Abstract
Aim
Although it is established that peri-implantitis is a bacterially induced disease, little is known about the bacterial profile of peri-implant communities in health and disease. The purpose of the present investigation was to examine the microbial signatures of the peri-implant microbiome in health and disease.
Materials and methods
Subgingival and submucosal plaque samples were collected from forty subjects with periodontitis, peri-implantitis, periodontal and peri-implant health and analyzed using 16S pyrosequencing.
Results
Peri-implant biofilms demonstrated significantly lower diversity than subgingival biofilms in both health and disease, however, several species, including previously unsuspected and unknown organisms, were unique to this niche. The predominant species in peri-implant communities belonged to the genera Butyrivibrio, Campylobacter, Eubacterium, Prevotella, Selenomonas, Streptococcus, Actinomyces, Leptotrichia, Propionibacterium, Peptococcus, Lactococcus and Treponema. Peri-implant disease was associated with lower levels of Prevotella and Leptotrichia and higher levels of Actinomyces, Peptococcus, Campylobacter, non-mutans Streptococcus, Butyrivibrio, and Streptococcus mutans than healthy implants. These communities also demonstrated lower levels of Prevotella, non-mutans Streptococcus, Lactobacillus, Selenomonas, Leptotrichia, Actinomyces and higher levels of Peptococcus, Mycoplasma, Eubacterium, Campylobacter, Butyrivibrio, Streptococcus mutans, and Treponema when compared to periodontitis-associated biofilms.
Conclusion
The peri-implant microbiome differs significantly from the periodontal community in both health and disease. Peri-implantitis is a microbially heterogeneous infection with predominantly gram-negative species, and is less complex than periodontitis.
Keywords: Bacteria, dental implant, peri-implantitis, DNA, pyrosequencing, 16S periodontitis
Introduction
Endosseous implants have become widely accepted treatment options for the replacement of missing teeth, with over a million implants being placed each year in the USA alone(2000). Although dental implants have a high survival rate, infections in the peri-implant crevice have been widely reported(Jepsen et al., 1996, Lindhe and Meyle, 2008, Renvert et al., 2007). Peri-implantitis is a bacterially induced inflammatory reaction that results in loss of supporting bone around an implant in function, which may eventually lead to loss of the implant fixture (implant failure). Recent evidence indicates that this condition is significantly prevalent; occurring in 56% of subjects and 12–46% of implant sites(Lindhe and Meyle, 2008).
It is known that bacteria colonize the peri-implant crevice soon after implant placement to establish polymicrobial communities in this space(Quirynen et al., 2006, Sanz et al., 1990), and that the biofilm associated with failing implants is significantly different from that of healthy implants(Botero et al., 2005, Hultin et al., 2002, Leonhardt et al., 1999, Mombelli and Mericske-Stern, 1990, Quirynen et al., 2006, Renvert et al., 2007, Sanz et al., 1990, Shibli et al., 2008). These microbial events mimic those occurring in the subgingival crevice; and previous investigations suggested that the peri-implant biofilm does not differ significantly from the subgingival biofilm, either in health or in disease(Botero et al., 2005, Hultin et al., 2002, Leonhardt et al., 1999, Mombelli and Mericske-Stern, 1990, Sanz et al., 1990, Shibli et al., 2008). Current treatment protocols of non-surgical debridement and antibiotic therapy have been based on this paradigm of microbial similarity, yet have demonstrated modest outcomes(Esposito et al., 2008, Renvert et al., 2009). Meanwhile, in vitro investigations have revealed that peri-implant community profiles are significantly influenced by the texture and composition of implant surfaces(Grimm et al., 2000, Grossner-Schreiber et al., 2009). Therefore, it is reasonable to expect that structural and topographical differences between implants and teeth will influence the composition of their respective microbiomes. In fact, emerging evidence indicates that periodontitis and peri-implantitis diverge significantly in their histopathological presentations; since the host response is dictated by these structural differences(Berglundh et al.). Thus, in order to advance our understanding of peri-implant and periodontal disease it is necessary to identify differences between implant and tooth-associated microbial communities.
Sequence analysis of the 16S ribosomal gene has provided new insights into the composition of the oral microbiome in health and disease, creating a paradigm shift in our understanding of these microbial communities(Li et al., Shchipkova et al., 2010). Pyrosequencing of PCR-amplified 16S rDNA (‘16S pyrotags’) is a next-generation sequencing methodology that generates thousands of sequences from several samples simultaneously. This unprecedented depth of coverage allows a more comprehensive examination of taxonomically heterogeneous community than was previously possible and has revealed a significantly greater level of microbial diversity than was previously apparent with Sanger sequencing(Charlson et al., 2010, Keijser et al., 2008, Li et al.). The purpose of the present investigation, therefore, was to examine microbial signatures associated with peri-implant health and disease and to identify differences between peri-implant and subgingival bacterial communities using 16S pyrotag sequencing and computational phylogenetics for bacterial identification.
Methods
Subject selection
Approval for this study was obtained from the Office of Responsible Research Practices at The Ohio State University. Ten subjects were identified in each of the following groups following clinical and radiographic examination: peri-implantitis, chronic periodontitis, periodontal and peri-implant health; and informed consent was obtained. Subjects were selected based on the criteria delineated by the Classification of Periodontal Diseases(Armitage, 1999) and the Consensus Report on Peri-implant Diseases(Lindhe and Meyle, 2008). Briefly, peri-implantitis was diagnosed when the implant demonstrated clinical signs of inflammation along with radiographic evidence of bone loss after at least one year in function, while implants not demonstrating bleeding on probing or suppuration with no radiographic evidence of marginal bone loss were classified as healthy. Similarly, subjects with probing depths ≤ 3mm and attachment loss ≤1mm were classified as healthy, while those exhibiting more than 30% of sites with attachment loss ≥ 4mm and were diagnosed with chronic periodontitis. Exclusion criteria for all groups included diabetes, pregnancy, HIV infection, use of immunosuppressant medications, bisphosphonates or steroids, antibiotic therapy or oral prophylactic procedures within the last three months and less than 20 teeth in the dentition. Inclusion criteria for the implant group were subjects with a single functioning implant bounded by teeth on either side.
Data collection
All subjects were examined by one and the same periodontist. Probing pocket depths (PPD) and clinical attachment levels (CAL) were recorded throughout the mouth on 6 sites per tooth using a PCP-UNC 15 probe. Bleeding on probing and plaque levels were recorded on a binary scale (presence/absence) for each surface.
Sample collection and DNA isolation
Samples were collected from each subject by inserting a total of 15 sterile endodontic paper points (Caulk-Dentsply) into the pockets, sulci or peri-implant crevice for 10 seconds, following isolation and supragingival plaque removal. From subjects with periodontitis, subgingival plaque samples were collected and pooled from four non-adjacent proximal sites demonstrating at least 6mm of clinical attachment loss and 5mm of probing pocket depths. From subjects with healthy or diseased implants, samples were collected from the selected peri-implant crevice. From periodontally healthy teeth, samples were collected and pooled from mesial sulci of 15 randomly selected teeth. Samples were placed in 1.5 ml microcentrifuge tubes and frozen at −80°C until further analysis. Bacteria were separated from the paper points by adding 200μl of phosphate buffered saline (PBS) to the tubes and vortexing. The points were then removed, and DNA was isolated with a Qiagen DNA MiniAmp kit (Qiagen, Valencia, CA) using the tissue protocol according to the manufacturer’s instructions.
Pyrosequencing
Multiplexed bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) was performed using the Titanium platform (Roche Applied Science, Indianapolis, IN) as previously described(Dowd et al., 2008) in a commercial facility (Research and Testing Laboratories, Lubbock, TX). Briefly, a single step PCR with broad-range universal primers and 22 cycles of amplification was used to amplify the 16S rRNA genes as well as to introduce adaptor sequences and sample-specific bar-code oligonucleotide tags into the DNA. Two regions of the 16S rRNA genes were sequenced: V1–V3 and V7–V9. The primers used for sequencing have been previously described (Kumar et al, submitted PlosONE). Adaptor sequences were trimmed from raw data with 98% or more of bases demonstrating a quality control of 30 and sequences binned into individual sample collections based on bar-code sequence tags, which were then trimmed. The resulting files were denoised with Pyronoise(Quince et al.) and depleted of chimeras using B2C2 (http://www.researchandtesting.com/B2C2.html). Sequences <300bp were discarded and the rest were clustered into species-level operational taxonomic units (s-OTUs) at 97% sequence similarity and assigned a taxonomic identity by alignment to locally hosted version of the Greengenes database(DeSantis et al., 2006) using the Blastn algorithm. Phylogenetic trees were generated and visualized using FastTree(Price et al., 2009). Unifrac and community diversity metrics were computed as previously described(Lozupone et al.). All analysis were conducted using the QIIME pipeline(Caporaso et al.).
Statistical analysis
Similarities between the 4 communities were quantified by computing weighted UniFrac distances, which were used for principal coordinate analysis (PCoA). Shannon diversity index was computed using s-OTU data(Shannon, 1997). A variance stabilizing transformation was used to create normal distribution of the data(Shchipkova et al., 2010). The proportion (p) of each s-OTU in the community of each subject was expressed as X = sin −1(√p) and ANOVA and 2-sample t-tests were used to compare the means of this transformed variable X across groups. Statistical analysis was carried out with JMP (SAS Institute Inc., Cary, NC) and graphics created using R (http://www.r-project.org/).
Results
Subgingival and submcosal samples were collected from 10 subjects in each group. Seven of the twenty subjects with implants had lost the tooth due to periodontitis and 5 due to root fractures or non-restorable coronal structure. The remaining presented with a long-standing edentulous space and the etiology of tooth loss was unknown. The groups were matched for age, gender, ethnicity and periodontal and peri-implant health status and hence, no differences were apparent between these two groups (Table 1). The clinical status of periodontally healthy subjects was significantly different from those with periodontitis; similarly, sites with peri-implantitis differed clinically from those with healthy implants.
Table 1.
Demographic and clinical characteristics of sample population.
| Healthy teeth | Healthy implants | Peri-implantitis | Periodontitis | |
|---|---|---|---|---|
| Age (years) | 35.5 ± 7.5 | 41 ± 5.4 | 39 ± 6.3 | 42.5 ± 8.2 |
| Sex (% males) | 72 | 65 | 60 | 75 |
| Implant type | ||||
| Astra (Osseospeed) | 5 | 5 | ||
| Zimmer (MTX) | 4 | 4 | ||
| Nobel (Replace Groovy) | 1 | 1 | ||
| Implant location | ||||
| Mandibular anterior | 2 | 2 | ||
| Maxillary anterior | 2 | 2 | ||
| Maxillary posterior | 4 | 4 | ||
| Mandibular posterior | 2 | 2 | ||
| Years in function | 5.3 ± 3.5 | 4.2 ± 3.8 | ||
| CAL (mm) | 0.8 ± 0.7 | 6.7 ± 2.9 | ||
| PPD (mm) | 2.4 ± 1.9 | 3.2 ± 1.7 | 6.0 ± 3.5 | 7.5 ± 3.5 |
| BOP (% frequency of detection) | 15 ± 3 | 10 ± 4 | 100 | 100 |
| Radiographic bone loss (%) | 0 | 0 | 65 ± 15 | 56 ± 12 |
CAL = Clinical Attachment Loss
PPD = Probing pocket depth
BOP = Bleeding on probing
A total of 397,286 chimera-depleted, denoised sequences were used for analysis. Overall, these sequences represented the phyla Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, Spirochaetes, TM7, OP11 and Synergistes; with Firmicutes accounting for 53.4% of all sequences. These sequences represented 84 genus level operational taxonomic units (OTUs) and 370 species-level OTUs. 3.2% of sequences were not classifiable at the genus level and were placed in higher order classifications. Uncultivated phylotypes accounted for an average of 52.6 ± 5.3% (mean±standard error) of the healthy subgingival biofilm and 44.6 ± 3.5%, 77.8 ± 3.9% and 48.4 ± 7.7% of the biofilms associated with periodontitis, healthy implants and peri-implantitis respectively.
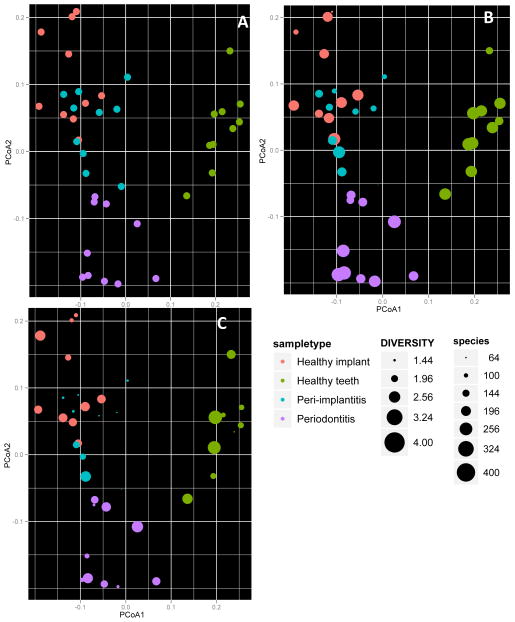
Principal component analysis of weighted Unifrac distances revealed a distinct partitioning of the bacterial communities associated with healthy teeth and implants (p< 0.01, PERMANOVA, Figure 1A). A similarly high level of partitioning was observed between the microbial profiles of periodontitis and peri-implantitis. A greater degree of similarity was observed between healthy and diseased implants than between healthy and diseased teeth. Both healthy and disease-associated subgingival microbial communities demonstrated significantly greater diversity than peri-implant communities (p<0.01, ANOVA, Figure 1B), however, the microbial profile of healthy implants was significantly more diverse than that of peri-implantitis (p<0.05, 2-sample t-test). The biofilm associated with peri-implantitis demonstrated significantly lower species richness when compared to healthy implants or diseased teeth (p<0.01, 2-sample t-test, Figure 1C). Species belonging to an average of 3 genera comprised 75% of the microbial community in each individual with peri-implantitis, while an average of 10 genera comprised the same proportion of the periodontitis-associated community in each individual (data not shown).
Figure 1.
Community characteristics of the peri-implant and subgingival microbiomes in health and disease. Panel A shows the Principal Co-ordinate Analysis of UniFrac distances, Panel B shows the Shannon diversity index for the 40 samples and Panel C shows the species richness of each sample.
The gram status of the peri-implant and subgingival communities is shown in Figure 2. As a whole, peri-implant communities contained significantly greater abundance of gram-negative bacteria than subgingival communities (p<0.01, ANOVA). Furthermore, healthy implants demonstrated the highest levels of gram-negative anaerobes and lowest abundance of gram-positive aerobes when compared to the other communities (p<0.01, ANOVA). The proportion of gram-negative anaerobes was also significantly different between peri-implantitis and periodontitis (p<0.01, 2-sample t-test).
Figure 2.
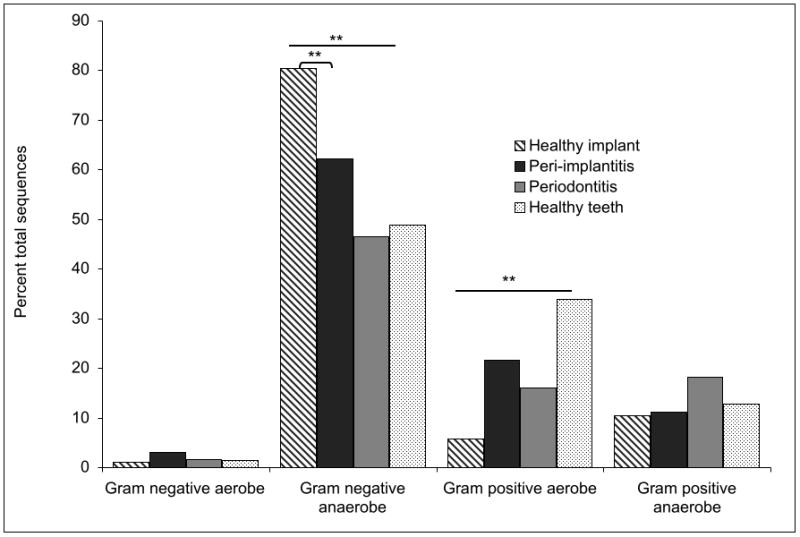
Gram staining characteristics of peri-implant and subgingival communities in health and disease. The gram characteristics of the uncultivated organisms was inferred from their nearest cultivated phylogenetic neighbor. (* p<0.05, ** p<0.01, ***p<0.001, ANOVA and 2-sample t-test on transformed variables).
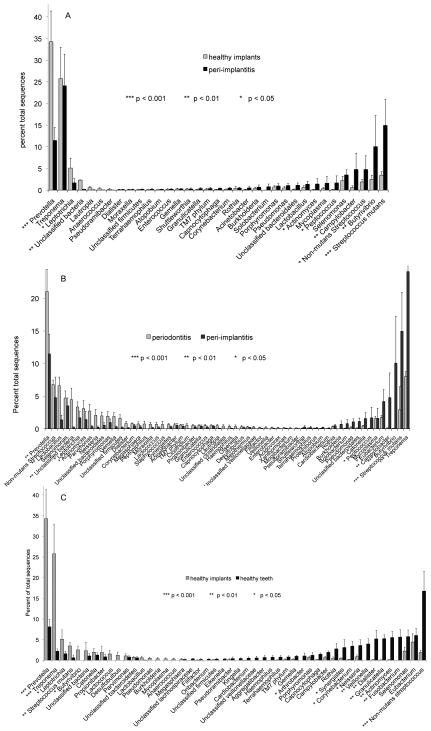
Peri-implantitis associated communities demonstrated significantly lower levels of the genera Leptotrichia, Propionibacter and Prevotella and higher levels of Actinomyces, Peptococcus, Campylobacter, non-mutans Streptococcus, Butyrivibrio, and Streptococcus mutans than healthy implants (p<0.05, 2-sample t-test on transformed variable, Figure 3A). These communities also demonstrated lower levels of Prevotella, non-mutans Streptococcus, Lactobacillus, Selenomonas, Leptotrichia, Actinomyces and higher levels of Peptococcus, Mycoplasma, Eubacterium, Campylobacter, Butyrivibrio, Streptococcus mutans, and Treponema when compared to periodontitis-associated biofilms (p<0.05, 2-sample t-test on transformed variable, Figure 3B). The greatest differences were observed between healthy implants and teeth (Figure 3C). Healthy implants demonstrated higher levels of Prevotella, Treponema, Leptotrichia, Streptococcus mutans, Butyrivibrio, Catonella, Propionibacter and Lactococcus and lower levels of Arthrobacter, Synergistes, Corynebacterium, Neisseria, Veillonella, Dialister, Granulicatella, Actinomyces, Fusobacterium and non-mutans Streptococcus when compared to teeth (p<0.05, 2-sample t-test on transformed variable).
Figure 3.
Distribution of sequences by genus. Comparisons between peri-implant communities in health and disease are shown in 3A, between peri-implantitis and periodontitis associated communities in #B and between healthy peri-implant and subgingival communities in 3C.
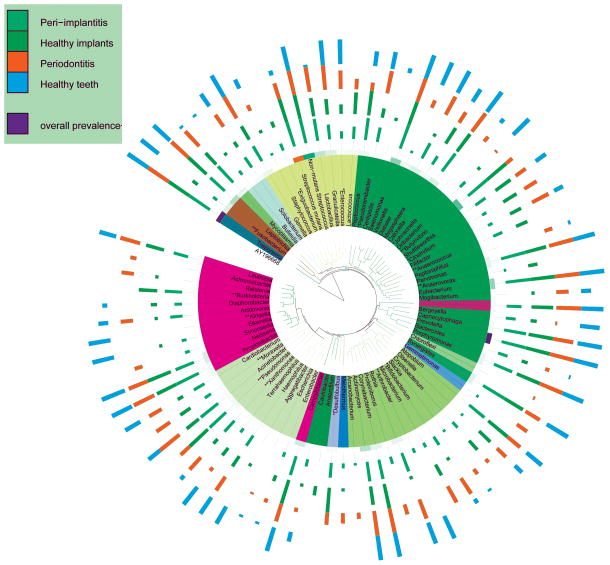
Several genera, e.g., Pseudoramibacter, Enterococcus, Moraxella, Desulfobulbus, Xanthomonas, Arthrobacter, Anaerococcus, Anaerovorax, Butyrivibrio, Exiguobacterium, Kingella, Fusobacterium, Pseudomonas, Burkholderia, Bifidobacterium, Clostridium and Johnsonella demonstrated significantly different detection frequencies between the groups (p<0.05, Fisher’s exact test). Certain genera, for example, Escherichia, Achromobacter, Peptoniphilus, Chloroflexi, Kingella and Johnsonella were detected only in the subgingival sulcus while the genera Anaerococcus, Anaerovorax, Anaerofilum, Exiguobacterium and Burkholderia were detected only in the peri-implant crevice (Figure 4).
Figure 4.
Detection frequency and overall abundances of genera in the four groups of samples. The phylogenetic tree was created using the Interactive Tree of Life (ITOL). (* p<0.05, ** p<0.01, ***p<0.001, Fisher’s exact test).
Discussion
Explorations of the peri-implant microbial community using cultivation and targeted molecular approaches have promulgated a paradigm of microbial similarity between subgingival and peri-implant communities. The present study harnessed the phylogenetic resolution of 16S deep sequencing and computational phylogenetics; which allowed a quantitative exploration of the peri-implant microbiome. Using this approach, we present the first comprehensive exploration of peri-implant microbial communities in health and disease and reveal a surprising picture of the peri-implant microbiome.
The main findings of the present investigation focus on the compositional differences between peri-implant and subgingival communities. Previous investigations sought to determine the extent of susceptibility conferred by pre-existing periodontitis to implant failure and therefore, compared the levels of known periodontal pathogens between teeth and implants(Botero et al., 2005, Hultin et al., 2002, Leonhardt et al., 1999, Mombelli and Mericske-Stern, 1990, Renvert et al., 2007, Sanz et al., 1990, Shibli et al., 2008). However, employing an open-ended, global approach to examining peri-implant and subgingival microbial communities revealed a significant partitioning between bacterial communities associated with teeth and implants (Figure 1A), suggesting that different bacterial lineages contributed to the composition of these communities. This is in contrast to the findings of previous investigations and may be attributed to differences in methodologies of plaque sampling and bacterial identification. In the present study, in an effort to obtain representative samples of disease and health, plaque samples were collected and pooled from sites with periodontitis or health in each subject, however implant samples were derived from a single implant. It is possible that this may have contributed to the differences observed between these samples and warrants further investigation. Unlike the aforementioned studies which were primarily focused on translocation of bacteria within intra-oral sites, the objective of the present study was an in-depth characterization of peri-implant communities and therefore, different subjects contributed subgingival and peri-implant samples. This was done for several reasons, the most important of which is that control of periodontal disease and successful maintenance of periodontal health is necessary before implant placement; making it difficult to collect samples of periodontitis-associated and peri-implantitis associated biofilms from the same subject. A more significant difference between the previous investigations and the present study is the different methodologies used for bacteria identification (anaerobic culturing and DNA-DNA checkerboard versus pyrosequencing). The limitations of using cultivation to identify organisms is well established, however, the drawbacks of directed DNA techniques in explorations of novel ecosystems is not as widely appreciated. These techniques employ primers and probes targeted to selected organisms; and therefore are not designed to detect unknown or unsuspected species in an ecosystem. Thus, this study underscores the importance of using a quantitative open-ended approach to comprehensively examine complex microbiomes.
The Shannon Diversity index provides a composite value that measures both the number of species (species richness) as well as the proportion of each species (species evenness)(Shannon, 1997). In an effort to minimize heterogeneity arising from number of sequences in each sample, all samples were analyzed by rarefaction and diversity estimated at a common sampling depth (9,000 sequences). Periodontitis-associated communities demonstrated greater diversity and richness when compared to healthy teeth (Figures 2B and 2C). This finding corroborates previously available evidence(Hutter et al., 2003, Kumar et al., 2005, Paster et al., 2001). By contrast, peri-implantitis associated communities demonstrated the least amount of diversity, with the fewest number of species, suggesting that peri-implantitis is a simple infection. This is further reinforced by the finding that in each individual, species belonging to no more than four genera comprised the majority (75%) of the flora. However, these dominant species were not the same in all individuals, suggesting that the disease is microbially heterogeneous.
The data suggest that peri-implantitis is similar to periodontitis in being a predominantly gram-negative infection, a finding that corroborates previous evidence(Shibli et al., 2008, Sanz et al., 1990, Leonhardt et al., 1999). However, a surprising finding was the large abundance of gram-negative anaerobes associated with healthy implants. This in contradistinction to prior investigations(Sanz et al., 1990, Apse et al., 1989, Mombelli and Mericske-Stern, 1990); nevertheless, taken together with the fact that a large fraction (77%) of the microbiome associated with peri-implant health is uncultivated, it is likely that, by circumventing the need for cultivation, the deep sequencing methodology provided a more representative picture than was previously possible using culturing.
Certain genera important in the etiology of periodontitis, namely, Treponema, Prevotella, Campylobacter and Eubacterium were also dominant members of the peri-implantitis associated flora (Figure 3). This is similar to earlier reports on abundances of some of these periodontopathogens in the peri-implant crevice(Leonhardt et al., 1999, Sanz et al., 1990, Shibli et al., 2008). However, the levels of Treponema, Campylobacter and Eubacterium were significantly higher in both peri-implant health and disease than in periodontitis. Additionally, some unusual lineages, for example, Butryvibrio fibrisolvens and Streptococcus mutans, were found in high abundance in the peri-implant crevice, as well as less numerous species belonging to the genera Peptococcus, Mycoplasma, Anaerococcus, Anaerovorax, Anaerofilum, Burkholderia and Exiguobacterium. B.fibrisolvens is a butyrate producer in the human gastrointestinal tract(Brown and Moore, 1960); however, its presence in the oral cavity has not been previously reported. This species was detected in every peri-implant crevice and in none of the subgingival sulci. The levels of these species were higher in peri-implantitis compared to healthy implants (Figure 2A). Further, a strong correlation was observed between levels of this organism and Treponema (r=0.69) irrespective of implant health status, suggesting a possible metabolic co-operation, since isobutyric acid is an important growth requirement for the treponemes(Hardy and Munro, 1966). A very interesting finding is that the organisms found only in the peri-implant crevice (Butyrivibrio, Anaerovorax, Anaerococcus, Anaerofilum, Exiguobacterium and Burkholderia) are known fermenters and of these, Butyrivibrio, Anaerovorax, Anaerococcus and Burkholderia are prolific butyrate producers(Matthies et al., 2000, Wang et al., 2003, Ezaki et al., 2001). The significance of this finding and its impact on therapy cannot be addressed by this study and an in-depth examination of butyrate-producers in the peri-implant crevice is warranted. Members of the genus Streptococcus have an interesting association with oral health; while most Streptococci are early colonizers of both supragingival and subgingival biofilms(Kolenbrander and Palmer, 2004), S.mutans has a strong association with dental caries(Gross et al., 2010). The present investigation identified high levels of S.mutans in peri-implant, but not periodontal, communities. S. mutans has been previously detected in sites with peri-implantitis; with a decrease in levels following therapy(Persson et al., 2010, Renvert et al., 2007), suggesting that this organism is important in peri-implant disease. Thus, the data suggest that while implant surfaces are colonized by periodontal bacteria, certain previously unknown and unsuspected bacterial species preferentially colonize this niche.
In summary, the data indicate that although certain periodontal bacteria can be found in the peri-implant crevice, the microbial fingerprint of peri-implant communities differ from subgingival profiles in several ways. Peri-implant biofilms demonstrate significantly lower diversity than subgingival biofilms in both health and disease; and several species, including previously unsuspected and unknown organisms, are unique to this niche. Further, peri-implantitis appears to be a simple, yet microbially heterogeneous infection, which is predominantly gram-negative.
Clinical relevance.
Scientific rationale
It is known that peri-implantitis is a bacterially induced disease, however, the composition of peri-implant microbial communities in health and disease is not well understood.
Findings
The profiles of the peri-implant and subgingival microbiomes were distinct and significantly different, with several previously unsuspected organisms detected around implants.
Clinical implications
Current protocols for treatment of peri-implantitis have been based on a paradigm of microbial similarity between implants and teeth and have demonstrated modest outcomes. The present study demonstrates critical differences between these communities, suggesting a probably different mechanism for the pathogenesis for peri-implantitis.
Acknowledgments
The research was supported by NIH grant 1R03DE018734-01A1
sources of funding: This study was funded by a start-up grant awarded to the corresponding author (PS Kumar) by the College of Dentistry, The Ohio State University.
Footnotes
Conflict of interest:
None of the authors have any conflicts of interest.
References
- Annual Industry Report. Implant Dentistry. 2000;9:192–194. [Google Scholar]
- Apse P, Ellen RP, Overall CM, Zarb GA. Microbiota and crevicular fluid collagenase activity in the osseointegrated dental implant sulcus: a comparison of sites in edentulous and partially edentulous patients. J Periodontal Res. 1989;24:96–105. doi: 10.1111/j.1600-0765.1989.tb00863.x. [DOI] [PubMed] [Google Scholar]
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- Berglundh T, Zitzmann NU, Donati M. Are peri-implantitis lesions different from periodontitis lesions? J Clin Periodontol. 38(Suppl 11):188–202. doi: 10.1111/j.1600-051X.2010.01672.x. [DOI] [PubMed] [Google Scholar]
- Botero JE, Gonzalez AM, Mercado RA, Olave G, Contreras A. Subgingival microbiota in peri-implant mucosa lesions and adjacent teeth in partially edentulous patients. J Periodontol. 2005;76:1490–1495. doi: 10.1902/jop.2005.76.9.1490. [DOI] [PubMed] [Google Scholar]
- Brown DW, Moore WEC. Distribution of Butyrivibrio Fibrisolvens in Nature. Journal of dairy science. 1960;43:1570–1574. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010;5:e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis. 2008;5:459–472. doi: 10.1089/fpd.2008.0107. [DOI] [PubMed] [Google Scholar]
- Esposito M, Grusovin MG, Coulthard P, Worthington HV. The efficacy of interventions to treat peri-implantitis: a Cochrane systematic review of randomised controlled clinical trials. Eur J Oral Implantol. 2008;1:111–125. [PubMed] [Google Scholar]
- Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. International Journal of Systematic and Evolutionary Microbiology. 2001;51:1521–1528. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
- Grimm WD, Cichon P, van der Hoeven H, Langendijk PS, Smith F, Worley MG, Schmitz I, Offenbacher S. The influence of sulfate-reducing bacteria colonization of 2 different bioresorbable barrier membranes for GTR. An 18-month case-controlled microbiologic and clinical study. Int J Periodontics Restorative Dent. 2000;20:91–99. [PubMed] [Google Scholar]
- Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232–10.. JCM.01232–10 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossner-Schreiber B, Teichmann J, Hannig M, Dorfer C, Wenderoth DF, Ott SJ. Modified implant surfaces show different biofilm compositions under in vivo conditions. Clin Oral Implants Res. 2009;20:817–826. doi: 10.1111/j.1600-0501.2009.01729.x. [DOI] [PubMed] [Google Scholar]
- Hardy PH, Jr, Munro CO. Nutritional Requirements of Anaerobic Spirochetes I. Demonstration of Isobutyrate and Bicarbonate as Growth Factors for a Strain of Treponema microdentium. J Bacteriol. 1966;91:27–32. doi: 10.1128/jb.91.1.27-32.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultin M, Gustafsson A, Hallstrom H, Johansson LA, Ekfeldt A, Klinge B. Microbiological findings and host response in patients with peri-implantitis. Clin Oral Implants Res. 2002;13:349–358. doi: 10.1034/j.1600-0501.2002.130402.x. [DOI] [PubMed] [Google Scholar]
- Hutter G, Schlagenhauf U, Valenza G, Horn M, Burgemeister S, Claus H, Vogel U. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology. 2003;149:67–75. doi: 10.1099/mic.0.25791-0. [DOI] [PubMed] [Google Scholar]
- Jepsen S, Ruhling A, Jepsen K, Ohlenbusch B, Albers HK. Progressive peri-implantitis. Incidence and prediction of peri-implant attachment loss. Clin Oral Implants Res. 1996;7:133–142. doi: 10.1034/j.1600-0501.1996.070207.x. [DOI] [PubMed] [Google Scholar]
- Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJJ. Human Oral Bacterial Biofilms. In: Ghannoum M, O’Toole G, editors. Microbial Biofilms. Washington D.C: ASM Press; 2004. pp. 85–117. [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt A, Renvert S, Dahlen G. Microbial findings at failing implants. Clin Oral Implants Res. 1999;10:339–345. doi: 10.1034/j.1600-0501.1999.100501.x. [DOI] [PubMed] [Google Scholar]
- Li L, Hsiao WW, Nandakumar R, Barbuto SM, Mongodin EF, Paster BJ, Fraser-Liggett CM, Fouad AF. Analyzing Endodontic Infections by Deep Coverage Pyrosequencing. J Dent Res. doi: 10.1177/0022034510370026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhe J, Meyle J. Peri-implant diseases: Consensus Report of the Sixth European Workshop on Periodontology. J Clin Periodontol. 2008;35:282–285. doi: 10.1111/j.1600-051X.2008.01283.x. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. Isme J. 5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies C, Evers S, Ludwig W, Schink B. Anaerovorax odorimutans gen. nov., sp. nov., a putrescine-fermenting, strictly anaerobic bacterium. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1591–1594. doi: 10.1099/00207713-50-4-1591. [DOI] [PubMed] [Google Scholar]
- Mombelli A, Mericske-Stern R. Microbiological features of stable osseointegrated implants used as abutments for overdentures. Clin Oral Implants Res. 1990;1:1–7. doi: 10.1034/j.1600-0501.1990.010101.x. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson GR, Samuelsson E, Lindahl C, Renvert S. Mechanical non-surgical treatment of peri-implantitis: a single-blinded randomized longitudinal clinical study. II. Microbiological results. J Clin Periodontol. 2010;37:563–573. doi: 10.1111/j.1600-051X.2010.01561.x. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Molecular Biology and Evolution. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. %R 1610.1093/molbev/msp1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirynen M, Vogels R, Peeters W, van Steenberghe D, Naert I, Haffajee A. Dynamics of initial subgingival colonization of ‘pristine’ peri-implant pockets. Clin Oral Implants Res. 2006;17:25–37. doi: 10.1111/j.1600-0501.2005.01194.x. [DOI] [PubMed] [Google Scholar]
- Renvert S, Roos-Jansaker AM, Lindahl C, Renvert H, Rutger Persson G. Infection at titanium implants with or without a clinical diagnosis of inflammation. Clin Oral Implants Res. 2007;18:509–516. doi: 10.1111/j.1600-0501.2007.01378.x. [DOI] [PubMed] [Google Scholar]
- Renvert S, Samuelsson E, Lindahl C, Persson GR. Mechanical non-surgical treatment of peri-implantitis: a double-blind randomized longitudinal clinical study. I: clinical results. J Clin Periodontol. 2009;36:604–609. doi: 10.1111/j.1600-051X.2009.01421.x. [DOI] [PubMed] [Google Scholar]
- Sanz M, Newman MG, Nachnani S, Holt R, Stewart R, Flemmig T. Characterization of the subgingival microbial flora around endosteal sapphire dental implants in partially edentulous patients. Int J Oral Maxillofac Implants. 1990;5:247–253. [PubMed] [Google Scholar]
- Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- Shchipkova AY, Nagaraja HN, Kumar PS. Subgingival Microbial Profiles of Smokers with Periodontitis. J Dent Res. 2010 doi: 10.1177/0022034510377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibli JA, Melo L, Ferrari DS, Figueiredo LC, Faveri M, Feres M. Composition of supra- and subgingival biofilm of subjects with healthy and diseased implants. Clin Oral Implants Res. 2008;19:975–982. doi: 10.1111/j.1600-0501.2008.01566.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Heazlewood SP, Krause DO, Florin THJ. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. Journal of Applied Microbiology. 2003;95:508–520. doi: 10.1046/j.1365-2672.2003.02005.x. [DOI] [PubMed] [Google Scholar]