure. A challenge in this emerging area is to determine mechanistically how these lncRNAs mediate the interplay between the enzymatic complexes that modify histones and the genome itself (Box 1 describes proposed models). ncRNA-mediated regulation of chromatin is not unique to Pol II genes; Box 2 describes an example for genes transcribed by RNA polymerase I in which the ncRNA recruits a chromatin modifying factor to specific loci by forming RNA/DNA triplexes (Box 2).
Box 1: Possible mechanisms for lncRNA-mediated chromatin modification.
Here we provide a summary of several proposed models for how lncRNAs could mediate the specific recruitment of chromatin modifying complexes to the genome, or control their activities (recently reviewed in [57]).
-
●
Proximity or regulation in cis. In this model, a ncRNA is transcribed from a locus adjacent to the region of the genome at which it functions to control chromatin modifications. The ncRNA, as it is being transcribed, can serve as a tether to recruit chromatin modifying complexes to that specific region of the genome. This mechanism of regulation might occur in systems in which an antisense transcript controls the expression of a sense transcript in a manner that is dependent on chromatin modifications and independent of the RNA interference (RNAi) machinery [58]. The current working model for the function of ANRIL involves cis regulation [19].
-
●
ncRNA-mediated looping. An ncRNA that is transcribed from one genomic locus could interact with chromatin modifying complexes bound to distal sites on the same chromosome, thereby creating an intrachromosomal loop to mediate long-range changes in chromatin structure. In this model, the three dimensional architecture of the assembly would be critical to controlling chromatin modification. HOTTIP RNA is thought to function via looping [21].
-
●
Formation of triple helixes. Specific sequences in ncRNAs could form triple helixes with specific DNA sequences, which might impart specificity in bringing a chromatin modifying complex bound to an ncRNA to sites on the genome. pRNA, which regulates transcription by Pol I (described in Box 2) is thought to function in this manner [59].
-
●
Tethering. Because various ncRNAs co-immunoprecipitate with chromatin modifying complexes from cellular extracts, ncRNAs might provide the link or tether to connect chromatin modifying complexes with each other and with DNA binding proteins or transcription factors that directly interact with chromatin.
-
●
Allosteric activation or repression of activity. It is possible that ncRNAs act as switches to either "turn-on" or "turn-off" the complexes that modify histone tails through a change in the conformation of the complex. The switch could impart changes both in the enzymatic activity and the specificity of DNA binding.
Box 2: Regulation of RNA polymerase I transcription by an ncRNA.
In mammalian genomes, rRNA genes exist in tandem arrays, many of which are transcriptionally silent and marked by heterochromatin and CpG methylation [60]. This silent state is maintained by NoRC, a multisubunit chromatin remodeling complex. NoRC associates with an ncRNA transcribed from the rDNA promoter region, known as pRNA (promoter-associated RNA) [61]. Depletion and overexpression experiments have shown that pRNA is required for the proper localization of NoRC, rDNA methylation, and a repressed chromatin state that prevents Pol I transcription.
Recently, ectopically expressed pRNA in mouse cells was found to trigger de novo methylation, recruitment of the DNA methyltransferase DNMT3b, and increased heterochromatic histone modifications (e.g. H4K20me3 and H3K27me3) at the rDNA promoters [59]. A specific sequence in pRNA was found to be both necessary and sufficient for targeting DNA methylation in the rDNA promoters. Intriguingly, this RNA sequence is similar to the sequence of an rDNA promoter element, the T0 region. When biotin-tagged pRNA variants were introduced into cells and subsequently recovered using streptavidin beads, endogenous rDNA co-purified, but only when the introduced pRNA variant contained the T0 region. In vitro experiments showed that the T0 region of pRNA could form a triplex with the T0 region of rDNA. Moreover, DNMT3b exhibits a preference for binding rDNA-pRNA triplexes compared to duplex rDNA. These data support a model in which the T0 region of pRNA forms a triplex with the rDNA promoter to recruit a DNA methytransferase. It remains to be determined whether DNA-RNA triplexes are a common mechanism that ncRNAs use for targeting epigenetic changes; this possibility is enticing given that triple-helix target sites are over-represented at human gene promoters [62].
HOTAIR
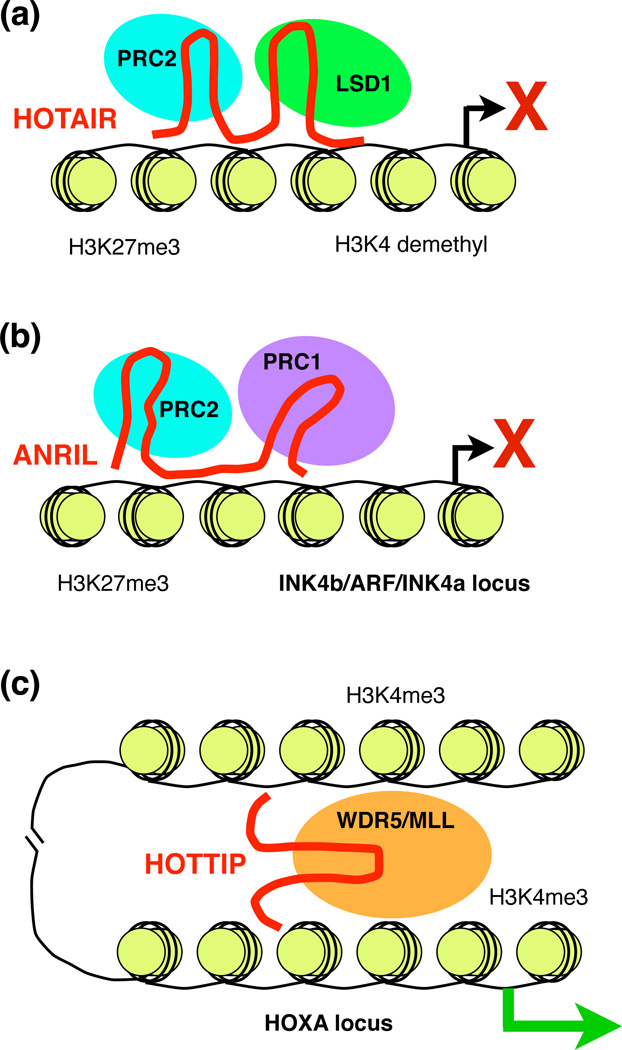
One of the earliest discovered and best characterized examples of an ncRNA that mediates changes in chromatin structure and gene expression is HOTAIR (Hox anti-sense intergenic RNA), a ~2.2 kb non-coding transcript made from the HOXC locus on chromosome 12 (Figure 1a). Hox loci contain groups of genes that together control the development of body segmentation in animals. HOTAIR was initially shown to act in trans (see Glossary) to silence transcription across the HOXD locus on chromosome 2 in human cells [12]. Knockdown of HOTAIR caused increased transcription, decreased H3K27me3, and decreased occupancy of the Polycomb group protein complex 2 (PRC2) at the HOXD locus. Moreover, HOTAIR co-immunoprecipitated with one of the subunits (SUZ12) of PRC2. These results led to a model in which HOTAIR is transcribed from the HOXC locus and recruits PRC2 to the HOXD locus via an unknown mechanism, leading to localized methylation of histone H3 (H3K27me3) and transcriptional silencing [12]. Knocking out HOTAIR in mice, however, resulted in no appreciable changes in expression or H3K27me3 occupancy patterns at the HOXD locus, suggesting either that mouse HOTAIR does not function similarly to human HOTAIR (the mouse and human HOTAIR sequences and proposed structures are poorly conserved) or that redundant mechanisms exist for controlling the epigenetic state and transcriptional activity of the HOXD locus in vivo [13]. Future mouse knockout experiments focusing on other lncRNAs will be important to test the in vivo significance of this new class of regulatory molecules (see Box 4).
Figure 1.
ncRNAs that regulate chromatin structure. (a) HOTAIR represses transcription by recruiting chromatin modifying complexes to specific regions of the genome. HOTAIR acts as a scaffold that can simultaneously bind PRC2 and LSD1, which methylate H3K27 and demethylate H3K4, respectively. (b) ANRIL represses transcription from the INK4b/ARF/INK4a locus by recruiting PRC1 and PRC2. The mechanism might involve an ANRIL transcript tethered to a Pol II elongation complex serving to recruit PRC1 and PRC2 [19]. (c) HOTTIP mediates long-range looping at one end of the HOXA locus, and activates transcription of genes by recruiting the histone modifier WDR5-MLL, which methylates H3K4.
Box4: Questions for future research.
How do lncRNAs regulate the proteins with which they interact? Although it is important to continue to identify new lncRNA regulators of transcription, it is equally important to better decipher the mechanisms by which the established ones function. Biochemical experiments are needed to confirm that individual lncRNAs bind directly to the proteins they likely regulate, and to determine how the association affects the activity of the protein.
What are the structures of lncRNAs bound to their protein targets? Currently there is a scarcity of structural information illustrating lncRNAs bound to their protein targets.. Crystal or NMR structures of lncRNA/protein complexes, even consisting of the minimal domains that interact, will provide highly valuable pictures of the complexes, which will enable experiments to directly test structure/function relationships and reveal similarities and differences between complexes.
Are lncRNAs that associate with histone modifying complexes localized with these complexes on chromatin genome-wide? Recently, two publications documented techniques to determine where across the genome specific lncRNAs are localized [64, 65]. The only mammalian lncRNA investigated that is pertinent to this article was HOTAIR, which was found to localized to >800 loci [64]. Although this technique has great potential to enable lncRNA research, questions about the connection between HOTAIR localization and function remain; for example, how HOTAIR affects gene expression at the loci to which it localized.
How can we best identify the functions of lncRNAs? With the growing accessibility of deep-sequencing technology, the number of reports identifying lncRNAs associated with a variety of proteins is rapidly increasing. The current challenge is to identify which of these interactions are important for biology and how the lncRNAs function. Knocking down lncRNAs is useful, however it is difficult to distinguish direct versus indirect effects. It will only be through the combination of multiple experimental techniques that the functions of individual lncRNAs will be understood.
What are the roles of lncRNAs in vivo? The vast majority of lncRNA studies are performed in cultured cells. The study in which HOTAIR was knocked out in mice revealed no appreciable changes at loci predicted to be under HOTAIR control [13]. This result raises many questions that need to be addressed by combining cell-based assays and experiments in model organisms. For example, do lncRNAs function differently in mice or is there redundancy built into lncRNA regulators?
Recently, it was proposed that HOTAIR serves as an RNA scaffold, with the ability to simultaneously bind multiple different histone modifying enzymes [14]. In addition to binding PRC2, HOTAIR was found to co-immunoprecipitate with LSD1, a demethylase that removes methyl groups from H3K4me2. LSD1 is part of the CoREST/REST repressor complex, which binds to regions flanking the HOXD locus, suggesting that HOTAIR might coordinate the targeting of both PRC2 and LSD1 to the same genomic region(s). In support of this, PRC2 was found to bind a 5' region of HOTAIR, whereas LSD1 bound a 3' region, and the two protein complexes co-immunoprecipitated from cells in a HOTAIR-dependent manner. Genome-wide experiments showed that knockdown of HOTAIR resulted in a coordinate loss of PRC2 and LSD1 at hundreds of genes, which also showed a corresponding increase in expression [14]. Because many lncRNAs are several kb in length and co-immunoprecipitate with chromatin modifying complexes [9], serving as scaffolds or platforms to coordinate the actions of multiple chromatin modifying complexes may prove to be a widely used mechanism of lncRNA function.
HOTAIR also functions to reprogram chromatin states during metastasis. A recent study showed that HOTAIR is systematically upregulated in breast carcinomas and its expression correlates with metastasis; moreover, overexpression of HOTAIR induced cellular invasiveness [15]. High HOTAIR expression also correlates with poor prognosis and metastasis in colorectal cancers [16]. To investigate the hypothesis that elevated HOTAIR levels could reprogram PRC2 occupancy in cancer, HOTAIR was overexpressed in breast cells and PRC2 occupancy was monitored by ChIP-chip [15]. Indeed, PRC2 occupied over 800 additional genes upon HOTAIR overexpression, and the pattern of occupancy changed from that typical of breast epithelial cells to a pattern that resembled that of embryonic fibroblast cells. Moreover, the genes that acquired PRC2 upon HOTAIR overexpression showed increased H3K27me3, decreased transcript levels, and are known to be downregulated in aggressive breast tumors.
ANRIL
ANRIL mediates changes in chromatin modifications that control gene expression across a locus critical to the progression of cancer, underscoring the impact of lncRNAs on cancer. The INK4b/ARF/INK4a locus in human cells encodes three important tumor suppressors that are deleted or silenced in a range of cancers [17]. Expression of these genes is controlled in part by PRC1, PRC2, and histone methylation [18]. An lncRNA called ANRIL (antisense noncoding RNA in the INK4 locus), which is transcribed antisense to the protein encoding genes in this locus, is upregulated in some cancer tissues [19]. ANRIL is a key player in controlling the epigenetic state of the INK4b/ARF/INK4a locus through interactions with subunits of PRC1 and PRC2 (Figure 1b). ANRIL was found associated with chromatin purified with an antibody against the CBX7 subunit of PRC1 [19], and ANRIL co-immunoprecipitated with SUZ12, a subunit of PRC2 [20]. Expression of antisense transcripts to ANRIL caused H3K27me3 and the occupancy of PRC1 and PRC2 to decrease, and levels of protein-encoding transcripts to increase at the INK4b/ARF/INK4a locus [19]. Expression of oncogenic Ras repressed expression of ANRIL and stimulated expression of tumor suppressor genes in the INK4b/ARF/INK4a locus [20].
The interplay between PRC1-RNA interactions and PRC1-histone interactions has been studied using pieces of ANRIL, the CBX7 subunit of PRC1, and methylated H4K27 peptides [19]. CBX7 uses distinct regions and residues to bind to either RNA or methylated H3K27. Although binding RNA or methylated peptide is not mutually exclusive, a slight negative cooperativity exists, therefore, the relative concentrations of the respective ligands at the locus likely dictates the preferred binding partner of CBX7. The expression of CBX7 variants containing point mutations that disrupt either binding to RNA or binding to methylated H3K27me result in repression of the protein-encoding genes in the INK4b/ARF/INK4a locus and impaired control of cellular senescence, with the mutation affecting binding to H3K27me having a more dramatic effect [19]. These studies serve as a model for the types of experiments that are needed to provide a deeper understanding of the mechanisms used by lncRNAs to regulate chromatin.
HOTTIP
In contrast with the previously described lncRNAs, HOTTIP (HOXA transcript at the distal tip) mediates transcriptional activation by controlling chromatin structure (Figure 1c). HOTTIP is a ~3.7 kb ncRNA transcribed from one end of the HOXA locus in mammalian cells [21]. Knockdown of HOTTIP decreased expression of genes within the HOXA locus with a potency that correlated with proximity to the HOTTIP gene, indicating that HOTTIP contributes to activating transcription of some, but not all, HOXA genes. At the affected genes, knockdown of HOTTIP led to a loss of the activating histone marks H3K4me3 and H3K4me2, as well as decreased occupancy of proteins contributing to establishing these marks: MLL1 (mixed lineage leukemia 1) and WDR5 (WD-repeat containing protein 5). Moreover, HOTTIP binds directly to WDR5 in vitro, and co-immunoprecipitates with WDR5 from cells. Chromosome conformation capture carbon copy assays detected abundant long range looping interactions localized to the active end of the HOXA locus from which HOTTIP is transcribed [21]. These data together support the model that intrachromosomal interactions bring transcribed HOTTIP in proximity to a cluster of genes in the HOXA locus where it recruits WDR5-MLL complexes, resulting in a region of H3K4me3 and transcriptional activation. The control of both higher order chromosome structure and lncRNA-mediated recruitment of histone modifiers provides a powerful means for localized activation of gene expression; is it possible that a similar mechanism might exist for other lncRNAs that control transcription in cis (Box 3).
Box 3: lncRNAs with enhancer-like function.
A group of human lncRNAs function in transcriptional activation in a manner reminiscent of enhancers — DNA elements found in mammalian promoters that can activate transcription from long distances. When these lncRNAs were knocked down, transcription of neighboring protein-encoding genes decreased [63]. The effects of knockdown on neighboring gene expression spanned hundreds of kb, but not all neighboring genes were affected. The ability of the lncRNAs to potentiate transcriptional activation of a neighboring gene could be recapitulated using an lncRNA and reporter gene contained on a transfected plasmid, suggesting that regulation occurs in cis. How widespread this means of regulation is across the genome and the mechanisms by which the lncRNAs function in gene-specific activation remain unknown.
ncRNAs that directly control the assembly or activity of transcription factor complexes
Multiple ncRNAs have been shown to control gene expression by regulating the activity of transcriptional activators, coregulators, and the general transcription machinery. These ncRNAs are diverse in their mechanisms of action and protein targets. The examples discussed below highlight the ability of ncRNAs to function as scaffolds for the docking of numerous proteins, to mimic functional DNA elements, and to directly regulate Pol II itself. It is possible these examples are representative of what will prove to be wide-spread mechanisms of transcriptional control as more regulatory lncRNAs are discovered.
Gas5 RNA
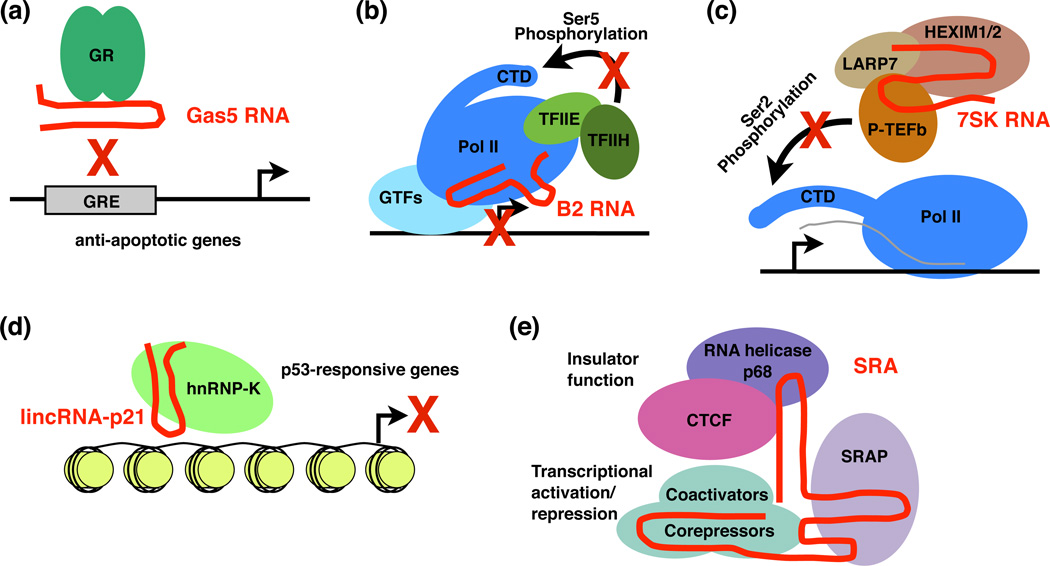
Gas5 (growth arrest-specific 5) is a ~600 nt ncRNA that is transcribed by Pol II, accumulates in growth-arrested cells, and sensitizes the cells to apoptosis by inhibiting glucocorticoid-mediated transcription of anti-apoptotic genes [22,23]. Glucocorticoids are steroid ligands that bind to and activate the glucocorticoid receptor (GR). GR is a transcription factor, that when bound by ligand, acts as a transcriptional activator that binds glucocorticoid response elements (GREs) in the promoters of target genes. The Gas5 ncRNA functions as a repressor of GR-mediated transcription by binding directly to the DNA binding domain of the ligand-activated GR [23]. Doing so blocks GR from associating with GREs (Figure 2a). Gas5 ncRNA folds into a structure that mimics a GRE; mutating this "decoy GRE" within Gas5 prevents it from competing with GREs for binding GR [23].
Figure 2.
ncRNAs that regulate transcription factors. (a) Gas5 ncRNA serves as a mimic of the DNA site to which GR binds. By keeping GR from binding GREs, Gas5 blocks the activation of GR-responsive genes. (b) B2 RNA binds directly to Pol II during the heat shock response. It represses transcription by building into complexes with Pol II at promoters and keeping the polymerase from engaging the DNA as well as blocking phosphorylation of the Pol II CTD by TFIIH. The schematic was adapted with permission from [34]. (c) 7SK RNA, which is found in a complex minimally with P-TEFb, LARP7, and HEXIM1 or HEXIM2, represses the P-TEFb kinase, which is responsible for phosphorylating Ser2 residues in the Pol II CTD during early elongation. (d) lincRNA-p21 recruits hnRNP-K to many p53-responsive genes and mediates repression of these genes. (e) SRA binds many proteins and macromolecular complexes (some of which are shown) to mediate transcriptional regulation. Moreover, through its association with the RNA helicase p68 and CTCF, SRA plays a crucial role in insulating genes from one another throughout the genome.
Mimicking DNA and blocking DNA binding represents a straightforward means by which an ncRNA can control the activity of a transcriptional activator. Indeed, RNA aptamers that have been selected to bind to transcription factors with high affinity typically bind to the proteins in the same region that DNA does, thereby blocking the DNA binding activity of the transcription factor [24–27]. This observation, together with the discovery and characterization of Gas5 ncRNA, raises the intriguing question of whether other cellular "decoy RNAs" might exist to specifically control the activity of DNA-binding transcription factors in response to a diversity of stimuli.
B2 and Alu RNAs
The most direct means to control transcription of mRNA genes is to target Pol II itself, which is the case for B2 and Alu RNAs that are transcribed from SINEs (short interspersed elements). SINEs are repeat elements that are widespread in mammalian genomes and have historically been considered as evolutionary junk left behind by retrotransposition [28]. However, RNA polymerase III (Pol III) transcribes some SINEs to produce ncRNAs of discrete length, such as mouse B2 RNA (180 nt) and human Alu RNA (280 nt) [29]. Interestingly, Pol III-transcribed B2 and Alu RNAs function as trans-regulators of Pol II transcription in response to heat shock [30,31]. These ncRNAs are upregulated in response to a variety of cell stresses and developmental signals [29]. After heat shock, B2 and Alu RNAs bind directly to Pol II and transiently repress general transcription [30–32]. The ncRNAs have been localized at the promoters of repressed genes along with Pol II, where they block productive transcription. In vitro, Pol II bound by the inhibitory ncRNAs is recruited to complexes on promoter DNA but it cannot properly engage the DNA, resulting in transcriptional inhibition [31,33] (Figure 2b).
In addition, B2 RNA represses the phosphorylation of serine 5 residues in the heptapeptide repeats found in the C-terminal domain (CTD) of the large subunit of Pol II [34]. Ser5 residues are phosphorylated by the kinase activity of TFIIH, a general transcription factor that is recruited to promoters with Pol II. The level of Ser5 phosphorylation on Pol II at repressed genes sharply decreased after heat shock, despite the continued presence of the TFIIH kinase. B2 RNA inhibits CTD phosphorylation by TFIIH, but only in the context of repressed complexes on promoter DNA, thereby invoking an allosteric model for repression. B2 and Alu RNA have the potential to control entire programs of gene expression, although how they do so with gene specificity remains unknown.
7SK RNA
7SK RNA is one of the best characterized ncRNA regulators of transcription, and serves as a paradigm for how the interplay between an ncRNA and the proteins with which it interacts provides multifaceted transcriptional control. 7SK RNA is an abundant Pol III transcript, ~340 nt in length, that serves as an RNA scaffold to regulate the activity and availability of the transcription factor P-TEFb (Figure 2c). P-TEFb is a cyclin-dependent kinase that phosphorylates serine 2 residues in the CTD of Pol II as well as the transcription elongation factors DSIF and NELF; these phosphorylations are crucial for the post-initiation regulation of transcription [35]. 7SK RNA forms the core of a small nuclear ribonucleoprotein (snRNP) complex that minimally contains P-TEFb, and the proteins HEXIM1/2 and LARP7, which control the activity of P-TEFb and stabilize 7SK RNA (as reviewed in [36]). In addition, mePCE, a methylphosphate capping enzyme that places a unique 5' cap on 7SK, serves a capping-independent function within the 7SK snRNP to facilitate the interaction between 7SK RNA and LARP7 [37].
The P-TEFb kinase is inactive when present in the 7SK snRNP complex and becomes active when it dissociates from the complex [38,39]. Thus, 7SK RNA controls the equilibrium between active and inactive P-TEFb, which is crucial for transcriptional regulation. Once P-TEFb dissociates and becomes active, then 7SK RNA is bound by a subset of heterogeneous nuclear ribonucleoproteins (hnRNPs) [40,41]. The release of P-TEFb from the 7SK snRNP can be triggered by several events, such as UV treatment and exposure to synthetic inhibitors of transcription, as well as by more physiological regulators such as kinase/phosphatase signaling pathways [38,39,42]. In addition, the transcriptional coactivator p300 can acetylate the cyclin T1 subunit of P-TEFb, thereby negatively regulating its association with 7SK RNA and HEXIM1 [43].
P-TEFb is a crucial elongation factor in regulating transcription of HIV genes, and the 7SK snRNP plays a unique role in this process. The HIV Tat protein interacts with 7SK RNA, and in doing so, remodels the 7SK snRNP to form a new inhibitory complex that is devoid of HEXIM1 and resistant to dissociation of PTEF-b in response to stress[44]. A new mechanistic model for transcriptional regulation of HIV genes has been proposed in which P-TEFb is first recruited to the HIV-1 promoter in an active state as part of the 7SK snRNP [45]. When transcription initiates and the 5' end of the primary transcript folds to form the so-called TAR element, Tat binds TAR and displaces the 7SK snRNP, thereby activating P-TEFb. Indeed, HEXIM1, LARP7 and Tat were found localized to the HIV-1 promoter in an RNA-dependent manner. Because P-TEFb acts globally to control transcription, it will be interesting to determine whether the 7SK snRNP is widely recruited to cellular promoters.
lincRNA-p21
A ~3 kb lncRNA, lincRNA-p21, which is transcriptionally activated by p53, mediates p53-dependent transcriptional repression in response to DNA damage [46] (Figure 2d). When lincRNA-p21 was knocked down, hundreds of genes were derepressed, the majority of which were also derepressed in response to p53 knockdown, indicating that lincRNA-p21 plays a functional role in mediating a p53-dependent program of transcriptional repression. The association of lincRNA-p21 with a particular hnRNP, hnRNP-K, has been revealed by mass spectrometry of proteins associated with in vitro transcribed lincRNA-p21 and by co-immunoprecipitation. It has been shown that lincRNA-p21 and hnRNP-K mediate repression of a common set of p53-responsive genes, hnRNP-K localizes to a significant fraction of these genes, and knockdown of lincRNA-p21 results in the loss of hnRNP-K occupancy at many of the genes [46]. These data support a model in which p53 directly activates transcription of lincRNA-p21, which in turn mediates, through its interaction with hnRNP-K, transcriptional repression of genes that are downregulated during the p53 response. It remains possible that lincRNA-p21 regulates gene expression through a post-transcriptional mechanism. Investigating changes in Pol II occupancy in the transcribed regions of genes that are derepressed in response to lincRNA-p21 knockdown would provide more direct test of the role of lincRNA-p21 in controlling transcription.
SRA
One of the first ncRNAs shown to control transcription, SRA regulates a repertoire of cellular pathways by serving as a platform on which many different proteins can interact. The gene encoding Steroid Receptor RNA Activator (SRA) is particularly interesting because its products include both coding and noncoding transcripts. The protein expressed from the SRA gene, named SRAP, functions as a transcriptional co-regulator [47,48]. SRA, a noncoding transcript, arises from alternative splicing of the SRA gene in a manner that disrupts the SRAP reading frame. Like the SRAP protein, the SRA ncRNA functions as a transcriptional co-regulator, working both in activation and repression, depending on its protein partners and the biological system (as reviewed in [49]). SRA co-immunoprecipitates with SRAP in mouse and human muscle samples, suggesting a complex regulatory interplay between these two products of a single gene [50].
First identified as an RNA coactivator for nuclear hormone receptors [51], SRA interacts with multiple proteins, including nuclear receptor coregulatory proteins, transcription factors, RNA helicases, and proteins that function in gene insulation (Figure 2e) [49]. Approximately 10 proteins bind directly to SRA, whereas other proteins associate with SRA as part of large macromolecular complexes. Thus, SRA likely functions as a scaffold to bring together multiple factors to regulate transcription. Additional complexity arises from the fact that SRA is post-transcriptionally modified by two pseudouridine synthases, which enhances its coactivation of nuclear receptor activity [52,53].
In addition, SRA mediates the interaction between the RNA helicase p68 and CTCF (CCCTC binding factor) [54]. CTCF is a DNA-binding protein important for the insulation of transcription in the genome [55]. CTCF binds to the RNA helicase p68, which also binds to SRA [56]. Consistent with its role as a scaffold RNA, knockdown of SRA inhibited the interaction between CTCF and p68 [54]. Both p68 and SRA are required for the proper function of CTCF in gene insulation. Approximately 20% of CTCF binding sites in the human genome are associated with p68, suggesting that SRA might have a broad role in regulating transcriptional insulation.
Concluding remarks
lncRNAs are becoming widely recognized as key regulators of mammalian transcription, denoting a fundamental shift in paradigm. It appears that lncRNAs evolved to provide an additional layer of control by regulating the protein factors that set transcriptional programs. To date, lncRNAs are known to control many different steps in the process of transcription. This diversity in function points at the potential for lncRNAs to act quite broadly as transcriptional regulators. It is possible that many lncRNAs will ultimately be found to participate in all levels of transcriptional control from the nuclear localization of transcription factors through transcriptional termination.
Multiple lncRNAs have been found to mediate changes in chromatin structure. This is currently the fastest growing class of lncRNAs known to regulate transcription. Altering the accessibility of the genome to Pol II and its associated factors is arguably the most efficient means to broadly activate or repress transcription. Perhaps this is why so many lncRNAs have evolved to regulate chromatin structure. lncRNAs involved in X chromosome inactivation and genomic imprinting also function by regulating chromatin [4–6].
The documented examples of lncRNAs controlling transcription will likely inspire and inform future discoveries in an ever expanding diversity of biological systems. Identifying additional transcriptional regulatory lncRNAs that share a function with characterized lncRNAs may be complicated because different RNAs can have similar functions in the absence of detectable sequence similarity (examples are B2 and Alu RNAs, as well as the variety of lncRNAs that interact with chromatin modifying complexes). Therefore, in many cases searching for conserved sequence motifs in lncRNAs may not yield insight into their functions. Although finding new lncRNAs that associate with transcriptional regulatory proteins is useful and important, the challenge lies in determining whether those lncRNA-protein interactions are biologically significant. Lastly, the possibility that lncRNAs might themselves have enzymatic activity should not be ignored.
Acknowledgements
This work was supported by a Public Health Service grant (R01 GM068414) from the National Institute of General Medical Sciences.
Glossary
- ChIP-chip
A technique to determine the sites across the genome that are occupied by a protein. An antibody is used to purify a protein and any associated chromatin from formaldehyde crosslinked cells (ChIP, chromatin immunoprecipitation). After reversing the crosslinks, the chromatin is hybridized to a microarray (chip) containing genomic sequences to determine the regions to which the protein was bound in cells. The chromatin can also be deep-sequenced (ChIP-seq) to determine the genome-wide sites of protein occupancy.
- Chromosome conformation capture carbon copy assay (5C)
A technique used to analyze the three-dimensional organization of a locus within cells. It enables researchers to assess long distance chromosomal interactions.
- Cis versus trans regulation
With respect to lncRNAs, a cis regulator controls transcription at or in close proximity to the region on the genome from which the lncRNA is transcribed. A trans regulator controls transcription at sites on the genome that are removed (often on a different chromosome(s)) from its site of transcription.
- Gene insulation
A process that protects genes from outside interactions that can lead to inappropriate activation or silencing. The need for gene insulation arises when adjacent regions on the genome have very different transcription patterns.
- Heterogeneous nuclear ribonucleoproteins (hnRNPs)
A family of proteins that bind to ncRNAs. hnRNP proteins are involved in several biological processes, including transcription, pre-mRNA processing, RNA transport, and translation.
- Polycomb group protein complex 2 (PRC2)
A multisubunit complex composed of Ezh2, Suz12, EED, and RbAp48. The Ezh2 subunit is a histone methyltransferase responsible primarily for trimethylating histone H3 on lysine 27 (i.e. H3K27me3). This histone modification is a mark for transcriptionally silent chromatin and PRC2 plays a predominant role in transcriptional repression across the genome.
- RNA aptamers
Small (most often non-natural) RNA molecules that bind very tightly to a target protein. Aptamers are usually created through the process of in vitro selection (also termed SELEX, systematic evolution of ligands by exponential enrichment), which iteratively selects and amplifies from a large pool of random sequences those that bind to a target.
- Small nuclear ribonucleoprotein (snRNP)
A complex that contains proteins bound to an snRNA (small nuclear RNA). For example, 7SK RNA, which is a snRNA, is found associated with proteins in a complex named the 7SK snRNP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, et al. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 4.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augui S, et al. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 6.Mohammad F, et al. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- 7.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mondal T, et al. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002071. e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kogo R, et al. Long non-coding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 17.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider C, et al. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 23.Kino T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3 doi: 10.1126/scisignal.2000568. ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, et al. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucl. Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh G, et al. Molecular mimicry of the NF-kappaB DNA target site by a selected RNA aptamer. Curr. Opin. Struct. Biol. 2004;14:21–27. doi: 10.1016/j.sbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Fan X, et al. Probing TBP interactions in transcription initiation and reinitiation with RNA aptamers that act in distinct modes. Proc. Natl. Acad. Sci. USA. 2004;101:6934–6939. doi: 10.1073/pnas.0401523101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhai G, et al. Characterization of RNA aptamer binding by the Wilms’ tumor suppressor protein WT1. Biochemistry. 2001;40:2032–2040. doi: 10.1021/bi001941r. [DOI] [PubMed] [Google Scholar]
- 28.Kramerov DA, Vassetzky NS. Short retroposons in eukaryotic genomes. Int. Rev. Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- 29.Liu WM, et al. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucl. Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen TA, et al. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 31.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Espinoza CA, et al. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 33.Yakovchuk P, et al. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakovchuk P, et al. B2 RNA represses TFIIH phosphorylation of RNA polymerase II. Transcr. 2011;2:45–49. doi: 10.4161/trns.2.1.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 37.Xue Y, et al. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucl. Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen VT, et al. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, et al. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 40.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrandon C, et al. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen R, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho S, et al. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009;28:1407–1417. doi: 10.1038/emboj.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobhian B, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawashima H, et al. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem. J. 2003;369:163–171. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emberley E, et al. Identification of new human coding steroid receptor RNA activator isoforms. Biochem. Biophys. Res. Commun. 2003;301:509–515. doi: 10.1016/s0006-291x(02)03070-x. [DOI] [PubMed] [Google Scholar]
- 49.Colley SM, Leedman PJ. Steroid Receptor RNA Activator - A nuclear receptor coregulator with multiple partners: Insights and challenges. Biochimie. 2011;93:1966–1972. doi: 10.1016/j.biochi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 50.Hube F, et al. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucl. Acids Res. 2011;39:513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 52.Zhao X, et al. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol. Endocrinol. 2007;21:686–699. doi: 10.1210/me.2006-0414. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X, et al. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell. 2004;15:549–558. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 54.Yao H, et al. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bell AC, et al. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe M, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitz KM, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santoro R. The silence of the ribosomal RNA genes. Cell. Mol. Life Sci. 2005;62:2067–2079. doi: 10.1007/s00018-005-5110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mayer C, et al. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 62.Goni JR, et al. Triplex-forming oligonucleotide target sequences in the human genome. Nucl. Acids Res. 2004;32:354–360. doi: 10.1093/nar/gkh188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu C, et al. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA. 2011 doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]