Abstract
The precise regulation of phosphoinositide lipids in cellular membranes is critical for cellular survival and function. Inositol 5-phosphatases have been implicated in a variety of disorders, including various cancers, obesity, Type II diabetes, neurodegenerative diseases and rare genetic conditions. Despite the obvious impact on human health, relatively little structural and biochemical information is available for this family. Here we review recent structural and mechanistic work on the 5-phosphatases with a focus on OCRL, whose loss of function results in OculoCerebroRenal syndrome of Lowe and Dent 2 disease. Studies of OCRL emphasize how the actions of 5-phosphatases rely on both intrinsic and extrinsic membrane recognition properties for full catalytic function. Additionally, structural analysis of missense mutations in the catalytic domain of OCRL provides insight into the phenotypic heterogeneity observed in Lowe Syndrome and Dent Disease.
Introduction
Phosphorylation of phosphatidylinositol at the 3, 4 or 5 position of the inositol ring generates seven phosphoinositides (PIs) that play key regulatory functions in cell physiology[1]. The related soluble inositol polyphosphates and pyrophosphates, generated from IP3 (a product of PI(4,5)P2 cleavage by phospholipases), are also important signaling molecules [2]. PIs function in diverse processes such as signal transduction, transport of ions and metabolites across membranes, exo-and endocytosis, regulation of the actin cytoskeleton, transcriptional regulation, and membrane trafficking. Their phosphorylated headgroups, which are localized on the cytosolic leaflets of membranes and help define membrane identity, interact with a variety of amino acid motifs or protein domains, and thus regulate protein-bilayer interactions. Key to their signaling function is their heterogeneous subcellular localization, which is achieved by differential localization of the enzymes that synthesize and metabolize them and by mechanisms to couple transport of a membrane from one compartment to another with a change of its phosphoinositide composition.
Inositol 5-phosphatases (originally defined as Types I-IV based on their biochemical properties) selectively dephosphorylate the 5 position of the inositol ring (Figure 1). Sequencing of the mammalian genome has identified ten of these enzymes. One of them, INPP5A (or type I inositol phosphatase), acts selectively on soluble inositol polyphosphates. The other nine enzymes (Types II-IV) act also, or preferentially, on the lipid bound phosphoinositides, primarily on PI(4,5)P2 and PI(3,4,5)P3 in an Mg2+-dependent manner. Each of the 5-phosphatases has specialized functions, due to unique cellular and subcellular distributions. They target distinct intracellular pools of PIs with differing global impacts on PI levels. This impact on overall cellular function depends on cell type. For example, Synaptojanin 1 is a major neuronal 5-phosphatase [3–5], whereas OCRL is a major 5-phosphatase in fibroblasts[6,7].
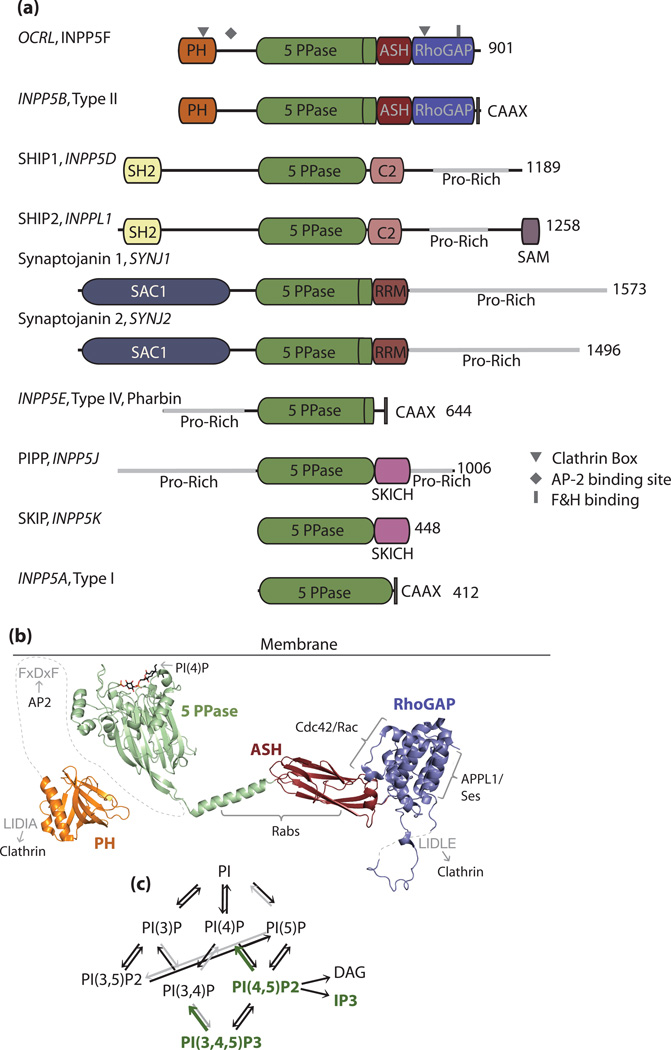
Figure 1. OCRL domain organization and interactions.
a). Domain organization of the inositol 5-phosphatases. The gene names are indicated in italics, and alternate names are also given. The proline rich domains contain multiple protein interaction sites, such as for SH3 domains, EH domains, and clathrin adaptors, which are not shown for simplicity. b). Structural representation of OCRL, colored according to domains (orange= PH domain, 2KIE.pdb; green= 5-phosphatase domain, 3MTC.pdb; red= ASH domain, 3QIS.pdb; and blue= RhoGAP domain, 2QV2.pdb). Interaction partners are indicated adjacent to their respective binding surface (black text). Motifs within OCRL that bind clathrin and AP2 are indicated in gray. All molecular graphics were generated with Pymol, www.pymol.org [81]. c). Metabolic interconversions of phosphoinositide species. Substrates of the 5-phosphatases are indicated in green.
5-phosphatases have been implicated in a broad spectrum of diseases and disorders (for review see[8]). Loss of function of OCRL results in the X-linked OculoCerebroRenal syndrome of Lowe and Type 2 Dent disease[9,10]. Mutations in INPP5E are found in a subset of patients with Joubert and MORM syndromes, two ciliopathies involving multiple organ dysfunction, commonly displaying mental impairment and physical deformations[11,12]. SHIP1 has restricted expression in cells of the hematopoietic lineage and mice lacking SHIP1 have myeloproliferative syndrome[13–15]. These facts have prompted exploration of SHIP1 function in relation to inflammation, immunity, and leukemias [16]. The close homologue SHIP2 is implicated in metabolic syndrome due to a link to obesity, insulin resistance, and hypertension[17–19]. Lastly, synaptojanin 1 activity (via its effects on brain PI(4,5)P2 levels) may contribute to the pathological manifestations of Down syndrome, in particular the early onset of Alzheimer’s Disease which affects those with Down syndrome[4,5].
Lowe Syndrome and Dent disease
Patients with Lowe Syndrome suffer primarily from congenital cataracts, neonatal hypotonia, intellectual disability (with distinct behaviors) and renal proximal tubule dysfunction (Fanconi syndrome). Mutations in OCRL are also found in a subset of patients with Dent disease (type 2) who selectively suffer from renal proximal tubular dysfunction. The majority of genetic abnormalities described to date for Lowe syndrome and Dent disease occur as deletions, frameshifts, and stop mutations, with a smaller fraction occurring as splicing and missense mutations[20]. Since a complete deletion of the OCRL gene results in Lowe syndrome, it is clear that lack of activity of this enzyme, and not a dominant effect due to truncated protein product, underlies disease.
5-phosphatase activity in soluble extracts of cultured skin fibroblasts derived from patients with both Lowe syndrome and Dent 2 disease is drastically reduced (less than 10% activity is found in affected individuals)[6,10]. Concurrently, PI(4,5)P2 levels are increased in patient fibroblasts relative to controls[7,21].
One of the important unsolved questions in the field regards penetrance, as OCRL loss of function results in a large phenotypic spectrum, even for mutations which completely abolish OCRL protein levels. One explanation may lie with INPP5B (also known as Type II 5-phosphatase), an OCRL homologue with ~45% sequence identity [22] and close structural similarity. INPP5B shares most interacting partners with OCRL, except for clathrin and the endocytic clathrin adaptor AP-2. Variations in the level of expression of INPP5B in the relevant tissues may affect the severity of the OCRL loss of function phenotype. INPP5B is not clearly linked to any human diseases.
Genetic studies in mice clearly demonstrate partial functional redundancy for OCRL and INPP5B[23]. Mice lacking OCRL are asymptomatic, whereas mice lacking INPP5B have testicular degeneration and male sterility. However, the double knockout animals are embryonic lethal. Splice site variation between species may explain why this compensation does not occur in humans [24,25]. Additional modifying elements contributing to this species variation could include alternative start sites[20], or variations in the activity or expression levels of interaction partners, PI(4,5)P2 effectors, or other phosphoinositide metabolizing enzymes.
Localization of OCRL
OCRL resides on vesicular structures throughout the endosomal system and the Golgi complex, and is also present at the plasma membrane in membrane ruffles and at late-stage endocytic clathrin-coated pits. This broad distribution is mediated by its numerous interactions (Figure 1). OCRL binds clathrin, clathrin adaptors, several GTPases, and the endocytic proteins APPL1 and Ses1/2. At least some of these interactions are not mutually exclusive, but APPL1 and Ses1/2 compete for binding to OCRL as vesicles progress along the endocytic pathway, suggesting interesting regulatory mechanisms through which membrane transport is coupled to the modification of binding partners.
The broad intracellular distribution of OCRL contrasts with the preferential distribution of its preferred substrates, PI(4,5)P2 and PI(3,4,5)P3, in the plasma membrane or in the earliest endocytic stations [26,27], as revealed by well-established fluorescent reporters for these PIs. This has three possible explanations. OCRL may i) counteract ectopic accumulation of spuriously produced PIs within intracellular membranes, ii) control pools of PI(4,5)P2 which are not detectable by currently available reporters or iii) act on small, yet physiologically important pools of PIs that undergo rapid turnover. Clearly, lack of OCRL results in an accumulation of intracellular PI(4,5)P2[7], and pathological phenotypes are thought to be the consequence of these changes. Studies of human cells deficient in OCRL function (patient cells or cells subjected to OCRL knock down) have revealed defects in endocytic trafficking[28–31], actin polymerization[32], establishment of cell polarity[33] and cytokinesis[28,34].
Domain organization of OCRL
The domain organization of OCRL and the structures of its folded domains are shown in Figure 1. INPP5B has the same domain organization, and, in fact, the structure of the catalytic domain shown in Figure 1b is of INPP5B (62% identical to the catalytic domain of OCRL which does not have a crystal structure available) (PDB code: 3MTC, Tresaugues et. al, unpublished).
The N-terminus of OCRL and INPP5B contain structurally similar PH (pleckstrin homology) domains despite poor sequence similarity in this region[35]. Both PH domains lack the basic patch needed for PI recognition and, accordingly, do not bind phosphoinositide-containing liposomes[35]. A loop within the PH domain of OCRL contains an unconventional clathrin box (an acidic residue in the 5th position of the motif is missing) which is absent from INPP5B as well as from homologs in invertebrates[35–37].
The PH domain connects to the 5-phosphatase domain, which has a Dnase I-like fold, by a region of approximately 100 amino acids, which in OCRL contains an AP-2 binding site. The 5-phosphatase domain of a Schizosaccharomyces pombe synaptojanin homolog (SPsynaptojanin) was the first structurally characterized member of this family[38]. This has recently been supplemented by structures (which have been solved and deposited into the protein data bank) of the catalytic domains of human INPP5B (PDB code: 3MTC, 3N9V), SHIP2 (PDB code: 3NR8), and INPP5E (PDB code: 2XSW), which have been solved and deposited into the Protein Data Bank (Tresaugues et. al, unpublished).
The 5-phosphatase domain of OCRL is followed by a short helix that connects it to the ASH(ASPM-SPD2-Hydin) and RhoGAP (Rho GTPase Activating) domains, which form a single folding module. The ASH domain has an immunoglobulin-like fold similar to Vesicle-Associated membrane Protein-Associated protein (VAP) and Major Sperm Protein (MSP) and is a domain often found in proteins resident in cilia or in the centrosomal area of the cell[39]. The RhoGAP domain lacks the catalytic arginine and is catalytically inactive (although slight GAP activity against Rac has been reported [40]). A loop in the Rho GAP domain contains a second clathrin box which is also not present in INPP5B[22,30,31]. The ASH-RhoGAP module regulates the majority of the protein-protein interactions currently described for OCRL and has a key role in membrane recruitment. Accordingly, patient missense mutations in this module that affect its interactions with binding partners (discussed below) (but do not affect inherent enzyme activity in-vitro) impair cellular localization and the stability of OCRL and are sufficient to cause disease[30,41].
The Inositol 5-phosphatase domain
The structure of the INPP5B catalytic domain was solved in complex with Mg2+ and diC8 PI(4)P (PBD code: 3mtc.pdb, Tresaugues et. al, unpublished), the product of the hydrolysis of a water-soluble short fatty acid chain PI(4,5)P2. The inositol ring differs in position as compared to that seen in the previously described structure of SPsynaptojanin with bound Ca2+ (instead of Mg2+) and IP2 (the product of IP3 hydrolysis) [38]. This may be due to the presence of a binding site in INPP5B for the acyl chain of PI(4)P, which may help to position the inositol ring. Alternatively, these differences may reflect different product release complexes, or the influence of the bound Ca2+ (which is inhibitory [42]) as apposed to the physiological Mg2+ ion.
Comparison of the crystal structures of INPP5B with the catalytic domains of SPsynaptojanin[38], SHIP2 (PDB code: 3NR8, Tresaugues et. al, unpublished), and INPP5E (PDB code: 2XSW, Tresaugues et. al, unpublished) show striking differences in the active site, most notably in the region corresponding to the acyl chain interacting site of INPP5B, highlighted in black boxes in Figure 2. These differences may explain the lipid specificities of these enzymes, such as the reported sensitivities to fatty acid composition and membrane curvature [38,43,44]. Further structural work on enzyme/lipid substrates will be most interesting in delineating how these enzymes “taste” specific membrane compartments, possibly even favoring recognition of substrates on membranes with positive curvature due to greater spacing of phospholipid head groups and partial exposure of their acyl chains. An impact of the bilayer on the catalytic activity of OCRL is consistent with reports that OCRL loss of function affects activity towards phosphoinositides more severely than the soluble inositols[6,7].
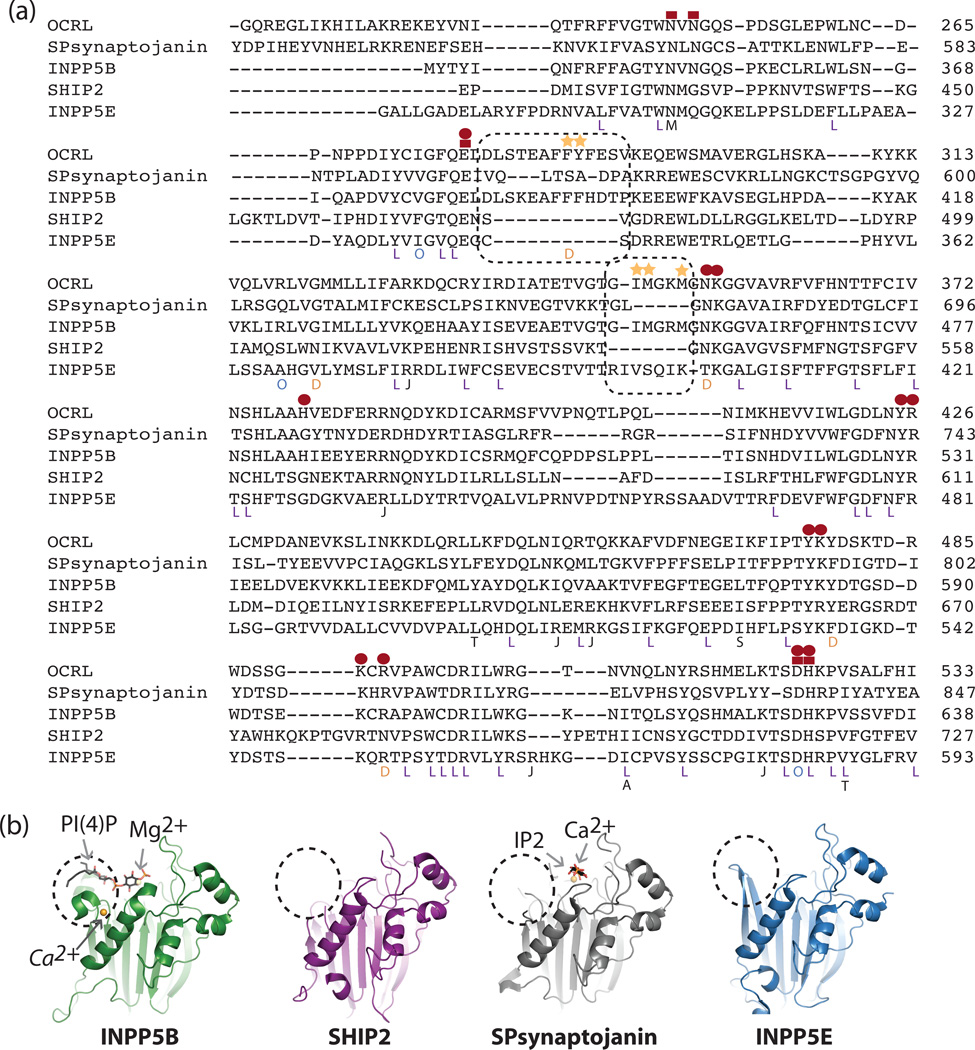
Figure 2. Structural analysis of the 5-phosphatase catalytic module.
a). Structure-based[82]sequence alignment of 5-phosphatases (PDB codes: 1I9ZA, 3MTCA, 3NR8B, 2XSWA). Active site residues are indicated by red symbols, red squares represent metal coordinating residues (either direct or via a water molecule), and red circles represent phosphoinositide interacting residues. The acyl chain binding site in INPP5B and corresponding residues in other 5-phosphatases are indicated by a dashed black box. Patient mutations are indicated underneath the residue: L= Lowe Syndrome, D= Dent 2, O = Both Lowe and Dent[20], J = mutations found in INPP5E in patients with JBTS1[12], T=mutations in SHIP2 associated with Type II Diabetes[63]. A = mutation in SHIP1 found in a patient with AML[66]. S = mutation in SHIP1 found in mice to produce a more severe phenotype than the knockout animal[83], M = Mutation in Synaptojanin 2 found to cause progressive hearing loss in the Mozart mouse[84]. b). Structural analysis of the 5-phosphatases. The region of most variation, which interacts with the acyl chain of PI(4)P when in complex with INPP5B, is indicated by a dashed black circle. The position of a second metal binding site in INPP5B, here a crystallographically identified Ca2+ ion (PDB code 3N9V.pdb, Tresaugues et. al, unpublished), which may have functional significance, is labeled in italics.
OCRL interactions
Clathrin
Clathrin coats mediate vesicular budding both from the plasma membrane (clathrin mediated endocytosis) as well as from intracellular membranes, namely endosomes and the trans-Golgi network. An important feature of OCRL, not shared by INPP5B and homologues in invertebrates, is the presence of multiple binding sites for clathrin coat components. OCRL is directed to late-stage endocytic clathrin-coated pits, and to endosomal/Golgi coats via two clathrin binding motifs, located in the PH and RhoGAP domains, and the binding motif for clathrin adaptors (Figure 1)[30,31,35,36]. The clathrin box containing loop on the RhoGAP domain undergoes alternative splicing to generate a long (A isoform, predominantly expressed in the nervous system[45]) and a short (B isoform) protein which differ in the presence of 8 amino acids adjacent to the clathrin box. This sequence enhances clathrin binding, likely by steric mechanisms[36].
Rab GTPases
Although additional interactions help define its precise localization, targeting of OCRL to intracellular membranes is critically dependent upon its interactions with GTP-bound Rab proteins[46]. Rabs are small GTPases whose reversible association with membranes help to spatiotemporally regulate intracellular membrane trafficking [47]. Rab GTPases also were reported to have a stimulatory effect on the activity of OCRL[46,48].
OCRL consistently displays a remarkably broad specificity for Rab GTPases in-vitro[34,46,49,50], capable of interacting with at least 16 different Rabs[49]. Some of the more prominent of these interactions, namely those with Rab5 (endosomes), 6 (Golgi and endosomes) and 35 (fast recycling route) were confirmed by functional and co-localization studies[34,46]. Interestingly, the interaction with Rab35 (not shared with INPP5B) targets OCRL to intercellular bridges in cultured cells, and lack of either OCRL or Rab35 impairs abscission through abnormal F-actin polymerization[34].
The Rab interaction surface on the ASH domain of OCRL was characterized in a recent structure of the co-complex of a fragment of OCRL with Rab8a [51]. In this structure, shown in Figure 3, Rab8 bridges the ASH domain and C-terminal catalytic domain helix. Rab effectors typically recognize a hydrophobic triad in the GTP bound Rab protein, which incorporates residues from its switch and interswitch regions. This triad, which was suggested to provide specificity for Rab/effector interactions, is not fully engaged in the OCRL/Rab8a complex (Figure 3). This unusual binding mode of OCRL for a Rab, which is likely to be shared by INPP5B, may explain the broad specificity of this enzyme for Rab proteins. [51]
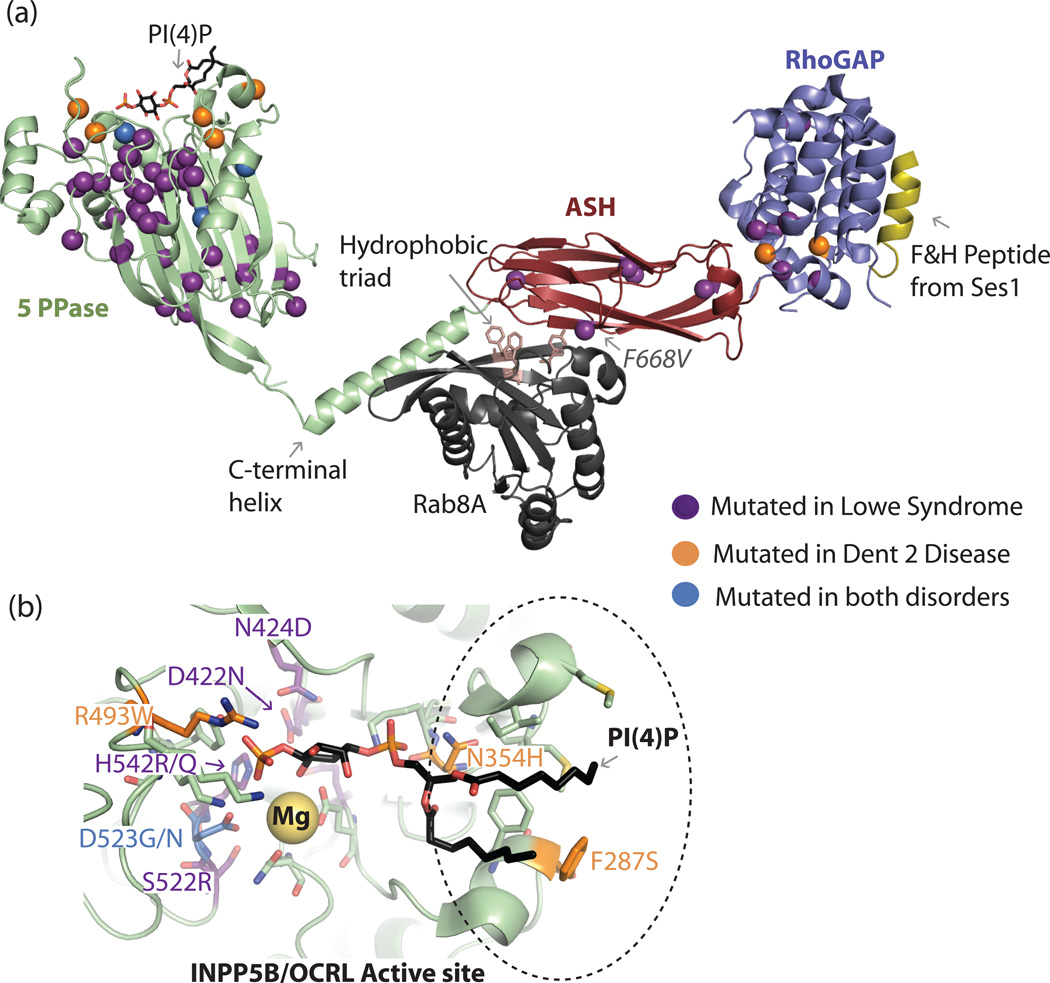
Figure 3. Structural analysis of missense mutations in OCRL.
a). Patient missense mutations are highlighted in the crystal structures of isolated OCRL domains in complex with binding partners, the PH domain and connecting region is not shown. The affected residues are represented as spheres. The bound PI(4)P diC8 lipid is represented as black/colored sticks on the INPP5B catalytic domain crystal structure (PDB code: 3MTC.pdb). The Mg2+ active site metal is a yellow sphere. Lowe syndrome associated mutations tend to cluster in the core folding modulus of the protein or in critical catalytic residues or loops and clearly affect the stability, loop structure, or catalytic activity of the enzyme. In contrast, the majority of mutations associated with a milder, Dent phenotype are surface residues that may not have a completely deleterious affect on the enzyme’s stability or activity. The structure of the ASH-RhoGAP module of OCRL in complex with Rab8A (black, PDB ID: 3QBT) and the F&H motif-containing peptide from Ses1 (yellow, PDB ID: 3QIS) shows the positions of patient missense mutations not present in the catalytic domain. The only clinical mutation known to affect protein-protein interactions and not protein stability (F668V) is indicated. The hydrophobic triad implicated in Rab specificity determination is shown as salmon-colored sticks. b). A closer view of INPP5B in complex with PI(4)P (PDB code: 3MTC) in a different orientation relative to a. Active site residues are shown as sticks, and waters in the active site are not shown for simplicity. Residues implicated in Lowe syndrome or Dent 2 disease are colored as indicated, numbering is for the corresponding residue in human OCRL.
RhoGAP interactions with other GTPases
The RhoGAP domains of OCRL and INPP5B interact with Rac and Cdc42, but only the Cdc42 interaction is GTP-dependent[30,40]. The interaction with Rac may be related to the localization of OCRL at membrane ruffles[52]. Binding to Cdc42 may at first seem surprising because PI(4,5)P2 and PI(3,4,5)P3 cooperate with Cdc42 in triggering actin nucleation. In fact, GTP-bound Cdc42 also binds PI4P 5-kinases (type I PIP kinases). Effective action of PI(4,5)P2 in actin regulation may require its turnover, thus explaining why both 5-kinases and 5-phosphatases are effectors of this GTPase. Additionally, OCRL also interacts with Arf1 and Arf6, which are also regulators of 5-kinases[53], and this interaction is disrupted by a patient mutation in the RhoGAP domain[54]. In INPP5B, but not in OCRL, the RhoGAP domain terminates with a CAAX motif that may cooperate with Rho family GTPases in its localization to the plasma membrane.
RhoGAP interactions with F&H Motif containing proteins
The RhoGAP domain of OCRL (and INPP5B) also interacts with three endocytic proteins containing the F&H motif: APPL1, Ses1 and Ses2. This motif is an approximately 12–13 amino acid sequence centered around phenylalanine and histidine residues essential for binding[30,55]. The crystal structure of the ASH-RhoGAP domain in complex with the F&H peptide of Ses2 shows that the peptide forms an amphipathic helix that recognizes an evolutionarily conserved surface on the RhoGAP domain present in OCRL and INPP5B but not in other RhoGAPs[56] Figure 3). Analysis of the evolutionary conservation of the F&H binding surface in OCRL suggests the existence of other F&H motif containing proteins, as this surface is conserved in organisms that do not have identifiable Ses or APPL homologs[56]. The identification of additional F&H motif-containing OCRL interactors may help shed new light onto the basic functions of this enzyme.
APPL1 is an endosomal Rab5 effector containing a BAR domain arranged in tandem with a PH domain, and binds receptors (for example growth factor receptors) through its PTB domain[57–59]. It also binds the endocytic adaptor GIPC through a C-terminal PDZ binding sequence[30,59–61]. APPL1, along with the closely homologous protein APPL2 (which does not contain an F&H motif but heterodimerizes with APPL1) is present on a subpopulation of peripheral early endosomes that function upstream of classical PI3P positive endosomes[26,57]. The APPL heterodimer, which also binds AKT (a PI(3,4,5)P3 effector), may coordinate endocytosis of receptors with modulation of their signaling. OCRL and INPP5B may contribute to this function via their property to dephosphorylate PI(3,4,5)P3, as demonstrated by a recent study on phagocytosis [27].
The highly similar Ses1 and Ses2 proteins contain a PH domain, a predicted coiled-coil responsible for oligomerization, and a C-terminal, predominantly unfolded, proline-rich region that contains the F&H motif [55,62]. Ses1 and Ses2 are localized on PI(3)P-positive classical early endosomes and the Golgi complex[55,62].
As endosomes mature and acquire PI(3)P, Ses proteins displace APPL1 from the F&H binding surface[55], while OCRL remains associated with endocytic vesicles at both the APPL1 and Ses stage, possibly via its Rab-dependent interactions. Adding further complexity, phosphorylation of APPL1 on two serine residues in the F&H motif abolish its interaction with OCRL[30,56]. Thus, the F&H proteins may help to fine tune the localization of OCRL on subdomains of the endosomal membrane possibly helping couple lipid turnover with sorting events.
Analysis of Lowe and Dent patient mutations
There are many ways in which patient mutations can affect function: complete or partial deletion leading to unstable fragments, abnormal levels due to defects in splicing, nonsense, frameshift, and missense mutations which cause loss of interactions, misfolding, or catalytic inactivity. What emerges from recent studies is that OCRL interactions are as important as catalytic activity for its action in cell physiology.
While some of these interactions may have regulatory functions, their main role is to precisely localize OCRL. This may represent a general strategy for 5-phosphatase function. For example, INPP5E contains a membrane anchoring CAAX motif. A premature stop codon in INPP5E just before the CAAX box was found in a patient with MORM syndrome (a ciliopathy)[11]. The cilliary localization of INPP5E was significantly affected by this mutation, although the enzyme retained its intrinsic catalytic activity[11]. As a second example, mutations in SHIP2 linked with either Type II diabetes [63], or found in a spontaneously hypertensive rat strain [64], have been mapped to the proline-rich region of the protein and could potentially affect protein-protein interactions, which are critical for SHIP2 localization [65].
Mutations in the PH domain
To date there exist no identified patient missense mutations in the PH domain, which is the least conserved region of the protein. Although it remains possible that these mutations could lead to embryonic lethality, this may reflect a non-critical role for the PH domain in OCRL function, possibly speaking to the redundancy of clathrin interaction sites. Furthermore, the occurrence of nonsense, frameshift, and in-frame deletions in this region are skewed, with nonsense and frameshift mutations occurring much more frequently in exons 1–7 (the PH domain is encoded in exons 2–5) in patients with Dent disease as compared to Lowe syndrome[20].
Missense mutations in the 5-phosphatase domain
The majority of missense mutations map to the 5-phosphatase domain, and were first analyzed using the SPsynaptojanin crystal structure as a model. This study showed that most of the missense mutations (14 at that time) found in Lowe syndrome patients affected conserved residues in 5-phosphatases, directly affecting either folding, substrate binding or catalytic activity[38].
Figure 3 depicts a ribbon representation of the INPP5B structure (closest homolog of OCRL), highlighting the residues identified to date as missense patient mutations in OCRL, which are all identical in INPP5B. The majority of mutations found in Lowe syndrome patients cluster in the hydrophobic core of the protein, suggesting that these mutant proteins would be destabilized. There are also a number of missense mutations clustered around the active site (Figure 3b), with either direct or structural impacts on catalytic activity. Interestingly, Dent disease mutations primarily localize to surface residues at or near the catalytic site, and do not typically occur in the core of the protein, whereas Lowe syndrome mutations predominantly affect the folding core Figure 3).
Disease-associated missense mutations found in the catalytic domains of other 5-phosphatases are shown in Figure 2a and Box 1. Interestingly, a somatic mutation found in SHIP1 and associated with Acute Myeloid Leukemia drastically decreased the catalytic efficiency of this enzyme in-vitro [66] and the same position in OCRL corresponds to a residue mutated in a Lowe syndrome patient.
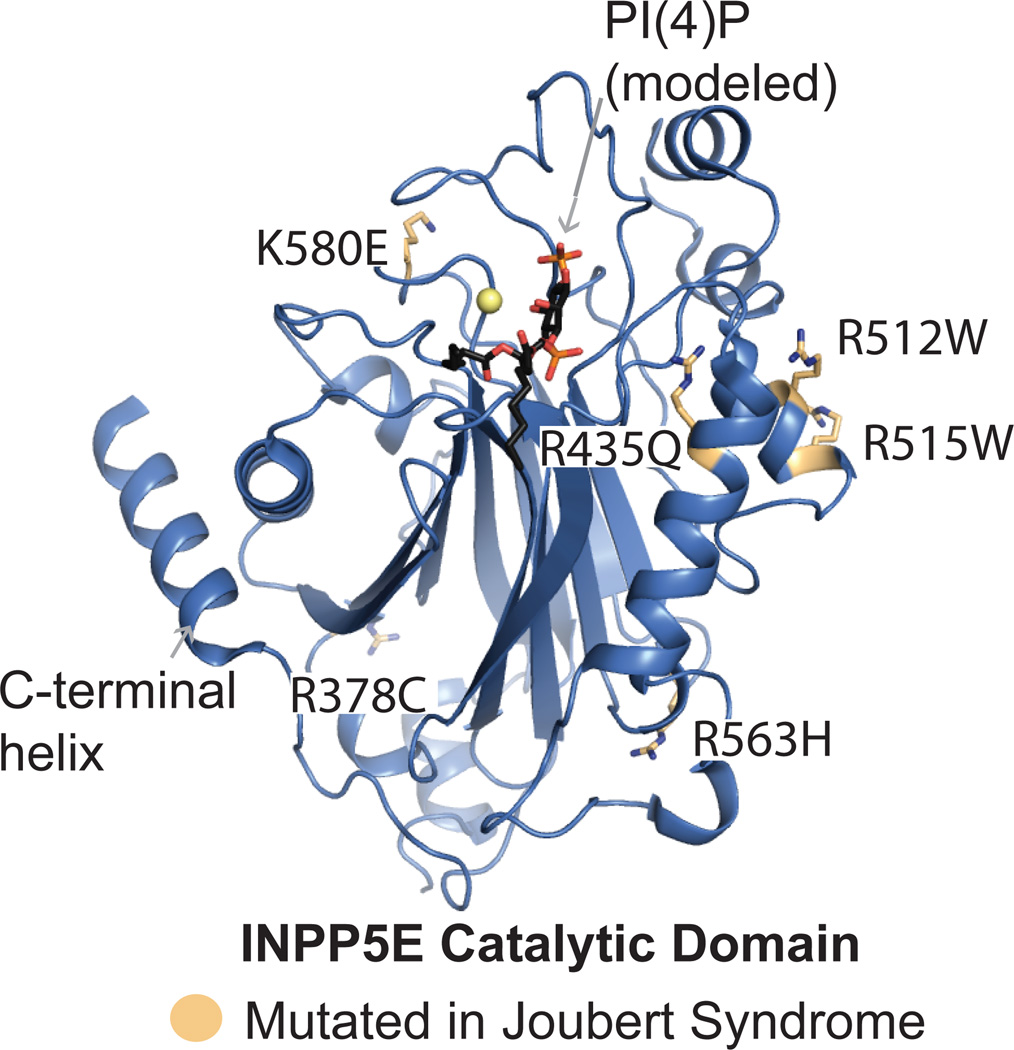
Box 1. Joubert syndrome associated missense mutations in INPP5E.
Missense mutations found in the INPP5E 5-phosphatase domain of patients with Joubert syndrome[12]. While some affected residues are in close proximity to the active site, none play an obvious direct role in catalysis, yet the catalytic activity towards PI(3,4,5)P3 is reported to be impaired by all mutations[12]. Two residues found mutated in patients (R378C and R563H) reside far from the active site and were reported to retain their activity towards PI(4,5)P2[12]. [12]Many of the affected residues are basic surface residues, suggesting a more complicated mechanism for disease than loss of catalytic function (Box Figure). Further understanding of the molecular mechanism for this loss of function awaits more thorough biochemical and structural analysis.
Missense mutations in the ASH-RhoGAP domain
The majority of missense mutations found in the ASH-RhoGAP module destabilize the protein, consistent with studies showing that patient cell lines harboring these mutations contain less OCRL protein[20], and produce a cytosolically localized enzyme when overexpressed[41]. Since formation of the F&H binding surface on the RhoGAP domain of OCRL is critically dependent on folding, these mutations also disrupt F&H motif binding[56] with varying affects on interactions with the Rho and Rab GTPases[41,54], and no effects on clathrin interactions (which relies on an unfolded clathrin box) [30,41,55]. One patient mutation in the ASH domain, however, does not destabilize the protein but directly impacts Rab binding [51,56]. These facts, coupled to the identification of deletions and insertions (including deletion of the majority of the RhoGAP domain) in Lowe syndrome patients[20], underscores the importance of OCRL interactions in its physiological function.
Conclusions and Future Perspectives
OCRL, the best structurally characterized inositol 5-phosphatase, is a major downregulator of intracellular PI(4,5)P2 and PI(3,4,5)P3 levels. As shown by the analysis of disease causing mutations, the function of its catalytic module is as important for its action as the function of the flanking regions that target the enzyme to specific intracellular sites. Such targeting occurs via interactions with a multiplicity of partner proteins in the endocytic pathway and in the Golgi complex that include clathrin, clathrin adaptors, Rabs, Rho family GTPases and F&H motif-containing endocytic proteins. At least some of these interactions are not energetically coupled [51], and structural modeling suggests that this enzyme can engage many of its binding partners simultaneously without steric constraint, as summarized in Figure 4. Given the structural variation observed in the active sites of the 5-phosphatases, it will be interesting to determine whether their active sites are optimally tuned to cooperate with these binding partners to direct specificity.
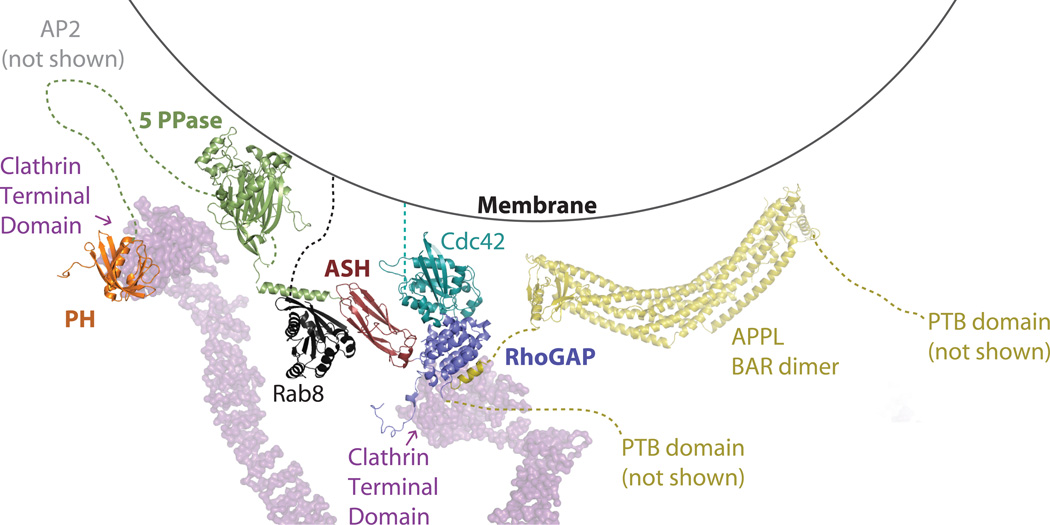
Figure 4. Model of OCRL at the membrane.
OCRL and interaction partners are modeled, utilizing currently available structural information. Based on this modeling, all known interaction surfaces can be engaged simultaneously without obvious steric conflict. Coloring is as shown as in previous figures, Cdc42 is teal. The membrane is depicted as a line, the legs and feet of the clathrin triskelion (PDB ID: 3IYV.pdb) as a purple surface. The interactions of the clathrin boxes of OCRL with the clathrin N-terminal domain were modeled after the structure of the clathrin N-terminal domain in complex with β-adaptin 3 (PDB ID: 1C9L, not shown). OCRL (PDB ID: 2KIE, 3MTC, 3QBT, 3QIS, 2QV2) and interaction partners (PDB ID: 3QBT, 1GRN, 2EJ8) are represented as ribbons. Based on structural homology, the interaction between OCRL and RhoGTPases is assumed to be similar to those seen for other RhoGAPs. The clathrin adaptor AP2 is not shown for simplicity.
The interaction of OCRL with clathrin coats (two binding sites for the heavy chain of clathrin and a binding site for clathrin adaptors) is particularly striking. Interestingly, other 5-phosphatases, such as SHIP2 and synaptojanin, interact with clathrin and its adaptors and are implicated in clathrin coat dynamics [65,67–69]. Furthermore PI4P 5-kinases interact with clathrin adaptors[70,71], thus emphasizing the importance of PI turnover for clathrin coat function.
Further emphasizing the link between membrane budding and PI metabolism is the partnership between OCRL and a BAR domain containing protein, APPL1. BAR domains, which are optimally suited to bind curved membranes[72], help coordinate membrane budding with other modifications that must occur in the bilayer. Another 5-phosphatase, synaptojanin, is highly dependent upon BAR domain containing proteins, primarily endophilin. Studies in multiple genetic models have shown that a lack of endophilin largely mimics the phenotype produced by the lack of synaptojanin [67,73,74]. Interestingly, the structural basis for the interaction between OCRL and APPL1 is completely different from that mediating the endophilin-synaptojanin interaction[56,75], revealing convergent evolution rather than evolutionary conservation.
Control of PI levels may provide therapeutic avenues for many common afflictions, such as obesity, cancer, Type II diabetes, inflammation, neurodegeneration, as well as the rare genetic diseases discussed here. However, there exists to date little means for pharmacological control of their levels, aside from the regulation of 3 phosphorylated PIs, which have a central role in cancer.
New tools for identifying small molecule modulators of 5-phosphatases have recently been developed and applied to SHIP1 and SHIP2 [76–79]. Indeed, according to the Aquinox website (http://www.aqxpharma.com/), an activator of SHIP1 is approved for Phase IIa clinical trials for treatment of a broad range of inflammation responses. Given success with these two targets, and structural variation seen in this enzyme family, the development of specific modulators is promising. Activators of INPP5B could potentially be useful for the treatment of Lowe syndrome and Dent disease. Inhibitors of PI4P 5-kinases could also be of benefit for conditions due to impaired 5-phosphatase activity.
The recent progress in producing an animal model to study OCRL loss of function [24] could have important practical applications in the design of treatment strategies for OCRL deficiency. Therapeutic approaches that target PI(4,5)P2 effectors may also be of benefit as inhibitors of actin polymerization partially counteracted the in-vitro defects in cytokinesis observed in OCRL deficient cells[34]. Along a related avenue, the recent report of a PI(4,5)P2 sensor, which titrates intracellular PI(4,5)P2 levels, may be used as a therapeutic tool for those affected with OCRL dysfunction[80]. Overall, it is clear that the further development of therapeutics geared towards modulating the 5-phosphorylated PIs will provide a multitude of benefits not restricted to basic research.
Box 2. Open Questions.
Molecular basis for phenotypic variations seen in patients with OCRL deficiency, including the difference between the Dent disease and Lowe syndrome phenotypes. This also includes the identification/characterization of modifying genes or genes whose mutations produce similar phenotypic effects.
Does the 5-phosphatase activity of OCRL act primarily to prevent the ectopic accumulation of inappropriate PIs (PI homeostasis) or to control rapidly turning over PI pools?
Precise cellular function of OCRL: a single dominant function or a multiplicity of functions (potential functions include vesicle coat dynamics and protein sorting, signaling, actin dynamics)?
Basis for the tissue specific defects produced by OCRL loss of function (endocytic defect in the kidney proximal tubules?) and mechanisms underlying intellectual disabilities in a subset of patients (i.e. Lowe syndrome patients).
Therapeutic strategies to counteract phenotypic manifestations.
Patient mutations in INPP5E are highlighted on the structure of INPP5E (PDB ID: 2xsw.pdb, Tresaugues et. al, unpublished), represented as light orange/atom colored sticks. PI(4)P is modeled to show the active site using the structure of INPP5B in complex with PI(4)P as a model (PDB ID: 3MTC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Di Paolo G, de Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Tsui MM, York JD. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzyme Regul. 2010;50:324–337. doi: 10.1016/j.advenzreg.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McPherson PS, et al. A presynaptic inositol-5-phosphatase. Nature. 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- 4.Voronov SV, et al. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down's syndrome. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9415–9420. doi: 10.1073/pnas.0803756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman DE, et al. Oligomeric amyloid-beta peptide disrupts phosphatidylinositol-4,5-bisphosphate metabolism. Nat. Neurosci. 2008;11:547–554. doi: 10.1038/nn.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suchy SF, et al. Lowe syndrome, a deficiency of phosphatidylinositol 4,5-bisphosphate 5-phosphatase in the Golgi apparatus. Hum. Mol. Genet. 1995;4:2245–2250. doi: 10.1093/hmg/4.12.2245. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, et al. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 1998;273:1574–1582. doi: 10.1074/jbc.273.3.1574. [DOI] [PubMed] [Google Scholar]
- 8.Astle MV, et al. Regulation of phosphoinositide signaling by the inositol polyphosphate 5-phosphatases. IUBMB Life. 2006;58:451–456. doi: 10.1080/15216540600871159. [DOI] [PubMed] [Google Scholar]
- 9.Attree O, et al. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- 10.Hoopes RRJ, et al. Dent Disease with mutations in OCRL1. Am. J. Hum. Genet. 2005;76:260–267. doi: 10.1086/427887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby M, et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 2009;41:1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- 12.Bielas SL, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helgason CD, et al. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazen AL, et al. SHIP is required for a functional hematopoietic stem cell niche. Blood. 2009;113:2924–2933. doi: 10.1182/blood-2008-02-138008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paraiso KHT, et al. Induced SHIP deficiency expands myeloid regulatory cells and abrogates graft-versus-host disease. J. Immunol. 2007;178:2893–2900. doi: 10.4049/jimmunol.178.5.2893. [DOI] [PubMed] [Google Scholar]
- 16.Kerr WG. Inhibitor and activator: dual functions for SHIP in immunity and cancer. Ann. N. Y. Acad. Sci. 2011;1217:1–17. doi: 10.1111/j.1749-6632.2010.05869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suwa A, et al. SHIP2 and its involvement in various diseases. Expert Opin. Ther. Targets. 2010;14:727–737. doi: 10.1517/14728222.2010.492780. [DOI] [PubMed] [Google Scholar]
- 18.Sleeman MW, et al. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat. Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- 19.Clément S, et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409:92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 20.Hichri H, et al. From Lowe syndrome to Dent disease: correlations between mutations of the OCRL1 gene and clinical and biochemical phenotypes. Hum Mutat. 2010;32:379–388. doi: 10.1002/humu.21391. [DOI] [PubMed] [Google Scholar]
- 21.Wenk MR, et al. Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat Biotechnol. 2003;21:813–817. doi: 10.1038/nbt837. [DOI] [PubMed] [Google Scholar]
- 22.Jefferson AB, Majerus PW. Properties of type II inositol polyphosphate 5-phosphatase. J. Biol. Chem. 1995;270:9370–9377. doi: 10.1074/jbc.270.16.9370. [DOI] [PubMed] [Google Scholar]
- 23.Janne PA, et al. Functional overlap between murine Inpp5b and Ocrl1 may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J Clin Invest. 1998;101:2042–2053. doi: 10.1172/JCI2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bothwell SP, et al. Mouse model for lowe syndrome/dent disease 2 renal tubulopathy. J. Am. Soc. Nephrol. 2011;22:443–448. doi: 10.1681/ASN.2010050565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bothwell SP, et al. Species-specific difference in expression and splice-site choice in Inpp5b, an inositol polyphosphate 5-phosphatase paralogous to the enzyme deficient in Lowe Syndrome. Mamm. Genome. 2010;21:458–466. doi: 10.1007/s00335-010-9281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoncu R, et al. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 2009;136:1110–1121. doi: 10.1016/j.cell.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohdanowicz M, et al. Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol. Biol. Cell. 2012;23:176–187. doi: 10.1091/mbc.E11-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben El Kadhi K, et al. The inositol 5-phosphatase dOCRL controls PI(4,5)P2 homeostasis and is necessary for cytokinesis. Curr. Biol. 2011;21:1074–1079. doi: 10.1016/j.cub.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 29.Vicinanza M, et al. OCRL controls trafficking through early endosomes via PtdIns4,5P(2)-dependent regulation of endosomal actin. EMBO J. 2011;30:4970–4985. doi: 10.1038/emboj.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erdmann KS, et al. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell. 2007;13:377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ungewickell A, et al. The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc Natl Acad Sci USA. 2004;101:13501–13506. doi: 10.1073/pnas.0405664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suchy SF, Nussbaum RL. The deficiency of PIP2 5-phosphatase in Lowe syndrome affects actin polymerization. Am. J. Hum. Genet. 2002;71:1420–1427. doi: 10.1086/344517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grieve AG, et al. Lowe Syndrome Protein OCRL1 Supports Maturation of Polarized Epithelial Cells. PLoS ONE. 2011;6:e24044. doi: 10.1371/journal.pone.0024044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dambournet D, et al. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 2011;13:981–988. doi: 10.1038/ncb2279. [DOI] [PubMed] [Google Scholar]
- 35.Mao Y, et al. A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J. 2009;28:1831–1842. doi: 10.1038/emboj.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhury R, et al. Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J. Biol. Chem. 2009;284:9965–9973. doi: 10.1074/jbc.M807442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafer EM. Clathrin-protein interactions. Traffic. 2002;3:513–520. doi: 10.1034/j.1600-0854.2002.30801.x. [DOI] [PubMed] [Google Scholar]
- 38.Tsujishita Y, et al. Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell. 2001;105:379–389. doi: 10.1016/s0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- 39.Ponting CP. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics. 2006;22:1031–1035. doi: 10.1093/bioinformatics/btl022. [DOI] [PubMed] [Google Scholar]
- 40.Faucherre A, et al. Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum. Mol. Genet. 2003;12:2449–2456. doi: 10.1093/hmg/ddg250. [DOI] [PubMed] [Google Scholar]
- 41.McCrea HJ, et al. All known patient mutations in the ASH-RhoGAP domains of OCRL affect targeting and APPL1 binding. Biochem. Biophys. Res. Commun. 2008;369:493–499. doi: 10.1016/j.bbrc.2008.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi Y, et al. Comparative mechanistic and substrate specificity study of inositol polyphosphate 5-phosphatase Schizosaccharomyces pombe Synaptojanin and SHIP2. J. Biol. Chem. 2004;279:44987–44995. doi: 10.1074/jbc.M406416200. [DOI] [PubMed] [Google Scholar]
- 43.Schmid AC, et al. Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett. 2004;576:9–13. doi: 10.1016/j.febslet.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 44.Chang-Ileto B, et al. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev. Cell. 2011;20:206–218. doi: 10.1016/j.devcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 46.Hyvola N, et al. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J. 2006;25:3750–3761. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 48.Shin HW, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukuda M, et al. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez-Gabin AG, et al. Interaction of Rab31 and OCRL-1 in oligodendrocytes: its role in transport of mannose 6-phosphate receptors. J. Neurosci. Res. 2010;88:589–604. doi: 10.1002/jnr.22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou X, et al. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J. 2011;30:1659–1670. doi: 10.1038/emboj.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faucherre A, et al. Lowe syndrome protein Ocrl1 is translocated to membrane ruffles upon Rac GTPase activation: a new perspective on Lowe syndrome pathophysiology. Hum. Mol. Genet. 2005;14:1441–1448. doi: 10.1093/hmg/ddi153. [DOI] [PubMed] [Google Scholar]
- 53.Krauss M, et al. ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J. Cell Biol. 2003;162:113–124. doi: 10.1083/jcb.200301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lichter-Konecki U, et al. The effect of missense mutations in the RhoGAP-homology domain on ocrl1 function. Mol. Genet. Metab. 2006;89:121–128. doi: 10.1016/j.ymgme.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 55.Swan LE, et al. Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1. Proc Natl Acad Sci USA. 2010;107:3511–3516. doi: 10.1073/pnas.0914658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pirruccello M, et al. Recognition of the F&H motif by the Lowe syndrome protein OCRL. Nat Struct Mol Biol. 2011;18:789–795. doi: 10.1038/nsmb.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miaczynska M, et al. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 58.Mao X, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat. Cell Biol. 2006;8:516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
- 59.Lin DC, et al. APPL1 associates with TrkA and GIPC1 and is required for nerve growth factor-mediated signal transduction. Mol. Cell. Biol. 2006;26:8928–8941. doi: 10.1128/MCB.00228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lou X, et al. GAIP, GIPC and Galphai3 are concentrated in endocytic compartments of proximal tubule cells: putative role in regulating megalin's function. J. Am. Soc. Nephrol. 2002;13:918–927. doi: 10.1681/ASN.V134918. [DOI] [PubMed] [Google Scholar]
- 61.Varsano T, et al. GIPC is recruited by APPL to peripheral TrkA endosomes and regulates TrkA trafficking and signaling. Mol. Cell. Biol. 2006;26:8942–8952. doi: 10.1128/MCB.00305-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noakes CJ, et al. The PH domain proteins IPIP27A and B link OCRL1 to receptor recycling in the endocytic pathway. Mol. Biol. Cell. 2011;22:606–623. doi: 10.1091/mbc.E10-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kagawa S, et al. Impact of SRC homology 2-containing inositol 5'-phosphatase 2 gene polymorphisms detected in a Japanese population on insulin signaling. J. Clin. Endocrinol. Metab. 2005;90:2911–2919. doi: 10.1210/jc.2004-1724. [DOI] [PubMed] [Google Scholar]
- 64.Marion E, et al. The gene INPPL1, encoding the lipid phosphatase SHIP2, is a candidate for type 2 diabetes in rat and man. Diabetes. 2002;51:2012–2017. doi: 10.2337/diabetes.51.7.2012. [DOI] [PubMed] [Google Scholar]
- 65.Nakatsu F, et al. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J. Cell Biol. 2010;190:307–315. doi: 10.1083/jcb.201005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo J-M, et al. Possible dominant-negative mutation of the SHIP gene in acute myeloid leukemia. Leukemia. 2003;17:1–8. doi: 10.1038/sj.leu.2402725. [DOI] [PubMed] [Google Scholar]
- 67.Milosevic I, et al. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 2011;72:587–601. doi: 10.1016/j.neuron.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perera RM, et al. Two synaptojanin 1 isoforms are recruited to clathrin-coated pits at different stages. Proc Natl Acad Sci USA. 2006;103:19332–19337. doi: 10.1073/pnas.0609795104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cremona O, et al. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- 70.Kahlfeldt N, et al. Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2. J. Biol. Chem. 2010;285:2734–2749. doi: 10.1074/jbc.M109.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thieman JR, et al. Clathrin regulates the association of PIPKIgamma661 with the AP-2 adaptor beta2 appendage. J. Biol. Chem. 2009;284:13924–13939. doi: 10.1074/jbc.M901017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 73.Schuske KR, et al. Endophilin is required for synaptic vesicle endocytosis by localizing synaptojanin. Neuron. 2003;40:749–762. doi: 10.1016/s0896-6273(03)00667-6. [DOI] [PubMed] [Google Scholar]
- 74.Verstreken P, et al. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- 75.Trempe J-F, et al. SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination. Mol. Cell. 2009;36:1034–1047. doi: 10.1016/j.molcel.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 76.Ong CJ, et al. Small-molecule agonists of SHIP1 inhibit the phosphoinositide 3-kinase pathway in hematopoietic cells. Blood. 2007;110:1942–1949. doi: 10.1182/blood-2007-03-079699. [DOI] [PubMed] [Google Scholar]
- 77.Brooks R, et al. SHIP1 inhibition increases immunoregulatory capacity and triggers apoptosis of hematopoietic cancer cells. J. Immunol. 2010;184:3582–3589. doi: 10.4049/jimmunol.0902844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suwa A, et al. Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br. J. Pharmacol. 2009;158:879–887. doi: 10.1111/j.1476-5381.2009.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rowe T, et al. A high-throughput microfluidic assay for SH2 domain-containing inositol 5-phosphatase 2. Assay Drug Dev Technol. 2006;4:175–183. doi: 10.1089/adt.2006.4.175. [DOI] [PubMed] [Google Scholar]
- 80.Mak LH, et al. A small molecule mimicking a phosphatidylinositol (4,5)-bisphosphate binding pleckstrin homology domain. ACS Chem Biol. 2011;6:1382–1390. doi: 10.1021/cb2003187. [DOI] [PubMed] [Google Scholar]
- 81.Schrödinger LLC The PyMOL Molecular Graphics System. 2010 [Google Scholar]
- 82.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen N-YN, et al. An ENU-induced mouse mutant of SHIP1 reveals a critical role of the stem cell isoform for suppression of macrophage activation. Blood. 2011;117:5362–5371. doi: 10.1182/blood-2011-01-331041. [DOI] [PubMed] [Google Scholar]
- 84.Manji SSM, et al. A mutation in synaptojanin 2 causes progressive hearing loss in the ENU-mutagenised mouse strain Mozart. PLoS ONE. 2011;6:e17607. doi: 10.1371/journal.pone.0017607. [DOI] [PMC free article] [PubMed] [Google Scholar]