Abstract
Escherichia coli cells lacking low molecular weight penicillin binding proteins (LMW PBPs) exhibit morphological alterations that also appear when the septal protein FtsZ is mislocalized, suggesting that peptidoglycan modification and division may work together to produce cell shape. We found that in strains lacking PBP5 and other LMW PBPs, higher FtsZ concentrations increased the frequency of branched cells and incorrectly oriented Z rings by 10- to 15-fold. Invagination of these rings produced improperly oriented septa, which in turn gave rise to asymmetric cell poles that eventually elongated into branches. Branches always originated from the remnants of abnormal septation events, cementing the relationship between aberrant cell division and branch formation. In the absence of PBP5, PBP6 and DacD localized to nascent septa, suggesting that these PBPs can partially substitute for the loss of PBP5. We propose that branching begins when mislocalized FtsZ triggers the insertion of inert peptidoglycan at unusual positions during cell division. Only later, after normal cell wall elongation separates the patches, do branches become visible. Thus, a relationship between the LMW PBPs and cytoplasmic FtsZ ultimately affects cell division and overall shape.
Keywords: cell shape; branching; FtsZ; PBPs; cell division; septation; D,D-carboxypeptidase
Introduction
Escherichia coli is a rod shaped bacterium with a uniform diameter and two hemispherical poles, a morphology that remains constant for most mutants and growth conditions. The mechanisms that generate and maintain this remarkable uniformity are the subject of intense recent scrutiny (Cabeen & Jacobs-Wagner, 2005, Cabeen & Jacobs-Wagner, 2007, Pichoff & Lutkenhaus, 2007, Singh & Montgomery, 2011, Young, 2003, Zapun et al., 2008). One morphological question has received much less attention since it was raised 18 years ago by Åkerlund et al (Åkerlund et al., 1993) and since re-addressed (Gullbrand et al., 1999, Young, 2003). That is the issue of why and how E. coli cells sometimes grow as branched forms.
Branched E. coli were observed more than 40 years ago (Suit et al., 1967), and since then E. coli and certain of its mutants have been seen to branch in many different conditions. For example, E. coli cells can take on Y (forked) or X shapes or can grow with amorphous kinks or buds of irregular diameter (Denome et al., 1999, Gullbrand et al., 1999, Nelson & Young, 2000, Woldringh et al., 1994, Young, 2003). Much evidence indicates that E. coli shape and branching is altered by treatments that inhibit cell division, DNA replication or peptidoglycan synthesis (Young, 2003), though the underlying mechanisms that create such cells remain unclear. Woldringh et al proposed that branches arise during division (Woldringh et al., 1994). Subsequent work has questioned this viewpoint because branch sites do not appear to coincide with sites where division is taking place and because cells branch even when the essential septation proteins FtsZ and FtsI are inactive (Gullbrand et al., 1999). They also point out that branching is exacerbated by mutations that affect aspects of peptidoglycan biochemistry which seem unrelated to septation (Gullbrand et al., 1999, Nelson & Young, 2000, Nelson & Young, 2001). Such results led Gullbrand et al to propose that branches develop from small asymmetries in the cell wall and not during the course of division (Gullbrand et al., 1999). The nature and origin of these asymmetries were not defined, and remain mysterious.
Notwithstanding the above arguments, a unifying theme among the disparate branching conditions is that they all, directly or indirectly, affect the structure, placement or function of the septation-specific FtsZ ring, which strongly suggests that FtsZ contributes to gross cell shape and branching (Young, 2003). Since bacterial morphology is tied to peptidoglycan (PG) synthesis, it seems reasonable that FtsZ would mediate its morphological effects by influencing this process. Consistent with this logic, cell shape depends on a relationship between FtsZ and the low molecular weight penicillin binding proteins (LMW PBPs), a set of enzymes that make minor modifications to peptidoglycan (Varma & Young, 2004). For example, inhibiting FtsZ in mutants lacking PBPs 5 and 7 causes a large proportion of the cells to grow as spiral filaments (Varma & Young, 2004). In addition, though FtsZ is best known for directing PG synthesis during cell division, it can also direct PG insertion into side walls near the poles in a process that is independent of septation (Varma et al., 2007, Varma & Young, 2009). Thus, there is ample phenomenological and experimental evidence that interfering with FtsZ alters bacterial morphology. What is not clear is how FtsZ performs this function.
Abundant evidence also indicates that modifying the bacterial cell wall affects cell shape and branching. In particular, an exuberant variety of morphological irregularities is exhibited by E. coli strains lacking one or more LMW PBPs that act as D,D-carboxypeptidases (D,D-CPases) or D,D-endopeptidases (D,D-EPases) (Denome et al., 1999, Nelson & Young, 2000, Nelson & Young, 2001, Young, 2003). Abnormal morphologies are also observed in mutants of Helicobacter pylori, Listeria monocytogenes and Streptococcus pneumoniae that lack similar D,D-CPase or D,D-EPase activities (Bonis et al., 2010, Pilgrim et al., 2003, Schuster et al., 1990, Sycuro et al., 2010). These observations suggest that PG-modifying enzymes help maintain proper cell shape in a variety of bacteria, but the reasons for these morphological effects are unclear. Why should a set of non-essential periplasmic enzymes that make relatively minor modifications to peptidoglycan have such a dramatic effect on bacterial shape, especially if a major morphological determinant is FtsZ, a cytoplasmic protein?
One hint for answering the above questions is that branched and abnormally shaped E. coli have patches of inert peptidoglycan and proteins at their poles, at the tips of each branch, and at sites of bending, kinking or blebbing (de Pedro et al., 2003). In fact, these aberrations share the structural and biochemical characteristics of wild type poles (de Pedro et al., 2004, de Pedro et al., 1997, Nilsen et al., 2004), implying that cell branches may be generated by a distortion of normal cell division (Young, 2003). If so, the new ectopic poles could be derived by splitting pre-existing poles, they might arise de novo from unrelated sites in the sidewall, or they could be generated during the course of normal cell division.
The above considerations leave us with the following conundrum: the ends of branched cells have the characteristics of normal poles, which originate, as far as we know, only during cell division; but cells can form branches even when normal cell division is inhibited. Here, we present data to resolve this paradox by showing that branching in E. coli is due to the aberrant localization of FtsZ during cell division, though this triggering event may not give rise to visible branches until well after division is complete. Furthermore, we show that the periplasmic LMW PBPs affect the orientation of the cytoplasmic FtsZ ring, and that the secondary D,D-carboxypeptidases PBP6 and DacD localize to the septal ring in the absence of PBP5. In the absence of these PBPs the Z ring is misplaced more often, which increases the frequency of branching. We conclude that cell branching, at least in E. coli, is a symptom of inaccurate septation brought about by aberrant Z ring localization or orientation.
Results
FtsZ overexpression enhances branching in PBP mutants
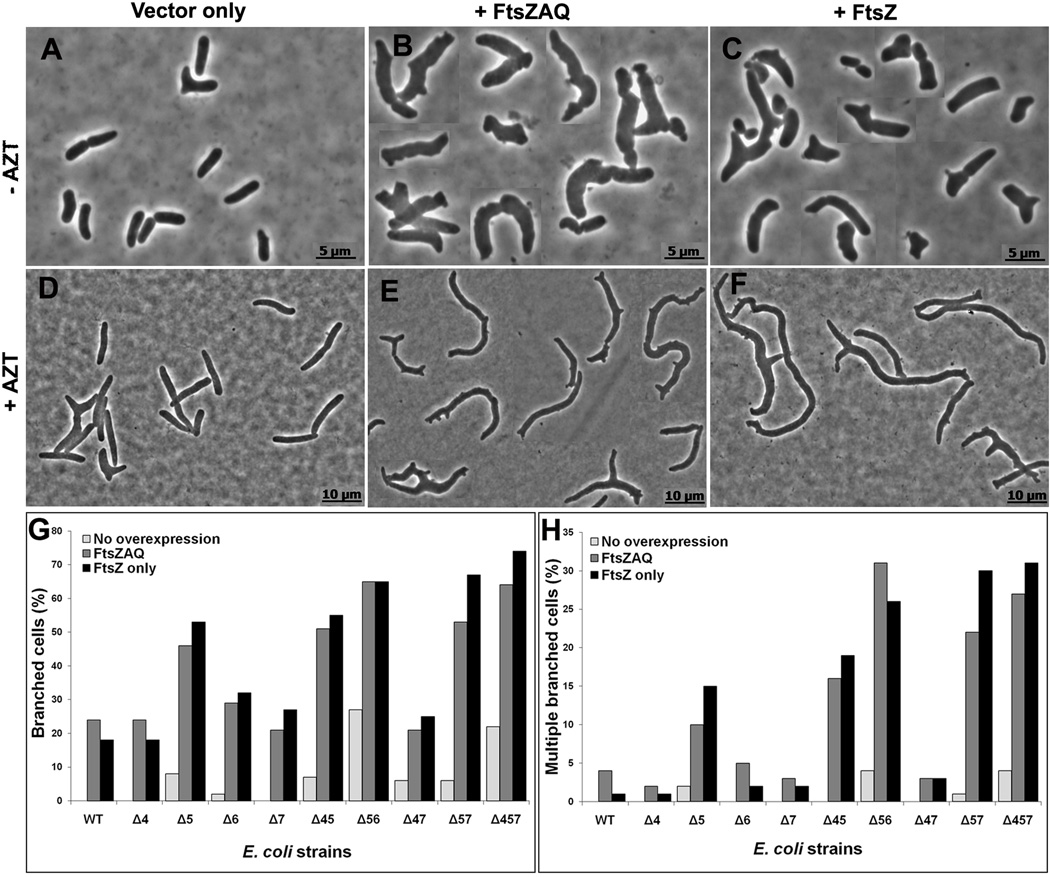
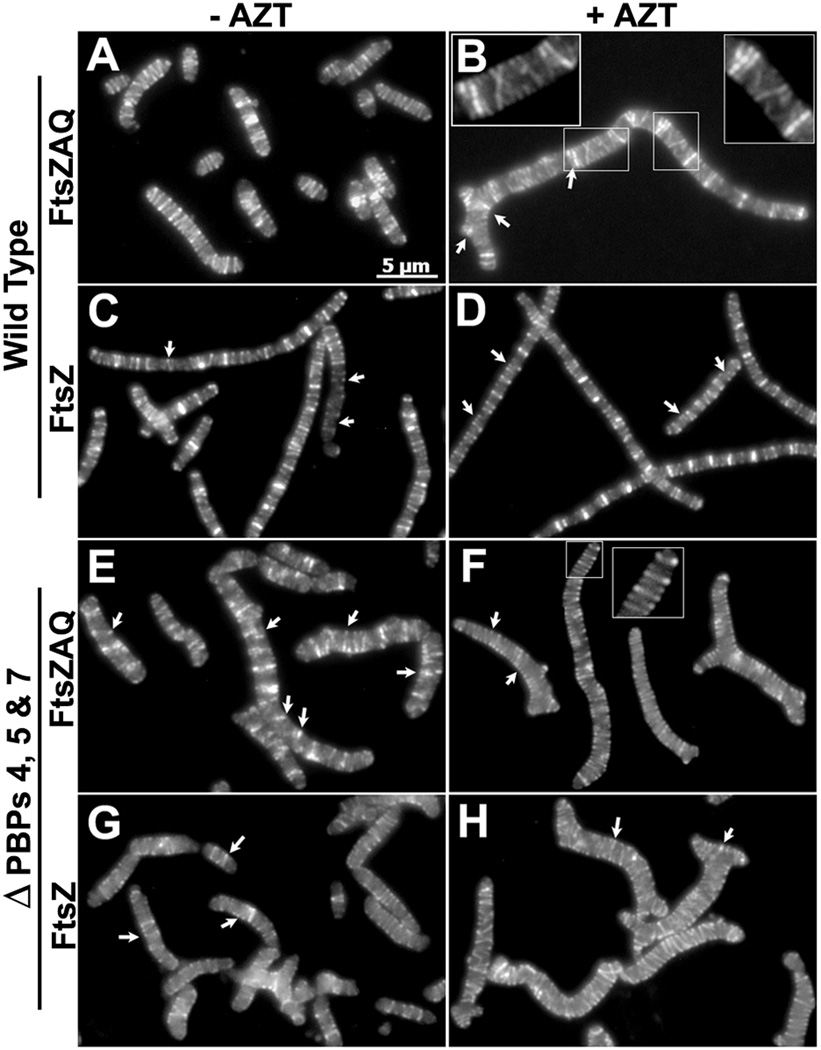
Some bacteria grow as branched cells when FtsZ levels are elevated (Dziadek et al., 2002, Ramos et al., 2005, Yu & Margolin, 2000), implying that this protein may be involved in the branching mechanism. Because similar morphological anomalies are observed in E. coli strains lacking certain LMW PBPs (Henderson et al., 1997, Meberg et al., 2004, Nelson et al., 2002, Nelson & Young, 2000, Nelson & Young, 2001), we reasoned that an increase in FtsZ might also exacerbate branching in these mutants. To test this idea, we overproduced FtsZ by transforming PBP mutants with plasmids pZAQ or pLPZ. pZAQ expresses the ftsQ, ftsA and ftsZ genes (Ward & Lutkenhaus, 1985), and pLPZ is a derivative of pZAQ that expresses only ftsZ (Experimental Procedures). In both cases gene expression was driven by native promoters so that FtsZ was expressed at a level approximately 3 times that of wild type (Fig. S1) (but note that the original authors measured the increase in FtsZ from pZAQ as being 7 times normal (Ward & Lutkenhaus, 1985)). When the FtsZAQ proteins were overexpressed in strains lacking different combinations of PBPs 4, 5, 6 and 7, overall cell shape was severely compromised and branching increased (Fig. 1). For example, when these proteins were overexpressed in E. coli CS315-1, a mutant lacking PBPs 4, 5 and 7, branched cells increased (compare Fig. 1B to Fig. 1A). In addition to cells with obvious branches, virtually all cells expressing excess FtsZAQ had rough and bumpy surfaces (e.g., Fig. 1B).
Fig. 1. FtsZ overexpression exacerbates branching in LMW PBP mutants.
E. coli CS315-1, which lacks PBPs 4, 5 and 7, harboring plasmids pBR322 (A, D) pZAQ (B, E) and pLPZ (C, F), were grown in LB-Tet medium until they reached an OD600 of 0.2. At this OD, cells were either harvested for microscopy (A, B, C) or were further grown for one mass doubling (MD) in the presence of 1 µg/ml of aztreonam to filament the cells (D, E, F). Panels A–C and D–F have different magnifications. The scale bars in panels A–C denote 5 µm and in panels D–F denote 10 µm. Panel G shows the percentage of cells with branches, including single and multiple protrusions. Panel H shows the percentage of cells with multiple branches (more than 3 poles per cell). In panels G and H, the percentage of branched cells in different strains harboring pBR322, pZAQ and pLPZ plasmids are represented by light, grey and black bars, respectively. At least 500 cells were counted for each strain. Panels G and H represent results averaged from two independent experiments.
In the foregoing experiments the extent of branching could not be quantified accurately because the cells were small, making it difficult to tell if morphological bulges were incipient branches or simply a reflection of uneven surface texture. However, buds and branches become more prominent when cell division is inhibited, making it easier to quantify the numbers of branched cells (Nelson & Young, 2000). Therefore, to distinguish between minor surface changes versus branching, cells were treated with aztreonam (AZT) for one mass doubling to inhibit PBP 3 and cell division. Branches (protrusions and buds) were more obvious in these elongated cells (Fig. 1E). In some PBP mutants (e.g., Δ56 and Δ457), almost all cells differed from the parental strain in length, diameter, and numbers of kinks or protrusions, but we scored filamentous cells only for the presence of branches and split poles. In elongated cells having at least one branch point, the percentage of branched cells in a mutant lacking only PBP5 increased from ~8% in the presence of wild type levels of FtsZAQ (Fig. 1G, Δ5 strain, light bar) to ~46% in the presence of additional FtsZAQ (Fig. 1G, Δ5 strain, grey bar). The numbers of branched cells continued to increase as additional LMW PBPs were deleted from this strain, so that when the FtsZAQ proteins were overexpressed in E. coli CS315-1 (Fig. 1E) the proportion of branched cells increased from ~22% to ~65% (Fig. 1G, strain Δ457, light vs grey bars). Similarly large increases in branching frequency occurred in all cells from which PBP5 had been deleted in combination with other LMW PBPs (Fig. 1G, all strains with Δ5). In the presence of excess FtsZAQ, mutants lacking only PBPs 4 or 7, alone or in combination, did not branch at higher rates than the parental strain, though the loss of these two PBPs did exacerbate branching in cells also lacking PBP5 (Fig. 1G). FtsZAQ-dependent branching increased from ~2% to ~29% in a mutant lacking only PBP6 (Fig. 1G, strain Δ6, light vs grey bars). The loss of both PBPs 5 and 6 raised the background level of branching to ~28% (Fig. 1G, Δ56 strain, light bar) and increased the rate of FtsZAQ-induced branching to ~65% (Fig. 1G, Δ56 strain, grey bar). Since excess FtsZAQ caused branching in 65–75% of cells from strains CS211-2 (lacking PBPs 5 and 6) and CS315-1 (lacking PBPs 4, 5 and 7), we did not investigate the phenotype in strains lacking even more PBPs.
Ectopic expression of FtsZAQ also increased the proportion of cells with severe branching (cells with two or more branch points) (Fig. 1H, all Δ5-containing strains, grey bars). In the extreme, when the FtsZAQ proteins were overexpressed in CS211-2 (Fig. 1H, Δ56 strain), in CS204-1 (Fig. 1H, Δ57 strain) or in CS315-1 (Fig. 1H, Δ457 strain), the percentage of multiply branched cells increased from <5% to ~28% (Fig. 1H, light vs grey bars). With one exception, the trends in severe branching were the same as those described for cells having at least one branch. For example, in mutants lacking only PBPs 4 or 7, the frequency of multiple branching was no greater than that seen in wild type cells (Fig. 1G, grey bars). These results indicated, once again, that PBP5 plays a major role in FtsZAQ-induced branching.
In the preceding experiments, increased branching was observed when all three division proteins (FtsZ, FtsA and FtsQ) were overproduced. To determine if branching was due to FtsZ alone or if it also required FtsA and FtsQ, we deleted ftsA and ftsQ from pZAQ without affecting the ftsZ promoters. The resulting plasmid, pLPZ, overproduced only FtsZ, and both pLPZ and pZAQ produced the same amount of FtsZ (Fig. S1). Cells overproducing FtsZ by itself branched at frequencies almost identical to those of strains harboring pZAQ (Fig. 1C and 1F; Fig. 1G and Fig. 1H, black bars), indicating that overexpression of FtsZ alone was sufficient to generate branches.
Finally, a surprising observation was that ~24% of parent strain cells exhibited branching when the FtsZAQ or FtsZ proteins were overexpressed (Fig. 1G, WT strain, grey and black bars). The pZAQ plasmid has been used to rescue the temperature sensitive filamentous phenotypes of several cell division proteins (Geissler et al., 2003, Geissler & Margolin, 2005), but there is no report of increased cell branching in such experiments (Begg et al., 1998, Dai & Lutkenhaus, 1992, Geissler & Margolin, 2005, Ward & Lutkenhaus, 1985). We found that E. coli MG1655 also exhibited similar branching frequencies in the presence of FtsZAQ (not shown), so the reason for the discrepancy does not seem to be that branching is strain specific. The best explanation is that branching was most clear when E. coli cells were elongated in the presence of aztreonam to accentuate morphological abnormalities. Also, the severity of branching was more obvious in mutants lacking several LMW PBPs. Thus, in the absence of these conditions, slight morphological changes may not have been apparent in previous investigations where FtsZ levels were elevated.
In short, moderate overproduction of FtsZ, alone or in combination with FtsA and FtsQ, exacerbated cell shape defects and enhanced the frequency of branching in E. coli. Importantly, morphological changes and branching were evident in wild type cells but were most apparent in mutants lacking PBP5 and other LMW PBPs. The results are consistent with previous reports that PBP5 plays an important role in cellular morphology (Nelson & Young, 2000, Nelson & Young, 2001) and suggests that cell shape is maintained by a relationship between division proteins and enzymes that modify the cell wall.
FtsZ overexpression does not change the muropeptide composition of peptidoglycan
Previous results lead to the idea that branches in LMW PBP mutants might be related to a change in the percentage of pentapeptides in peptidoglycan (Meberg et al., 2004, Varma et al., 2007), suggesting a connection between the state of FtsZ and cell wall synthesis. Therefore, we speculated that FtsZ overproduction might change the number of pentapeptides and affect branching frequency. To test this possibility, we analyzed the muropeptide composition of peptidoglycan from PBP mutants harboring vector plasmid and from cells expressing FtsZ from pZAQ or pLPZ. No major changes in muropeptide composition were observed among these strains (Table 1). Consistent with previous results, the number of pentapeptides and peptide crossbridges increased in PBP mutants, and did not depend on the presence or absence of plasmid vector (Table 1 and data not shown). Thus, branching was not caused by an obvious change in pentapeptide concentration or muropeptide composition.
Table 1.
FtsZ overexpression does not change peptidoglycan composition.
| Molecular percentage | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PG Composition |
Wild Type | Δ 4, 7 | Δ 5 | Δ 4, 5, 7 | ||||||||
| - | FtsZ, A, Q | FtsZ | - | FtsZ, A, Q | FtsZ | - | FtsZ, A, Q | FtsZ | - | FtsZ, A, Q | FtsZ | |
| Pentapetides | 1.5 | 1.1 | 1.0 | 0.7 | 1.4 | 1.1 | 7.4 | 7.2 | 6.9 | 21.7 | 25 | 23.6 |
| Cross linkage | 30.9 | 30.1 | 30.0 | 39.4 | 37.7 | 38.6 | 31.8 | 30.4 | 31.8 | 38.7 | 39.1 | 39.6 |
| Monomers | 69.0 | 69.6 | 69.6 | 61.0 | 62.3 | 61.5 | 67.0 | 68.4 | 66.8 | 62.0 | 61.9 | 62.2 |
| Dimers | 26.3 | 26.0 | 26.4 | 36.0 | 34.3 | 35.4 | 29.1 | 27.9 | 28.6 | 34.9 | 34.8 | 33.9 |
| Trimers | 2.3 | 2.1 | 1.8 | 1.7 | 1.7 | 1.6 | 1.3 | 1.3 | 1.6 | 1.9 | 2.1 | 2.8 |
| Lipoprotein | 11.4 | 9.3 | 9.5 | 9.8 | 10.2 | 9.5 | 9.3 | 8.5 | 10.4 | 7.5 | 7.2 | 7.0 |
| Anhydro | 1.9 | 1.7 | 1.5 | 1.0 | 2.1 | 1.6 | 1.7 | 0.9 | 2.9 | 1.7 | 1.9 | 0.8 |
| DAP-DAP | 3.2 | 2.5 | 2.6 | 4.0 | 3.5 | 2.8 | 3.2 | 2.7 | 3.3 | 1.8 | 2.3 | 2.0 |
Peptidoglycan was isolated from E. coli strains harboring pLP322 (−), pZAQ (FtsZAQ) or pLPZ (FtsZ), and the murein composition was analyzed by HPLC as described in Experimental Procedures. The molecular percentage of each muropeptide is reported. Strains that did not harbor any plasmid showed similar PG compositions (not shown). The results represent the average PG composition derived from two independent experiments.
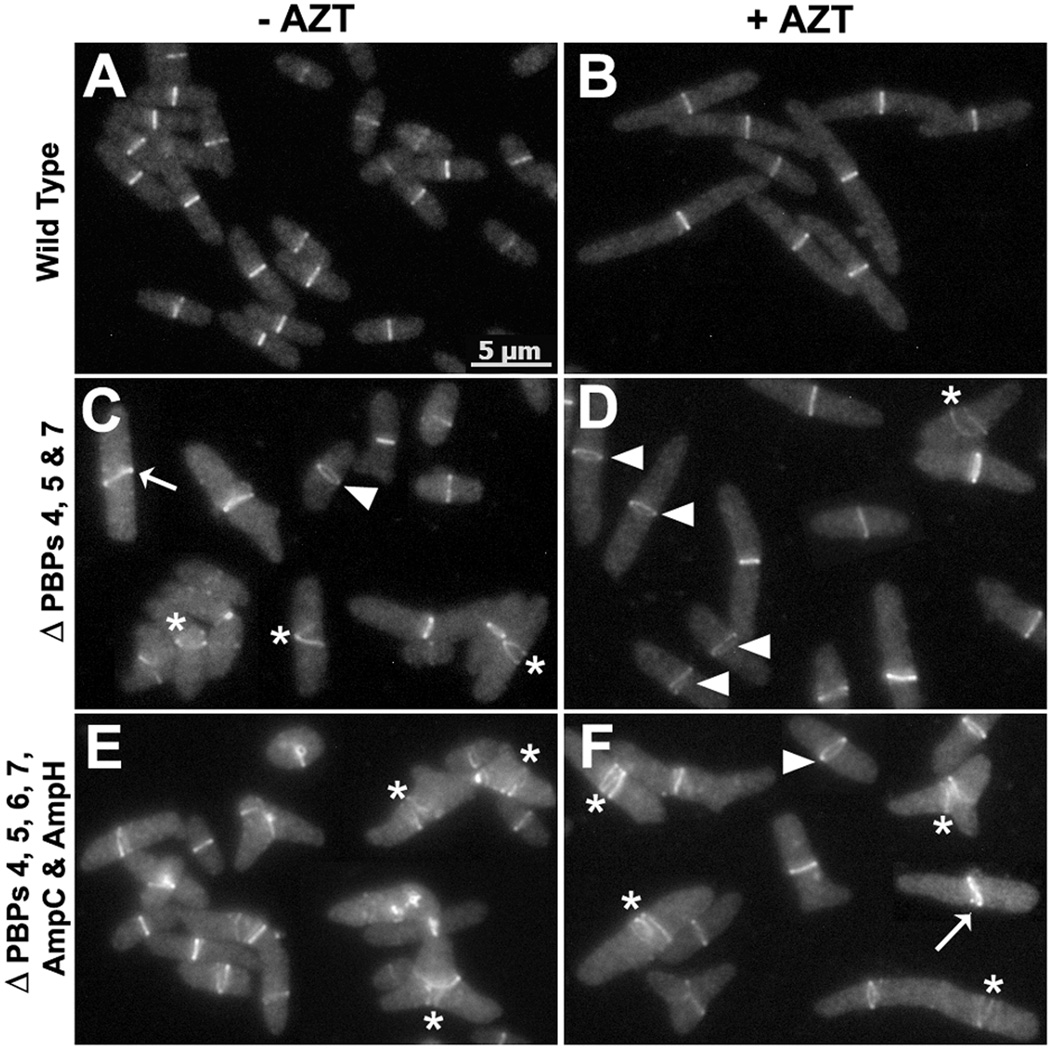
Branches arise from sites of abnormal septation
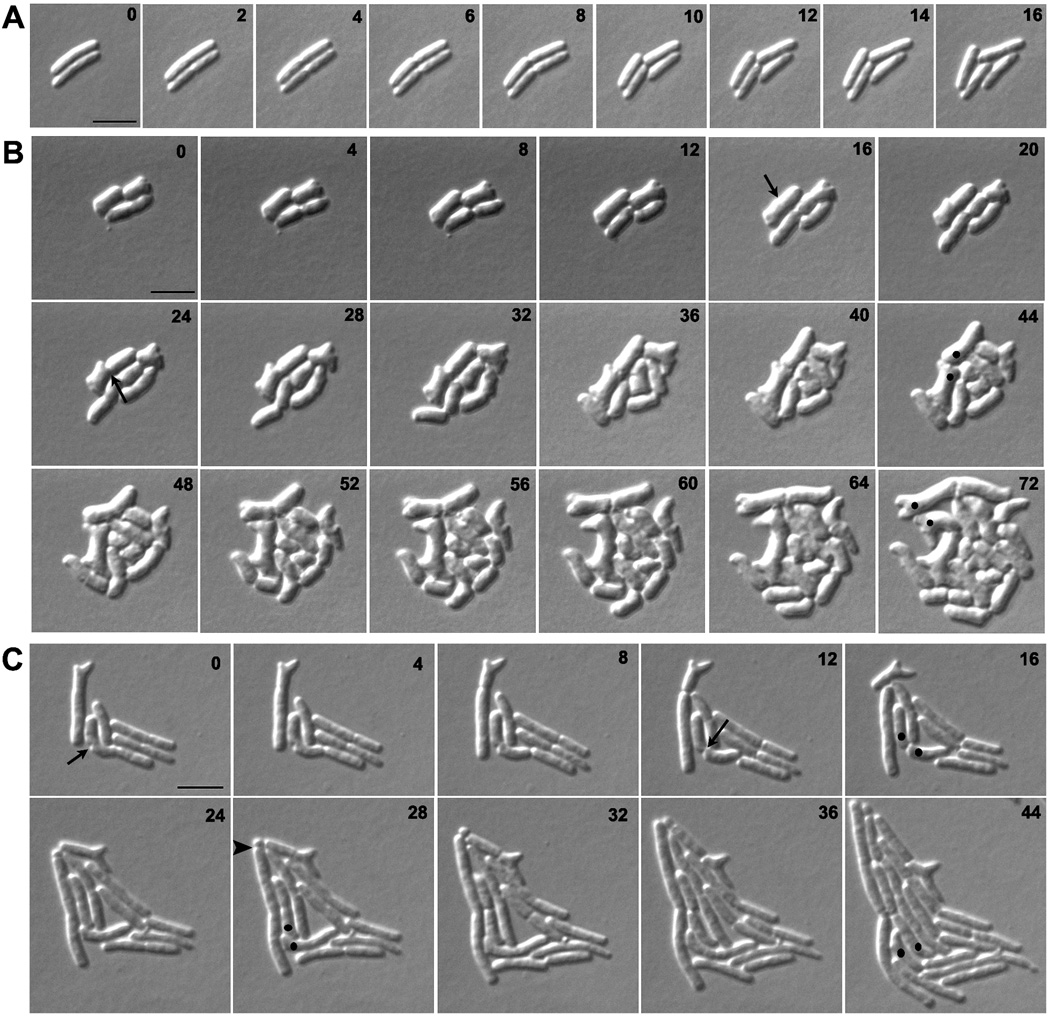
To visualize the origin of branched cells, we followed PBP mutants by time-lapse microscopy. Normal cell division occurs by uniform circumferential constriction of the septum (Fig. 2A), but mutants lacking multiple LMW PBPs displayed numerous episodes of abnormal and uneven septal constriction (e.g., Fig. 2B). During subsequent cell growth, branches arose from sites created by these latter events. The typical morphological progression is illustrated by the sequence of events occurring in E. coli CS612-1 (lacking PBPs 4, 5, 6, 7, AmpC and AmpH). Septal invagination first began on one side of the cell (Fig. 2B, arrow at 16 min) and proceeded for several minutes before the opposite side began to invaginate (Fig. 2B, arrow at 24 min). These abnormal events gave rise to topological changes in the poles of newly generated daughter cells (Fig. 2B, dots at 44 min). In wild type cells, division produces two symmetrical hemispherical cell poles, whereas in these PBP mutants squared and asymmetric poles were created at high frequency (e.g., 48% of poles in CS612-1 and 35% of poles in CS315-1). These abnormal poles eventually gave rise to buds that elongated into branches during subsequent growth (e.g. Fig. 2B, dots at 72 min). In some cells, branches emerged immediately after the abnormal septation event (e.g., Fig. 2B, from the 44 min to the 72 min time points). In other cells, blebs produced by asymmetric invagination remained relatively unchanged for several generations so that multiple buds accumulated during repeated rounds of abnormal septation. Eventually, elongation of these morphological abnormalities also produced cells with multiple branches (not shown).
Fig. 2. Branches arise from abnormal septation.
E. coli strains A) CS109 (wild type), B) CS612-1 (Δ PBPs 4, 5, 6, 7, AmpC and AmpH), and C) CS109 harboring pLPZ, were grown and imaged as described in Experimental Procedures. Arrows in panels B and C represent abnormal cell constrictions. In panels B and C, dots were used to follow abnormal cell poles produced by abnormal cell constrictions and which gave rise to branches. The arrow head in panel C marks a mini-cell. The numbers on each image indicates the time in minutes. The scale bar equals 5 µm.
In cells with wild type levels of FtsZ, branching occurred most frequently in strains lacking several LMW PBPs. However, abnormal septation was observed even in a mutant lacking only PBP5 (CS12-7) (Fig. S2) and in a mutant lacking PBPs 4, 5 and 7 (CS315-1) (Fig. S3A). The branching severity was only 8% and 22%, respectively, in these two strains (Fig. 1G, Δ5 and Δ457 strains, light bars). In each, subtle changes during constriction gave rise to abnormal septa, and branching was always associated with a morphological abnormality that could be traced to a previous aberrant division event.
Excess FtsZ increased the frequency of abnormal septation, leading directly to an increase in the number of branched cells. This occurred even when FtsZ was overproduced in wild type cells (Fig. 2C, arrows at 0 and 12 min; and Fig. 1G and 1H, WT strain, grey and black bars). However, FtsZ overexpression increased the frequency of aberrant invaginations to a much higher degree in strains lacking multiple LMW PBPs (when PBP 5 was also absent). For example, 85% of the FtsZ-induced septation events were abnormal in E. coli CS315-1, a strain lacking PBPs 4, 5 and 7 (e.g., Fig. S3B). In addition, strains with excess FtsZ accumulated a larger number of cells having multiple buds and blebs, which eventually produced cells with multiple branches (e.g., Fig 1H, grey and black bars). FtsZ overproduction also resulted in minicell formation (Fig. 2C, arrow head at 28 min), in agreement with previous reports (Ward & Lutkenhaus, 1985).
If aberrant cell division generates abnormalities leading to branching, then no new branch points should be generated if cell division cannot be completed. To test this prediction, we interrupted division by inhibiting invagination in strain S703-1, which lacks seven PBPs and branches at a rate even higher than that of CS315-1 (not shown) (Meberg et al., 2001, Nelson & Young, 2000, Nelson & Young, 2001). Cells were incubated on an agar pad plus aztreonam to inhibit PBP3, and their growth was monitored by time-lapse microscopy. Branches arose only from buds or abnormalities that preexisted before division was inhibited, and no new buds or branches were generated while septation was prevented (as exemplified by the results presented in Figures 2, 6, S2, S3 and S6, and as confirmed by the results discussed in Figure 3). More importantly, cells that were morphologically normal at the beginning of the experiment grew as filaments but did not branch (not shown).
Fig. 6. Abnormal FtsZ polymers are responsible for abnormal cell constriction.
E. coli strains LP18-1 (wild type) (A) and LP1 (Δ PBPs 4, 5 and 7) (B) were grown and imaged as described in Experimental Procedures. White arrow heads in panel B (at time 20 and 25 min) represent abnormal FtsZ polymers that formed on one side of the cell which eventually resulted in abnormal cell constriction (black arrowhead at 35 min). These events gave rise to abnormal poles, leading to the formation of a branch (white dots are inserted to mark and follow the development of selected abnormal cell poles). Abnormal FtsZ polymers are denoted by symbols described in Fig. 5. The numbers in each panel indicate time in minutes. Only selected time points were included in this figure for clarity (see Fig. S6 for a compilation of all time points). The scale bar in panels A and B equals 5 µm.
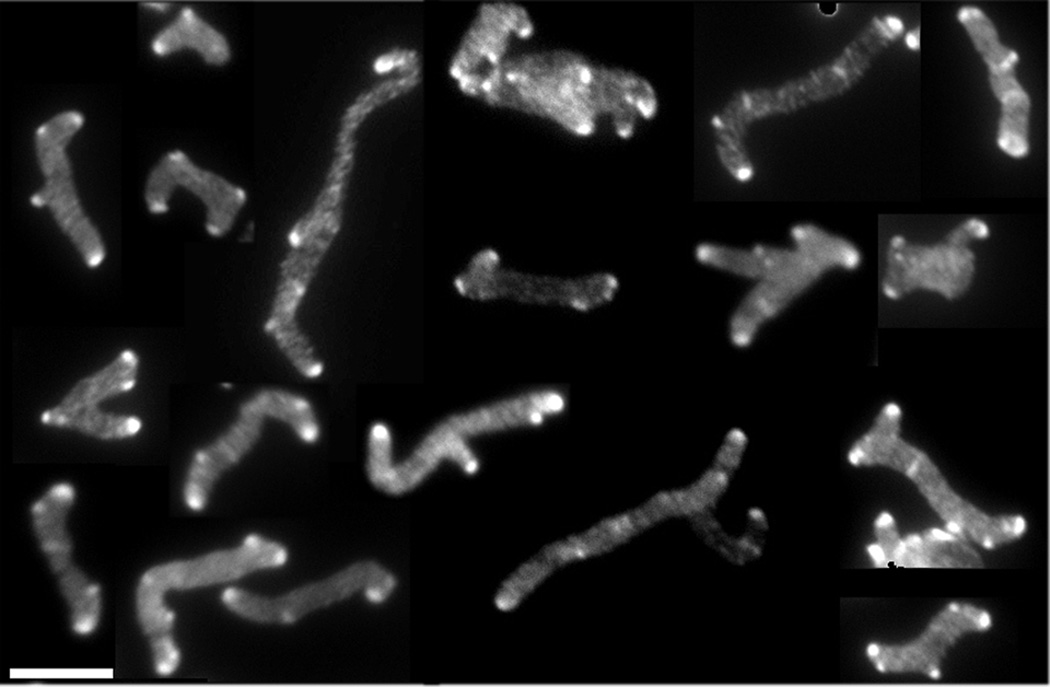
Fig. 3. All branches have inert PG at their poles.
E. coli CS703-1, a mutant lacking PBPs 1a, 4, 5, 6, 7, AmpC and AmpH, was grown in LB containing 100 µg/ml of D-cys for three generations, after which cells were chased in the absence of D-cys by growing them in LB plus 1 µg/ml of aztreonam (to inhibit cell division) for 2 h (~3 mass doublings). After the chase, sacculi were isolated and immunolabelled to detect D-cys residues, as described (de Pedro et al., 1997). This figure is a mosaic of images taken from different fields of view. The scale bar equals 5 µM.
An alternate possibility was that branches arose de novo from the side walls instead of from aberrant sites created during abnormal septation. To test this, we incorporated D-cysteine evenly into the peptidoglycan of strain CS703-1 and then grew the cells for 2 h (~3 mass doublings) in the absence of D-cysteine but in the presence of aztreonam to inhibit cell division. In these conditions, the D-cysteine label disappears from the side walls but is retained at the poles (de Pedro et al., 1997). Therefore, if branches arose only from sites where abnormal septation had previously created asymmetric poles, then all branches in the resulting cell filaments would retain the original D-cysteine label. However, if branches also arose de novo from the cylindrical cell wall, then the polar ends of at least some branches would have little or no D-cysteine label. The results were quite clear: the poles of all cells and branches were brightly labeled and none were devoid of D-cysteine (Fig. 3), indicating that all branches originated from preexisting poles.
The preceding results support the interpretation that cell branches arise from residual morphological scars created during abnormal septation. In addition, the frequency of these aberrant scars and branches increases in the absence of certain LMW PBPs (especially PBP5) and in the presence of excess FtsZ.
LMW PBP mutants have abnormal FtsZ rings
Time-lapse microscopy indicated that abnormal constrictions in PBP mutants appeared very early during invagination (Fig. 2B). Because FtsZ is the first protein to localize to the division site, we speculated that abnormal Z rings might be responsible for the asymmetric constrictions. To test this idea, we visualized the localization of FtsZ by expressing FtsZ-GFP from a single copy of Plac::ftsZ-gfp integrated at the chromosomal lambda attachment site.
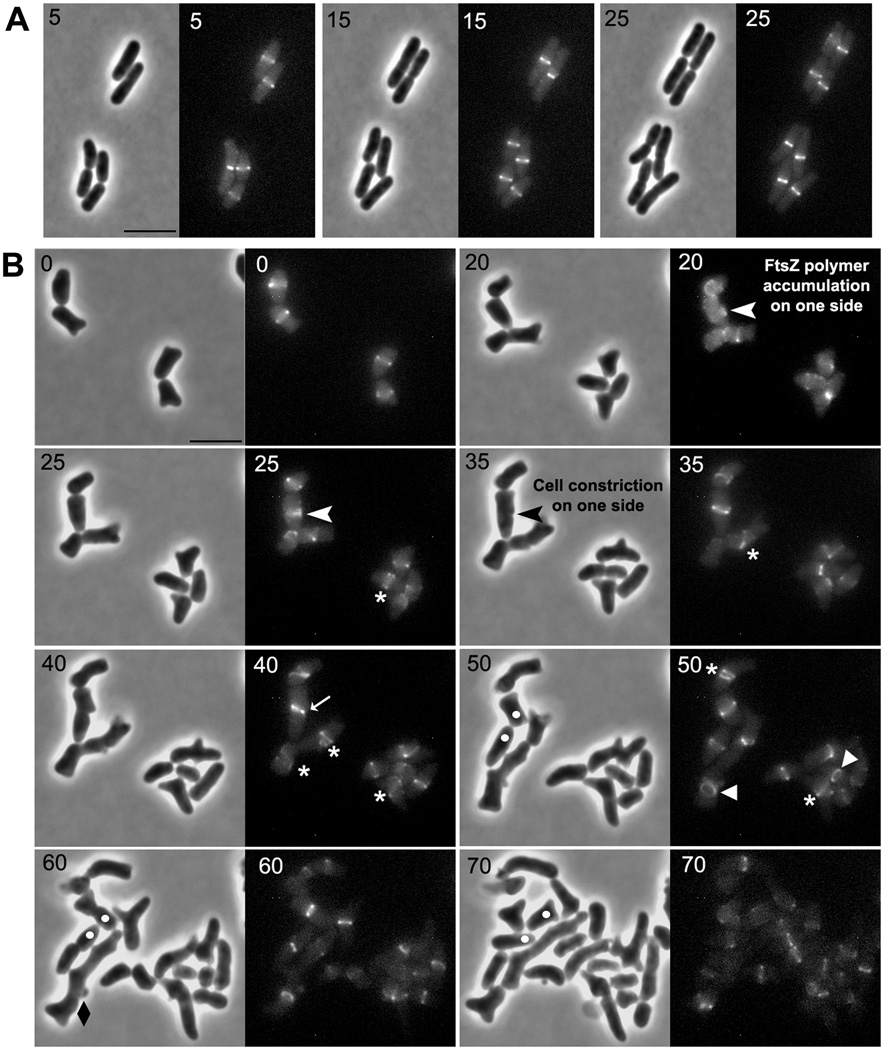
As expected, wild type cells overexpressing FtsZAQ (Fig. 4A) or FtsZ (Fig. 4C) grew as elongated cells that had multiple Z rings. These rings were also prominent in cells grown in the presence of aztreonam (Fig. 4B and 4D). Some Z rings appeared to be interconnected, forming short helices or spirals (e.g., Fig. 4B, inset), though others formed incomplete arcs that might be ring fragments or parts of a helix or spiral (e.g., Fig. 4, arrows). In almost all wild type cells, FtsZ was distributed along the cell length as a series of relatively evenly spaced, brightly labeled bands in a background of more lightly labeled rings, spirals or arcs (Fig. 4A and 4B). In contrast, in most cells of a mutant lacking PBPs 4, 5 and 7 (E. coli LP1), excess FtsZAQ or FtsZ produced localization patterns that were less well organized (Fig. 4E–4H). Most striking was that many fewer cells exhibited evenly spaced FtsZ bands, and more FtsZ rings were non-standard (incomplete rings, spirals or arcs) (Fig. 4E–4H).
Fig. 4. FtsZ overexpression produces spiral and helical FtsZ rings and bands.
E. coli LP18-1 (wild type) harboring pZAQ (A, B) or pLPZ (C, D), and E. coli LP1 (ΔPBPs 4, 5 and 7) harboring pZAQ (E, F) or pLPZ (G, H), were grown and imaged as described in Experimental Procedures. Cells in panels B, D, F and H were grown for 1 mass doubling in the presence of 1 µg/ml of aztreonam. The inset in panel B is an enlarged portion of the figure to show the FtsZ spirals more clearly. White arrows represent incomplete Z-rings and FtsZ arcs. All images have same magnification; the scale bar in panel A equals 5 µm.
Overproducing FtsZ in LMW PBP mutants produced large numbers of unusual FtsZ structures. Therefore, we thought that in the presence of wild type levels of FtsZ the PBP mutants themselves might also contain abnormal Z rings. To determine if this was true, we expressed low levels of FtsZ-GFP (20% or less of wild type levels, Fig. S4) in the parent strain and in PBP mutants known to branch frequently. These cells did not elongate, indicating that this level of FtsZ expression did not seriously affect division (not shown). As expected, each parent cell had a single Z ring located at mid-cell and oriented approximately perpendicular to the long axis (Fig. 5A). These bands persisted even when cell division was inhibited with aztreonam (Fig. 5B). In contrast, a mutant lacking PBPs 4, 5 and 7 (LP1) (Fig. 5C–D) or a mutant lacking six PBPs (LP17-1) (Fig. 5E–F), had large numbers of aberrant FtsZ bands that appeared to be spirals and incomplete or slanted rings (Fig. 5C–F, marked by asterisks and arrows). In addition, some Z rings were broad and oriented at an angle other than perpendicular to the long axis of the cell (Fig. 5C–F, marked by triangles). Similar results were observed when division was inhibited (Fig. 5D and 5F). The frequencies of these abnormal Z structures increased in strains lacking additional PBPs (Table 2). For example, in a strain lacking PBPs 4, 5 and 7 (LP1), ~19% of the cells had abnormal FtsZ polymers. This population increased to ~42% of cells in a strain lacking six PBPs (LP17-1) (Table 2). In short, the number of abnormal FtsZ structures increased in proportion to the frequency with which each strain formed branches.
Fig. 5. LMW PBP mutants have aberrant Z rings.
E. coli strains LP18-1 (wild type) (A, B), LP1 (ΔPBPs 4, 5 and 7) (C, D) and LP17-1 (ΔPBPs 4, 5, 6, 7, ampC and ampH) (E, F) were grown and imaged as described in Experimental Procedures. Cells in panels B, D and F were incubated for ~1 mass doubling in the presence of 1 µg/ml of aztreonam. FtsZ polymers that appear to be spirals or helices, incomplete Z rings, arcs and FtsZ polymers that did not form compact rings were considered to be spirals, and are illustrated by the Z ring marked by asterisks (*). Rings that are broader (take up more space than normal along the long axis of the cell) are represented by a triangle (▲); these Z-rings may be slanted but appear as rings instead of bands, probably because they are oriented differently. Slanted rings that are not perpendicular to the long axis of the cell are illustrated by those marked with white arrows. All images have the same magnification; the scale bar in panel A equals 5 µm.
Table 2.
The frequency of abnormal FtsZ rings increases with the loss of multiple PBPs.
| Percentage of FtsZ polymers | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | PBPs Deleted |
Normal | Spirals | No Rings |
Broad Rings |
Slanted Rings |
Total Abnormal Z-polymers |
No. of Cells counted |
| LP18-1 | - | 87 | - | 13 | - | - | - | 554 |
| LP1 | 4, 5 & 7 | 65.5 | 11 | 15.8 | 2.5 | 5.2 | 18.7 | 557 |
| LP16-1 | 4, 5, 6 & 7 | 62.5 | 14.4 | 13.2 | 5.3 | 4.6 | 24.3 | 560 |
| LP17-1 | 4, 5, 6, 7, AmpC & AmpH | 39.7 | 28 | 18.4 | 3.4 | 10.5 | 42.0 | 554 |
FtsZ-GFP was visualized as described in Experimental Procedures. The percentages represent cells with one perpendicular FtsZ ring (Normal), cells that had no rings or spirals (No-rings), and cells with other forms of FtsZ (e.g., spiral, helix, incomplete Z-rings, arcs and polymers not forming compact rings) (Spirals). Representatives of spiral forms are marked with asterisks (*) in Fig. 5. In addition, other cells were classified as having FtsZ rings that were extended or tilted in a non-native orientation (Wide Rings), and representatives of this type are marked by triangles (▲) in Fig. 5. Finally, some cells had FtsZ rings oriented at an angle other than 90° to the cell axis (Slanted Rings), and representatives are marked with white arrows in Fig. 5.
Cells of PBP mutants (e.g., Fig. 5, C–F) were visibly wider than were cells of the parent strain (Fig. 5, A and B). Thus, the primary effect of removing these LMW PBPs might be to increase cell diameter, which in turn might be the proximal cause of aberrant Z ring orientation. We measured the widths of at least 200–300 cells of each strain in two separate experiments and found that, compared to the parent (LP18-1), the diameters of three branching mutants increased by 27–40% (LP1, Δ457) (Fig. 5, C–D), by 29–35% (LP16-1, Δ4567) (not shown) and by 30–34% (LP17-1, Δ4567 ΔAmpC ΔAmpH) (Fig. 5, E–F) (also, see Supplemental Fig. S5). In parallel, the number of cells having abnormal Z rings increased from zero in the parent strain to 19–25%, 24–29% and 28–42%, respectively, for the three mutants (Table 2 and Fig. S5). When plotted by diameter, there was a tendency for wider cells to have a higher percentage of abnormal Z rings, ranging from ~20% in smaller cells to ~30–50% in wider cells (Fig. S5). Even so, in one mutant (LP17-1), 18% of Z rings were abnormal in those cells whose widths overlapped those of the parental strain (LP18-1) (Fig. S5). This latter observation suggests that PBP loss can affect Z ring orientation even in the absence of large diameter changes. However, the data cannot yet distinguish between a purely direct affect of PBPs on Z ring orientation versus an indirect effect mediated by increased cell width, especially in larger cells.
Branch points are nucleated by aberrant FtsZ rings
To determine if abnormal FtsZ rings created the aberrant septation that lead to branching, we followed Z ring formation by time-lapse microscopy in E. coli LP18-1 (parent) and LP1 (lacking PBPs 4, 5 and 7). Cells were imaged every 5 min to minimize bleaching and to prevent excessive UV exposure. In the parent, Z rings were perpendicular to the long axis in all cells (Fig. 6A), though FtsZ polymers might be diffuse during the initial stages of Z ring formation (e.g., Fig. S6A, at 0, 25 and 35 min). However, in the mutant lacking PBPs 4, 5 and 7 (LP1), abnormal FtsZ structures (slanted rings, incomplete rings, spirals and wide rings) occurred much more often (e.g., Fig. 6B, white arrows, arrow heads, asterisks and triangles). Formation of abnormal FtsZ rings preceded abnormal septation. In a typical example, FtsZ polymers first accumulated on one side of a cell (Fig. 6B, white arrow heads at 20 and 25 min) and eventually formed a slanted FtsZ ring (Fig. 6B, white arrow at 40 min). The cell began to constrict only from one side (Fig. 6B, black arrow head at 35 min), which produced a slanted septum. This slanted septum produced daughter cells with abnormal cell poles, which eventually elongated into branches (Fig. 6B, marked by white dots at 50, 60 and 70 min). In a similar series of steps, aberrant Z rings gave rise to abnormal constrictions that produced kinks and bends in other cells. These aberrant structures persisted within the cell cylinder during subsequent growth and only later developed into branches. Without time-lapse observations these branches would seem to arise from the lateral wall, when in fact all branches elongated from sites previously created by inaccurate septation (e.g., Fig. 6B, 60 min, the kinked cell denoted by the diamond symbol). The results indicated that aberrant Z ring formation leads to asymmetric invagination, which, upon further growth, creates cells with branches and other morphological abnormalities.
Cellular localization of LMW PBPs
Because branching in these mutants was triggered by frequent rounds of abnormal septation, we speculated that one or more LMW PBPs might be involved in orienting the division plane. Such a possibility is credible because PBP5, the protein whose loss coincides most strongly with branching, localizes to the septum in a substrate dependent manner (Potluri et al., 2010). Other LMW PBPs play auxiliary roles in shape maintenance (Meberg et al., 2004, Nelson & Young, 2001), and the frequency of branching and morphological oddities increased as these were deleted in combination with PBP5. Thus, additional LMW PBPs might localize to the septum before or during division to augment or partially replace the function of PBP5. To investigate this possibility, we determined the cellular localization of LMW PBPs 6, 7 and DacD.
We replaced the natural N-terminal signal sequences of PBPs 4, 5, 6, 7 and DacD with dsbA-SS-sfgfp, so that the resulting fusion proteins were translocated to the periplasm via the co-translational SRP pathway instead of by the Sec pathway. This method successfully exports functional sfGFP-PBP5 and other proteins to the periplasm (Dinh & Bernhardt, 2011, Paradis-Bleau et al., 2010, Peters et al., 2011, Potluri et al., 2010, Uehara et al., 2010). To verify that these proteins reached the periplasm, we removed the carboxy terminal amphipathic helixes from each PBP so that the fusion proteins could not bind to the inner membrane; we then assayed the soluble periplasmic fractions for the presence of these proteins (Fig. S7). DsbA-sfGFP versions of PBPs 5, 6, 7 and DacD were exported successfully into the periplasm (Fig. S7A–B), though for unknown reasons PBP4 was not (Fig. S7A). In addition, all the PBPs except sfGFP-DacD were able to bind the fluorescent penicillin substrate, Bocillin-650 (Fig. S7A–B, S7C–D, and S8), indicating that the proteins were folded correctly and were enzymatically active. A portion of each sfGFP-PBP was observed in non-periplasmic fractions (Fig. S7A and S7B), probably because some fraction of the proteins were still attached to peptidoglycan fragments or were caught in the process of being synthesized or exported.
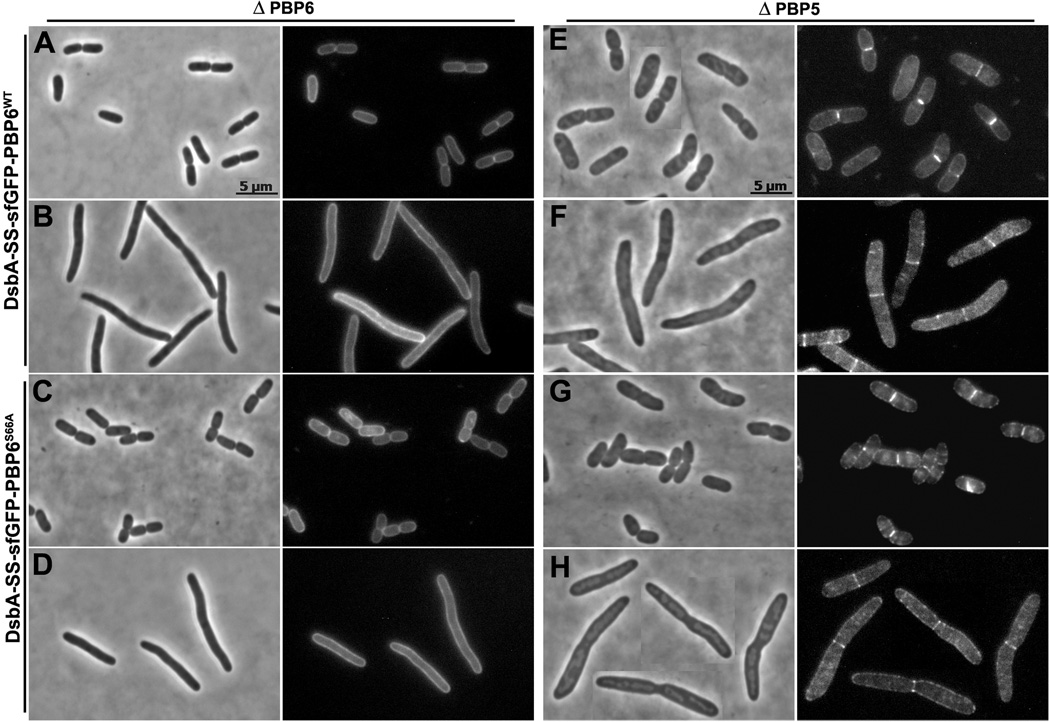
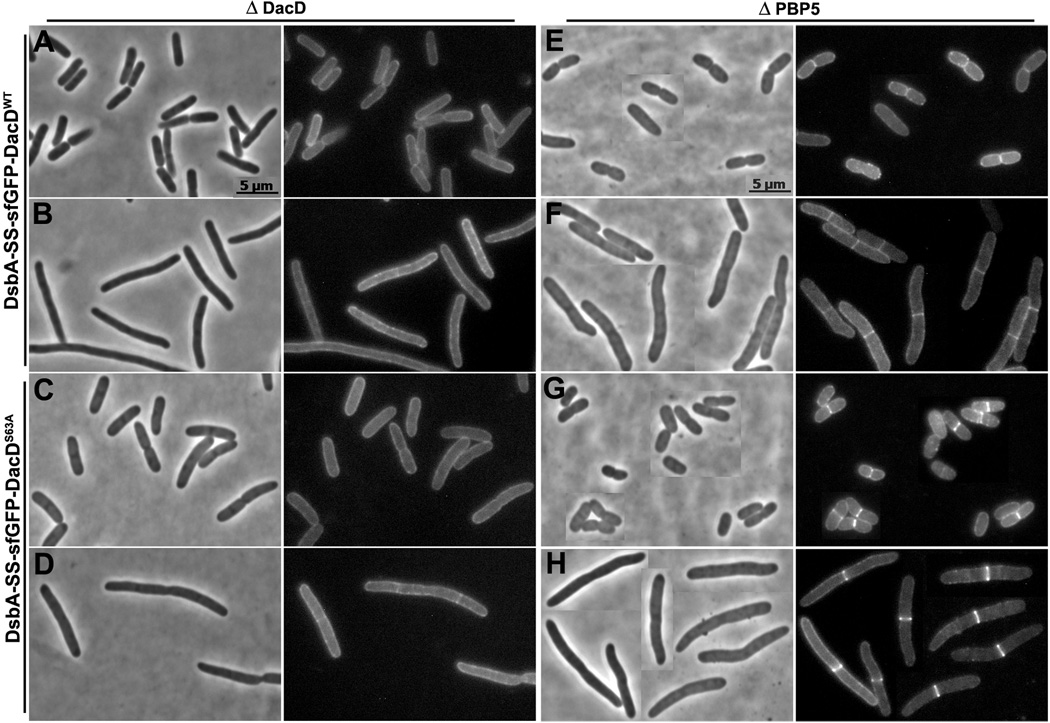
PBP6 and DacD localize to the septum in the absence of PBP5
When sfGFP-PBP6 and sfGFP-DacD were expressed in E. coli strains lacking PBP6 or DacD, respectively, the fusion proteins were dispersed evenly throughout the periplasm (Fig. 7A and 8A). However, close observation revealed that in some cases faint fluorescent bands appeared at potential septation sites in cells filamented in the presence of aztreonam (Fig. 7B and 8B). Similar patterns were observed when the fusion proteins were expressed in a wild type strain (not shown). In contrast, in a strain lacking PBP5 (CS12-7), sfGFP-PBP6 localized to incipient septa of constricting cells (Fig. 7E), a pattern that was clearer in cell filaments where division was inhibited (Fig. 7F). sfGFP-DacD localized to potential septal sites in the absence of PBP5 (Fig. 8E), though not quite as well as did PBP6 (Fig. 7E), but DacD did localize clearly to septa in division-inhibited filaments (Fig. 8F). The endopeptidase PBP7 was dispersed evenly throughout the periplasm in wild type E. coli (not shown), in a strain lacking wild type PBP7 (Fig. S9), and in mutants lacking PBP5 or multiple PBPs (not shown). The uniform localization of sfGFP-PBP7 served as an internal control, indicating that septal localization was not an artifact of all sfGFP fusion proteins. In addition, sfGFP exported by itself via this route was dispersed evenly throughout the periplasm, with no preference for the septum (Fig. S10).
Fig. 7. sfGFP-PBP6 localizes to the septum in cells lacking PBP5.
E. coli CS17-1 (ΔPBP6) expressing DsbA-SS-sfGFP-PBP6 from pLP602 (A and B), or DsbA-SS-sfGFP-PBP6S66A from pLP607 (C and D). E. coli CS12-7 (Δ PBP5) expressing DsbA-SS-sfGFP-PBP6 from pLP602 (E and F), or DsbA-SS-sfGFP-PBP6S66A from pLP607 (G and H). Cells were grown and imaged as described in Experimental Procedures. Cells in panels B, D, F and H were filamented by treating with 1 µg/ml of aztreonam for 1 mass doubling. Each panel has a phase contrast image on the left and the corresponding fluorescence image on the right. All images have same magnification; the scale bars in panels A and E equal 5 µm.
Fig. 8. sfGFP-DacD localizes to the septum in cells lacking PBP5.
E. coli CS18-1 (ΔDacD) expressing DsbA-SS-sfGFP-DacD from pLP652 (A and B), or DsbA-SS-sfGFP-DacDS63A from pLP656 (C and D). E. coli CS12-7 (ΔPBP5) expressing DsbA-SS-sfGFP-DacD from pLP652 (E and F), or DsbA-SS-sfGFP-DacDS63A from pLP656 (G and H). Cells in panels B, D, F and H were filamented by treating with 1 µg/ml of aztreonam for 1 MD. Each panel has a phase contrast image on the left and the corresponding fluorescence image on the right. The fluorescence images in panels E, F, G and H were processed similarly. All images have same magnification, and the scale bars in panels A and E equal 5 µm.
The fact that PBP6 and DacD localized to the septum only when PBP5 was absent suggested that localization might depend on the presence of cell wall pentapeptides, since PBP5 actively removes these compounds. To test if localization depended on enzymatic activity, we changed the active-site serine to alanine in PBP6, PBP7 and DacD. This procedure removes a single hydroxyl group and eliminates enzymatic activity because the carboxypeptidase and endopeptidase reactions require the –OH group of this serine. As predicted, the mutation prevented binding of Bocillin-650 by PBPs 6 and 7 (Fig. S7C and S7D).
In an E. coli strain lacking PBP5, the PBP6 mutant (sfGFP-PBP6S66A) localized to the septum to the same degree as did wild type PBP6 (Fig. 7G and 7H). Even more surprising, septal localization of the DacD mutant (sfGFP-DacDS63A) became more prominent than that of wild type DacD (Fig. 8G and 8H). Also, PBP6S66A formed septal rings in E. coli CS211-2 (Δ PBPs 5 and 6), and DacDS63A formed equivalent rings in strain CS235-1 (Δ PBP5 and DacD) (not shown). These results indicated that septal localization of the mutant proteins did not require interactions with endogenous wild type PBP6 or DacD. sfGFP-PBP7S67A continued to be distributed evenly throughout the periplasm in a strain lacking PBP7 (not shown).
Even though the serine-to-alanine mutation eliminated the penicillin binding ability of PBP6, the mutant proteins might retain the ability to bind pentapeptide substrate or they might have residual D,D-CPase activity (though this was unlikely). Overproducing DacDS63A (with its own signal sequence and no sfGFP domain) resulted in the lysis of E. coli cells at a rate comparable to that induced by overproduction of wild type DacD (Fig. S11B), implying that DacD retained at least one of these activities. To determine if the active-site mutants of PBP 6 and DacD were enzymatically competent, we expressed the wild type and mutant proteins in a pentapeptide-rich E. coli strain (CS704-1) and analyzed the muropeptide composition of its peptidoglycan. Wild type DacD decreased the number of pentapeptides by a drastic amount, from 75% to 3% (Table 3), but mutant DacDS63A had an insignificant effect on pentapeptide levels, which decreased from 75% to 70% (Table 3). These results indicated that DacDS63A had no, or very little, D,D-CPase activity. Expressing wild type PBP6 decreased the amount of pentapeptides only slightly, from 75% to 67% (Table 3), an observation consistent with the lower D,D-CPase activity of PBP6 compared to PBP5 (Chowdhury et al., 2010). Overproducing PBP6S66A did not reduce pentapeptide levels at all (Table 3), confirming that this mutant had no D,D-CPase activity. Overall, though we could not confirm that PBP6 and DacD localization required the presence of pentapeptides at the septum, the results strongly support the interpretation that enzymatically inactive PBP6S66A and DacDS63A can still localize to pentapeptide-rich septa.
Table 3.
PBP6 and DacD active site mutants do not have D, D-CPase activity.
| Molecular Percentage | |||||
|---|---|---|---|---|---|
| PG Composition |
- | PBP6 | PBP6S66A | DacD | DacDS63A |
| pLP18-Kan | pLPKC604 | pLP606 | pLPKC653 | pLP655 | |
| Monomers | 57 | 56.7 | 55.8 | 58.5 | 60.9 |
| Dimers | 39.5 | 39.8 | 40.5 | 39.0 | 36.2 |
| Trimers | 3.5 | 3.6 | 3.7 | 2.5 | 2.9 |
| Lipoprotein | 2.6 | 3.6 | 2.6 | 8.4 | 3.8 |
| Anhydro | 4.8 | 5.0 | 5.3 | 3.8 | 4.4 |
| DAP-DAP | 0.31 | 0.2 | 0.3 | 2.8 | 0.3 |
| Pentapeptide | 75.1 | 66.6 | 75.8 | 3.1 | 70.1 |
| Penta-glycine | 5.8 | 6.2 | 6.8 | 0.3 | 7.3 |
| Cross-Linkage | 46.5 | 47.0 | 48.0 | 44.0 | 42.0 |
| Average Length | 20.8 | 20.1 | 19.0 | 26.1 | 22.5 |
Peptidoglycan was isolated from E. coli CS704-1 harboring pLP18-Kan, pLPKC604, pLP606, pLPKC653 or pLP655. The murein composition of each sample was analyzed by HPLC as described in Experimental Procedures. The molecular percentage of each muropeptide is reported.
Septal localization of PBP6 and DacD depends on the presence of a membrane anchor
Septal localization of PBP5 depends the availability of pentapeptide substrates, but removing its amphipathic membrane anchor eliminates this localization and its morphological functions (Potluri et al., 2010). To determine if septal localization of PBP6 and DacD also depended on their membrane anchors, we removed the carboxy-terminal amphipathic helices from the sfGFP fusion versions of PBP6 and DacD. In a strain lacking PBP5 (CS12-7), both mutant proteins failed to localize to septa but were, instead, dispersed evenly throughout the periplasm (Fig. S12). Because the active site mutant DacDS63A bound to septa better than did wild type DacD (Fig. 8), we thought this protein might localize to septa even in the absence of its amphipathic helix. Contrary to this expectation, the sfGFP-DacDS63A-ΔC13 protein, which was deficient in D,D-CPase activity and lacked the C-terminal amphipathic helix, failed to localize to septa and was dispersed evenly in the periplasm of a strain lacking PBP5 (not shown). Amphipathic helices anchor the LMW PBPs to the outer surface of the inner membrane, so the results indicate that PBP6 and DacD, like PBP5, must be oriented correctly if they are to find and bind their substrates.
Discussion
Understanding how bacteria branch is not merely a matter of curiosity but is a way to investigate morphological mechanisms, the control of cell division, and the existence of interactions between cellular compartments. The results reported here link all three of these elements. By examining a set of PBP mutants that branch with high frequency, we show that PBP5 and other LMW PBPs help orient FtsZ rings at mid-cell. Mutants lacking such fine-tuning exhibit skewed Z ring geometries that lead to asymmetric invaginations, which give rise to aberrantly shaped and branched cells. This means that the periplasmic PBPs influence septal localization of the cytoplasmic Z ring, by either direct or indirect means. The results emphasize the growing realization that cell elongation, cell division and peptidoglycan synthesis mutually affect one another.
Z ring orientation in LMW PBP mutants
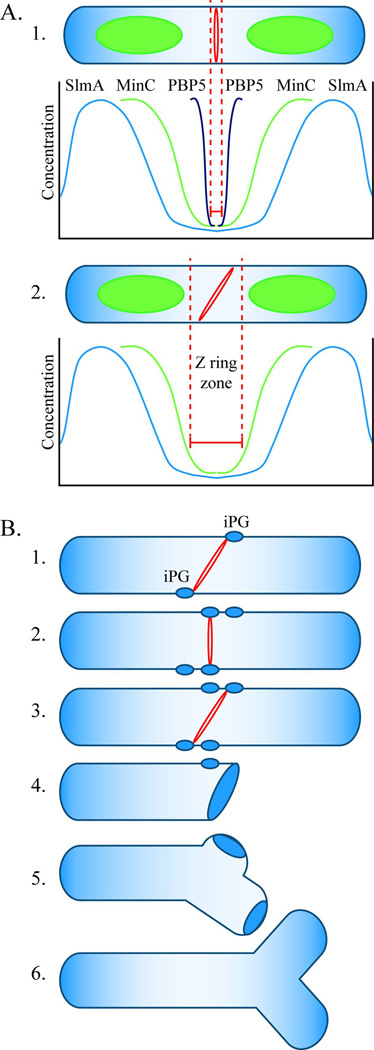
There is strong evidence that mis-oriented FtsZ rings or divisomes generate aberrant morphologies in a variety of circumstances, suggesting that cell division is a critical point at which gross morphological decisions are made (Young, 2003, and references therein). Normally, Z ring formation is restricted to the midcell of E. coli by the negative regulators MinC and SlmA, each of which inhibits FtsZ polymerization (Dajkovic et al., 2008, Lutkenhaus, 2007, Rothfield et al., 2005). The MinCDE proteins oscillate from one cell pole to the other and create a gradient of MinC that is low enough near the cell center to permit Z ring formation (Fig. 9A, 1), and SlmA binds to specific sites on the chromosome to prevent FtsZ polymerization over the nucleoid (Bernhardt & de Boer, 2005, Cho et al., 2011, Rothfield et al., 2005, Tonthat et al., 2011). However, in mutants lacking both systems almost all Z rings are still oriented perpendicular to the long axis of the cell (Bernhardt & de Boer, 2005). Also, there seems to be some latitude in Z ring formation even in wild type cells, where there is evidence for a fairly wide inhibitor-free midcell zone in which FtsZ can polymerize. For example, FtsZ polymers sometimes appear as closely spaced rings with neighboring polymers separated by as much as 50–400 nm (Fu et al., 2010), and Z rings often split and move apart before reforming a single ring prior to division (Erickson et al., 2010, Fu et al., 2010). All these observations imply that at least one other mechanism helps orient Z rings properly. Here, our results indicate that the LMW PBPs, and PBP5 in particular, influence the geometric organization of Z rings such that they form in more places and at unnatural angles, leading to malformed daughter cells.
Fig. 9. Model for FtsZ ring localization and cell branching in E. coli.
A. FtsZ ring localization. A1. In wild type E. coli, the FtsZ ring (red ring) is restricted to the center of an elongating cell (“Z ring zone” between dotted red lines) by three mechanisms. The SlmA protein is associated with nucleoids (green ovoids), and its concentration is lowest near the cell’s center and near the poles (blue line). This FtsZ inhibitor restricts Z ring formation to a relatively broad area at these three sites. The MinC protein prevents polymerization of FtsZ at the poles and restricts Z ring formation to a small region near the cell’s midpoint, where the concentration of this inhibitor is lowest (green line). PBP5 (black line) fine tunes the location of Z rings to an even narrower zone at the cell’s center. The action of PBP5 is probably aided by other LMW PBPs. A2. In the absence of PBP5, FtsZ can polymerize in a larger midcell zone to form a slanted Z ring (shown) or multiple Z rings (not shown). Before invagination begins, the orientation of midcell Z rings can vary within the permissive zone. B. Mechanism of cell branching. B1. In the absence of PBP5, slanted FtsZ rings can form at the center of the cell and initiate synthesis of inert peptidoglycan (iPG) (blue ovals). B2. Before invagination begins, the Z ring can reorient and initiate the synthesis of iPG at sites well separated from the first. B3. A non-perpendicular Z ring triggers invagination. B4. The daughter cells have one pole consisting of iPG, plus at least one other patch of iPG some distance away. B5. During subsequent cell growth, insertion of peptidoglycan into the sidewall separates the true pole from the patch of iPG. B6. Cell wall elongation around the iPG patch creates a branch and an ectopic pole.
The mechanism by which the LMW PBPs influence Z ring orientation and cell shape may be direct or indirect, though the two possibilities need not be mutually exclusive. The simplest indirect mechanism would be that the loss of PBP5 and other LMW PBPs increases cell diameter, which then leads to impaired Z ring formation and a higher frequency of aberrant divisions and branching. In the present study the average diameter of PBP mutants at the point of the Z ring increased by 27–42%, and increasing cell width was accompanied by an increasing fraction of abnormal Z rings. However, in one mutant, about a fifth of the cells with wild type diameters exhibited abnormal Z rings, implying that the PBP loss might affect Z ring orientation even in cells of normal diameter. Nonetheless, we cannot rule out the possibility that relatively small changes in diameter are sufficient to alter Z ring dynamics. Thus, the question becomes whether the increase in cell diameter precedes and causes Z ring abnormalities or whether abnormal Z rings occur first and lead to inaccurate septation that increases cell diameter.
If PBP5 influences Z ring orientation more directly, the effect would require the relay of information between PBP5 in the periplasm and FtsZ in the cytoplasm. PBP5 could be in contact with one or more intermediary proteins that are themselves in contact with FtsZ, or else PBP5 might influence Z ring placement by removing the terminal D-alanine residue from each peptidoglycan side chain. There seems little likelihood that PBP5 makes contact with FtsZ through one or more intermediary proteins. The morphological effects of PBP5 depend on a short amino acid sequence located in and near the active site, and changing just eight residues within this pocket is enough to create a PBP6 variant that can replace the morphological function of PBP5 (Ghosh & Young, 2003). Because these residues are buried or in close contact with the peptidoglycan substrate, it is unlikely they form part of a protein-protein interaction domain. This argues that the D,D-carboxypeptidase activity of PBP5 drives its ability to influence cell shape. Such an interpretation is consistent with the levels of pentapeptides in PBP mutants. In wild type cells PBP5 removes almost all terminal D-alanine residues so that the fraction of cell wall pentapeptides is less than 0.5% (Höltje, 1998, Li et al., 2004, Varma et al., 2007). However, removing progressively more LMW PBPs increases the pentapeptide fraction to 5–20% (Table 1) (Li et al., 2004, Varma et al., 2007), and we show here that the frequency of misaligned Z rings and the number of branched cells both rise in direct proportion to these increases. Thus, if PBP5 directly affects the orientation of FtsZ, then the most likely mechanism would be one that depends on the presence or absence of terminal D-alanine residues in the cell wall. In any case, whether the PBPs affect Z ring orientation directly or indirectly, substantial changes in gross cell morphology affect the oscillation dynamics of MinCDE, alter the size and orientation of the inhibitor-free zones in which Z rings can form, and lead to oddly placed rings and septation (Varma et al., 2008). Thus, any PBP-dependent effects on cell width or shape will also disturb the Min system and so create a secondary cascade that would exacerbate morphological problems.
Regardless of the specific mechanism, the results indicate that PBP5 and other LMW PBPs help orient Z rings and, by extension, affect the geometry of cell division and bacterial shape. An indirect effect via alteration of cell diameter would signal the existence of a mechanism in which subtle changes in cell morphology affect cytoplasmic events. A direct mechanism would place the LMW PBPs in a protein interaction pathway that connects them to FtsZ and would represent a novel FtsZ-dependent regulation of cell morphology.
Model for cell branching in E. coli
Branches can arise in only a few ways. First, an unusual form of peptidoglycan synthesis might split normally inert cell poles. Continued cell wall synthesis could separate these polar fragments, and further elongation would produce observable branches. A problem with this alternative is that it requires the existence of a novel biochemical pathway that inserts material into inert peptidoglycan. Although not impossible, no such pathway has been detected. Nevertheless, as but one recent example, investigators have observed cells with oddly shaped poles that seem to split and generate branched cells (Zaritsky et al., 2011). However, the misshapen cells observed in this particular case were produced by perturbing chromosomal replication (Zaritsky et al., 2011), which itself disrupts FtsZ localization and may create asymmetric poles by leaving a scar of misplaced inert peptidoglycan (Young, 2003). Because the ends of such cells are deformed they are liable to branch, but need not do so by splitting a normal cell pole (see following discussion).
A second possible branching mechanism is that new poles might arise de novo from the cell sidewall. This occurs in the genus Streptomyces, where the DivIVA protein accumulates at sites along the sidewall and initiates peptidoglycan synthesis perpendicular to the existing cell axis (Flärdh & Buttner, 2009, Hempel et al., 2008). Similarly, when overproduced, the TipN protein mislocalizes to the lateral walls of Caulobacter crescentus, where it directs the synthesis of new cell wall and leads to branching (Lam et al., 2006, Schofield et al., 2010). Such a mechanism would explain branch formation in the absence of cell division and would be consistent with the interpretation proposed by Gullbrand et al (Gullbrand et al., 1999). However, E. coli has no DivIVA or TipN homologue, and no analogous sidewall branching has been observed in this organism. In addition, whereas E. coli elongates by diffuse insertion of peptidoglycan into its cylindrical side walls, Streptomyces elongates by inserting new peptidoglycan apically at its poles (Flärdh, 2010, Flärdh & Buttner, 2009) and C. crescentus synthesizes its stalk in an apical manner (Divakaruni et al., 2007, Schmidt & Stanier, 1966). The ability to grow apically sets these bacteria apart and may indicate that they have a branching mechanism that is unavailable to E. coli.
A third possibility is that branches originate because of inaccurate cell division events. We believe this alternative is most consistent with our current results and with the branching literature, and we propose the following simplified model (Fig. 9). In this scenario, an early triggering event (misplaced Z rings) generates a signal (inert peptidoglycan, iPG) that is embedded into the bacterial sidewall at some distance from what will eventually become the invaginating septum and final pole (Fig. 9B, 1–4). Later, as the wall elongates with cell growth, new peptidoglycan is inserted between patches of iPG by means of routine sidewall biochemistry (Fig. 9B, 5). At random intervals, as determined by the distribution and activity of peptidoglycan synthases, a patch of iPG is extruded by lateral wall synthesis, giving rise to a visible branch (Fig. 9B, 6). These postulated events are consistent with past reports and with the known biochemical and functional characteristics of poles. For example, in this model iPG remains refractory to the insertion of new material, and normally stable poles are not split by some new method of peptidoglycan synthesis or turnover. Also, branch creation requires only that cell wall synthesis continue to act on sidewall peptidoglycan, with no need to invoke other novel biochemistry.
Observations made 20 years ago by Bi and Lutkenhaus indicate that FtsZ can behave in a way consistent with this proposed model (Bi & Lutkenhaus, 1992). These authors found that an FtsZ mutant (FtsZ26) had altered Z ring geometry and produced cells with blunt and protruding poles as well as branches. They also observed that numerous division events occurred at acute angles instead of perpendicular to the long axis of the cell, and these included many cells with two offset V-shaped invagination sites (Bi & Lutkenhaus, 1992). Because only one of these paired sites went to completion, the blunt or angled end of at least one daughter cell retained a patch of peptidoglycan that had been synthesized during an aborted constriction event. In effect, one end (the nominal pole) of this daughter was a combination of iPG synthesized during invagination plus at least one other patch of iPG, separated from the invaginated patch by an expanse of intervening side wall, just as we now propose occurs in all cases of branching (Fig. 9B).
Although this third possibility is most consistent with our current findings, at first glance it is in conflict with previous observations that seemingly rule out a role for division in branch generation (Gullbrand et al., 1999). Gullbrand et al concluded that cell division was not involved in this process because the FtsZ84 protein did not localize consistently at branch sites (Gullbrand et al., 1999). However, their observations can be reconciled easily with our model by recognizing that the nucleation of a branch point depends on a transient activity of FtsZ before invagination begins. Since Z rings disassemble after invagination, and since we observed that branches may elongate from these abnormal sites later during the course of normal cell elongation, it is understandable why Gullbrand et al did not observe FtsZ at the base of new branches. Thus, superficially conflicting interpretations are resolved by the simple expedient of separating the initial triggering event (inaccuracies during cell division) from subsequent visible outgrowth of the branch, which is independent of ongoing division. As an added benefit, the patches of iPG predicted by our model would constitute the “wall asymmetries” previously posited as being the source of branch points (Gullbrand et al., 1999).
Finally, regarding the role of inert peptidoglycan (iPG), the model incorporates the knowledge that patches of iPG are always associated with the original poles and with the tip ends of branches (Fig. 3) (de Pedro et al., 2003, Nilsen et al., 2004). That is, when division is inhibited and the cells are allowed to grow into filaments, every resulting branch is capped by a fragment of iPG that was present at the beginning of the experiment. This means that no new branch points are nucleated during subsequent cell elongation and none are nucleated de novo from the lateral walls. Since, as far as is known, iPG is generated only by an FtsZ-dependent reaction during cell division, the failure to find branches with no iPG at their ends is strong evidence that branching is nucleated by previous imprecise division events. The remaining question, then, is whether mislocalized Z rings can nucleate the formation of separate patches of iPG. This is quite reasonable because the first observable cell wall modification during septal development is the FtsZ-dependent synthesis of a hoop of iPG in the complete absence of invagination (de Pedro et al., 1997, Rothfield, 2003). Synthesis of this material occurs even when FtsI or FtsA are inactivated (de Pedro et al., 1997, de Pedro et al., 2003), indicating that iPG is created early in the first stage of divisome development (Aarsman et al., 2005). Because incomplete Z rings or arcs can initiate invagination (Erickson et al., 2010, Li et al., 2007), it follows that such rings must also be able to nucleate the formation of iPG. The persistence of iPG rings, arcs and patches in the sidewalls of PBP mutants (de Pedro et al., 2003) is consistent with a scenario in which multiple misplaced Z rings produce misplaced iPG fragments that can develop into branches later.
Role of alternative LMW PBPs
PBP5 is the major and most active E. coli D,D-carboxypeptidase (D,D-CPase) and localizes to the septum in a substrate-dependent manner (Potluri et al., 2010). We now show that two other D,D-CPases, PBP6 and DacD, localize efficiently to the septum only in cells lacking active PBP5. Even so, these latter enzymes do not replace the physiological function of PBP5 because neither completely complements the shape defects of mutants lacking PBP5 (Nelson & Young, 2001), probably because neither is as enzymatically active as PBP5 (Table 3) (Chowdhury et al., 2010). Nonetheless, PBP6 and DacD contribute to this process because in their absence the pentapeptide fraction increases (Table 1) (Li et al., 2004, Varma et al., 2007). Because most pentapeptides are present in newly synthesized peptidoglycan (Vollmer & Bertsche, 2008) and because during division most new synthesis is concentrated at the invagination site (Wientjes & Nanninga, 1989), the effect of deleting additional D,D-CPases is consistent with the idea that removing extraneous pentapeptides affects Z ring orientation either indirectly, by altering cell width, or more directly, by limiting an FtsZ-directed pathway to a narrow zone around future septation sites.
Summary
In E. coli, the most obvious phenotype associated with the absence of PBP5 is the generation of cells with highly aberrant shapes and multiple branches. Our present results suggest that these phenotypes arise because such mutants position their septal rings less accurately. Therefore, helping orient the FtsZ ring appears to be a major physiological function of PBP5, supplemented by the activities of other LMW PBPs. More work is required to determine whether PBP5 produces these morphological effects via direct interaction with FtsZ or indirectly by altering cell diameter. Nonetheless, the actions of these periplasmic PBPs must be coordinated in some way with cytoplasmic proteins if the cell is to orient its division site accurately. Finally, the results suggest that branched cells arise as the result of any growth condition, treatment or mutation that interrupts or reorients the Z ring, which, in turn, nucleates the formation of iPG at inappropriate sites. In short, this means that cell branching can be viewed as an overt visual symptom of impaired FtsZ assembly or localization.
Experimental Procedures
Bacterial strains and growth conditions
E. coli strains used are listed in Table 4. Bacteria were grown in Luria-Bertani medium (Difco, Detroit, Mi.) with appropriate antibiotics where required: tetracyclin (10 µg/ml), kanamycin (50 µg/ml), chloramphenicol (20 µg/ml), or ampicillin (50 µg/ml). Unless otherwise specified, all chemicals and antibiotics were from Sigma Chemical Co. (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.). E. coli strains LP1, LP16-1, LP17-1 and LP18-1 were constructed by P1 transducing the λ(attL-lom)::bla lacIq P207-ftsZ-gfp into strains CS315-1, CS446-1, CS612-1 and CS109, respectively, using phage lysates prepared from E. coli strain EC448 (Weiss et al., 1999). E. coli strains CS235-1, CS395-1, CS476-1 and CS704-1 were derived from CS235-1K, CS395-1K, CS476-1K and CS704-1K, respectively, by transient expression of RP4 ParA resolvase to excise the kanamycin resistance cassette from the dacD deletion sites, using the protocol described previously (Denome et al., 1999). During the resolvase curing process, 0.1 mM histidine was added to the minimal medium because the CS476-1K and CS704-1K strains carried a hisG4 auxotrophic mutation.
Table 4.
Bacterial Strains
| Strain | Description | Reference / Source |
|---|---|---|
| CS109 | W1485, F− thi glnV (supE) rph-1 rpoS | (Denome et al., 1999) |
| CS9-19 | CS109 ΔpbpG | (Denome et al., 1999) |
| CS11-2 | CS109 ΔdacB | (Denome et al., 1999) |
| CS12-7 | CS109 ΔdacA | (Denome et al., 1999) |
| CS17-1 | CS109 ΔdacC | (Denome et al., 1999) |
| CS18-3 | CS109 ΔdacD | B. Meberg, unpublished |
| CS203-1B | CS109 ΔpbpG ΔdacB | (Denome et al., 1999) |
| CS204-1 | CS109 ΔpbpG ΔdacA | (Denome et al., 1999) |
| CS211-2 | CS109 ΔdacA ΔdacC | (Denome et al., 1999) |
| CS219-1 | CS109 ΔdacA ΔdacB | (Denome et al., 1999) |
| CS235-1 | CS109 ΔdacA ΔdacD (KanR cured from CS235-1K) | This work |
| CS315-1 | CS109 ΔdacA ΔdacB ΔpbpG | (Denome et al., 1999) |
| CS395-1 | CS109 ΔdacA ΔdacC ΔdacD (KanR cured from CS395-1K) | This work |
| CS446-1 | CS109 ΔdacA ΔdacB ΔdacC ΔpbpG | (Denome et al., 1999) |
| CS476-1 | CS109 ΔdacA ΔdacB ΔdacC ΔdacD hisG4 (KanR cured from CS476-1K) | This work |
| CS612-1 | CS109 ΔdacA ΔdacB ΔdacC ΔpbpG ΔampC ΔampH | (Denome et al., 1999) |
| CS703-1 | CS109 ΔmrcA ΔdacA ΔdacB ΔdacC ΔpbpG ΔampC ΔampH | (Denome et al., 1999) |
| CS704-1 | CS109 ΔdacA ΔdacB ΔdacC ΔdacD ΔpbpG ΔampC ΔampH hisG4 (KanR cured from CS704-1K) | This work |
| LP1 | CS315-1 Δ(λattL-lom)::bla lacIq P207-ftsZ-gfp | This work |
| LP16-1 | CS446-1 Δ(λattL-lom)::bla lacIq P207-ftsZ-gfp | This work |
| LP17-1 | CS612-1 Δ(λattL-lom)::bla lacIq P207-ftsZ-gfp | This work |
| LP18-1 | CS109 Δ(λattL-lom)::bla lacIq P207-ftsZ-gfp | This work |
| DH5-alpha | Φ80dlacZDM15 Δ(lacZYA-argF)U169 deoR recA1 endA hsdR17(rk−,mk+) phoA supE44 thi-1 gyrA96 relA1 | Lab collection |
| EC100 | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL (StrR) nupG | Epicentre Biotechnologies |
| XL-1-Blue | recA1 lac endA1 gyrA96 thi1 hsdR17 supE44 relA1 [F' proAB lacIq lacZΔM15 Tn10] (TetR) | Stratagene |
| XL-10-Gold | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F´ proAB lacIqZΔM15 Tn10 (TetR) Amy CamR] | Stratagene |
Plasmid constructions
Plasmids used are listed in Table 5, and their construction details are provided in the Supplemental Experimental Procedures. Unless otherwise specified, PfuUltra II Fusion HS DNA polymerase (Stratagene) was used to amplify PCR fragments for cloning. PCR amplification was performed in an MJ mini gradient thermal cycler (Bio-Rad, Hercules, CA). Standard DNA manipulation and molecular biology techniques were used for cloning genes into plasmids, as described previously (Priyadarshini et al., 2006). The sequence of each cloned gene was confirmed by DNA sequencing (MWG Biotech or by the UAMS DNA Sequencing Facility).
Table 5.
Plasmids
| Plasmid | Description | Reference |
|---|---|---|
| pBR322 | TetR, AmpR | (Gallant et al., 2005) |
| pZAQ | PftsQAZ-ftsQAZ, TetR, pBR322 background | (Ward & Lutkenhaus, 1985) |
| pLPZ | PftsZ-ftsZ, TetR, pBR322 background | This work |
| pBAD18-Kan | pBAD, KanR | (Guzman et al., 1995) |
| pLP18-Kan | pBAD, KanR (no HindIII in KanR) | This work |
| pPJ4 | pBAD-dacB, CamR | (Nelson & Young, 2001) |
| pAG6 | pBAD-dacC, CamR | (Nelson et al., 2002) |
| pPJ7 | pBAD-pbpG, CamR | (Nelson & Young, 2001) |
| pPJDacD | pBAD-dacD, CamR | (Nelson & Young, 2001) |
| pLP8 | Plac lacIq, KanR, pMLB1113 background | (Potluri et al., 2010) |
| pLP9 | Plac-dsbA(SS)-sfgfp lacIq, KanR | (Potluri et al., 2010) |
| pLP402 | Plac-dsbA(SS)-sfgfp-dacB, lacIq, KanR | This work |
| pLPKC403 | pBAD-dacB, KanR | This work |
| pLPKC404 | PBAD-dsbA(SS)-sfgfp-dacB, KanR | This work |
| pLP405 | PBAD-dacBS62A, KanR | This work |
| pLP406 | Plac-dsbA(SS)-sfgfp-dacBS62A, lacIq, KanR | This work |
| pLPKC407 | Plac-dsbA(SS)-sfgfp-dacBΔC17, lacIq, KanR | This work |
| pLP602 | Plac-dsbA(SS)-sfgfp-dacC, lacIq, KanR | This work |
| pLPKC604 | pBAD-dacC, KanR | This work |
| pLPKC605 | PBAD-dsbA(SS)-sfgfp-dacC, KanR | This work |
| pLP606 | PBAD-dacCS66A, KanR | This work |
| pLP607 | Plac-dsbA(SS)-sfgfp-dacCS66A, lacIq, KanR | This work |
| pLPKC608 | Plac-dsbA(SS)-sfgfp-dacCΔC22, lacIq, KanR | This work |
| pLP652 | Plac-dsbA(SS)-sfgfp-dacD, lacIq, KanR | This work |
| pLPKC653 | pBAD-dacD, KanR | This work |
| pLPKC654 | PBAD-dsbA(SS)-sfgfp-dacD, KanR | This work |
| pLP655 | PBAD-dacDS63A, KanR | This work |
| pLP656 | Plac-dsbA(SS)-sfgfp-dacDS63A, lacIq, KanR | This work |
| pLPKC657 | Plac-dsbA(SS)-sfgfp-dacDΔC13, lacIq, KanR | This work |
| pLP659 | Plac-dsbA(SS)-sfgfp-dacDS63AΔC13, lacIq, KanR | This work |
| pLP702 | Plac-dsbA(SS)-sfgfp-pbpG, lacIq, KanR | This work |
| pLPKC704 | pBAD-pbpG, KanR | This work |
| pLPKC705 | PBAD-dsbA(SS)-sfgfp-pbpG, KanR | This work |
| pLP706 | PBAD-pbpGS67A, KanR | This work |
| pLP707 | Plac-dsbA(SS)-sfgfp-pbpGS67A, lacIq, KanR | This work |
| pKC2 | PBAD- dsbA(SS)-sfgfp, KanR | This work |
| pLP525 | PBAD-dsbA(SS)-sfgfp-dacA, KanR | This work |
| pLP516 | pBAD-dacA, KanR | This work |
| pLP517 | pBAD-dacAS44G, KanR | This work |
| pBAD18-Cam | pBAD, CamR | (Guzman et al., 1995) |
| pPJ5 | pBAD-dacA, CamR | (Nelson & Young, 2000) |
| pLP513 | pBAD-dacAS44G, CamR | This work |
| pLP4 | Plac, lacIq, KanR | (Potluri et al., 2010) |
| pLP514 | Plac-dacAS44G, lacIq, KanR | (Potluri et al., 2010) |
| pLP515 | Plac-dacA, lacIq, KanR | (Potluri et al., 2010) |
| pLP528 | Plac-dacAΔC18, lacIq, KanR | (Potluri et al., 2010) |
Cell fixation and sample preparation for microscopy
Live cells were examined in most experiments. However, sometimes cells were chemically fixed with paraformaldehyde (2.40%), glutaraldehyde (0.04%) [Electron Microscopy Sciences], and sodium phosphate buffer (30.2 mM, pH 7.5). When required, DAPI was added to the fixative at a final concentration of 0.25 µg/ml. One ml of culture grown in LB was mixed with fixative and incubated at room temperature for 20 min, after which the cells were further incubated on ice for 1 h. After incubation on ice, cells were washed 3 times with PBS, resuspended in 100–200 µl PBS and stored at 4°C until they were imaged. Fixed or live cells (5 µl) were spotted onto a microscope slide coated with 1% agarose and incubated at room temperature for 10 min to immobilize the cells, after which they were imaged.
Microscopy
Cells and fluorescent fusion proteins were visualized by using a Zeiss Axio Imager.Z1 microscope fitted with a 100× EC Plan-Neofluar oil immersion (phase) objective with a numerical aperture of 1.3, or with a 100× differential interference contrast (DIC) objective with a 1.45 numerical aperture. Filter cubes for fluorescence image acquisition were from Zeiss. GFP was visualized with the 38HE filter set and DAPI was visualized with filter set 49. Photographs were collected with an Axiocam MRm cold charge-coupled device camera, using a typical exposure time of 2–3 seconds (fluorescence images) or 6–20 millisec (phase and DIC images). Images were processed to adjust brightness and contrast by using Adobe Photoshop.
Microscopic inspection of branched cells
E. coli strains harboring pBR322, pLPZ or pZAQ were grown overnight in LB-Tet at 37°C. The following morning, the cultures were diluted 1:250 into LB-Tet and incubated at 37°C until they reached an OD600 of 0.2. At this point, 1 ml of culture was fixed for microscopic analysis, and aztreonam (1 µg/ml, final) was added to the culture to inhibit cell division. The cultures were grown for one mass doubling, harvested and fixed for microscopic analysis. Aztreonam-treated cultures were used to determine the percentage of branched cells because some strains had such aberrant cell shapes that it was difficult to judge the number of branches or buds per cell. Elongating the cells allowed branches to be observed more clearly. Cells were inspected for the presence of branches and buds and were classified as single or multiple branched cells. Branches and buds were defined as clearly elongated protrusions along the axis or as split poles. All other abnormalities, such as bends, kinks, blebs, and imperfections, were not counted as branches. At least 500 cells were counted for each sample.
FtsZ-GFP expression
For imaging FtsZ-GFP in cells not overproducing FtsZ, E. coli strains LP1, LP16-1, LP17-1 and LP18-1 were grown overnight at 30°C in LB + 0.1% glucose, and then diluted into in LB + 25 µM IPTG, to give an OD600 of ~ 0.015. Two sets of each culture were inoculated: one was grown for 4 h, harvested and fixed; the other was grown for 3.2 h, at which time aztreonam was added (1 µg/ml final concentration) and the cultures were allowed to grow for another 40 min, after which the cultures were harvested and fixed. DAPI was included in the fixative solution to stain DNA. For localizing FtsZ-GFP in cells harboring either pZAQ or pLPZ, cells were grown overnight in LB-Tet at 37°C. The next morning the cultures were diluted 1:500 and grown for 1 h at 37°C, after which 200 µM IPTG was added to induce FtsZ-GFP production, and incubated for 2.45 h more. When required, after 2.15 h of induction, aztreonam (1 µg/ml final) was added and the cultures were allowed to grow for another 30 min. In this case, live cells were imaged because fixing the cells resulted in the loss of helical FtsZ polymers, probably because spiral or helical FtsZ polymers are unstable when cells are fixed with formaldehyde (Peters et al., 2007).
sfGFP-PBP expression
To mimic endogenous PBP levels, different concentrations of IPTG were used to determine the optimal PBP expression level. Adding 25 µM IPTG produced sfGFP-PBP fusion proteins similar to the endogenous PBP levels (Fig S8), and this IPTG concentration was used to express PBPs for localization studies.
Localizing PBP6
E. coli strains harboring appropriate plasmids were grown overnight, then diluted 1:500 into LB-Kan broth and grown at 30°C for 2 h. Two cultures were inoculated for each experiment. After 2 h growth, IPTG (25 µM final) was added to both sets. One culture was grown for 2.3 h with IPTG, harvested and live cells were imaged. The second culture was grown for 2 h with IPTG, at which time aztreonam (1 µg/ ml final) was added and the cells incubated for another 40 min (1 mass doubling). Similar localization conditions were used to express and localize other PBP fusions.
DacD localization
Overnight cultures were diluted 1:250 and grown for 1 h, after which IPTG (25 µM final) was added and the cultures were grown for another for 2.3 h. When required, aztreonam was added after 2.2 h of IPTG induction, and the cells were grown for one more mass doubling.
PBP7 localization
Overnight cultures were diluted 1:200 and grown for 1.3 h, after which 25 µM IPTG was added and further incubated for 2.3 h. When required, aztreonam was added after 2 h of IPTG induction and the cultures were grown for one more mass doubling.
Time-lapse microscopy
For time-lapse microscopy, a custom-designed, heated incubation chamber was constructed around the microscope, and the chamber temperature was maintained at 37°C by circulating hot air into the incubator with an Air-Therm ATX temperature controlled air-heater (World Precision Instruments). For growing cultures on the microscope stage, agar pads were prepared by transferring 120 µl of LB plus 0.6% agar into each of the wells of a chambered microscope slide (Lab-Tek chamber slide, Cat # 177402). To follow cell growth and branch formation, cells were grown in LB at 37°C until they reached exponential growth phase (OD600 ~0.2–0.4), at which time 2 µl of culture was transferred to the surface of the solidified agar pad, and the cells allowed to settle by incubating the slides at 37°C for 10–15 min before imaging. Images were captured automatically every 2 min using the time-lapse option in the AxioVision software, and the samples were focused either manually or automatically. For imaging FtsZ-GFP, overnight cultures of E. coli LP1 or LP18-1 were grown at 37°C in LB plus 0.1% glucose. The cultures were diluted 1:1,500 into LB plus 25 or 30 µM IPTG and incubated for 3.5 h, placed on agar pads containing 12.5 µM IPTG, and the cells were photographed manually every 5 min.
Supplementary Material
Acknowledgements
We thank Katherine Claire Mowrer for constructing the pLPKC series of plasmids, Sarah Vestal for analyzing diameter and Z ring correlations, Christopher Mosley for helping with FtsZ-GFP time-lapse experiments, and Mary Laubacher for figure formatting. We also thank Piet de Boer (Case Western Reserve University) for providing FtsZ anti-serum. This work was supported by Grant R01-GM061019 from the US National Institutes of Health, and by the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
References
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Åkerlund T, Nordström K, Bernander R. Branched Escherichia coli cells. Mol. Microbiol. 1993;10:849–858. doi: 10.1111/j.1365-2958.1993.tb00955.x. [DOI] [PubMed] [Google Scholar]
- Begg K, Nikolaichik Y, Crossland N, Donachie WD. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol. 1998;180:881–884. doi: 10.1128/jb.180.4.881-884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Molec. Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. Isolation and characterization of ftsZ alleles that affect septal morphology. J. Bacteriol. 1992;174:5414–5423. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonis M, Ecobichon C, Guadagnini S, Prevost MC, Boneca IG. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol. Microbiol. 2010;78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat. Rev. Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Skin and bones: the bacterial cytoskeleton, cell wall, and cell morphogenesis. J Cell Biol. 2007;179:381–387. doi: 10.1083/jcb.200708001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Nat. Acad. Sci. USA. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Nayak TR, Young KD, Ghosh AS. A weak DD-carboxypeptidase activity explains the inability of PBP 6 to substitute for PBP 5 in maintaining normal cell shape in Escherichia coli. FEMS Microbiol. Lett. 2010;303:76–83. doi: 10.1111/j.1574-6968.2009.01863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 2008;18:235–244. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- de Pedro MA, Grunfelder CG, Schwarz H. Restricted mobility of cell surface proteins in the polar regions of Escherichia coli. J. Bacteriol. 2004;186:2594–2602. doi: 10.1128/JB.186.9.2594-2602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli. J. Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Young KD, Höltje JV, Schwarz H. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 2003;185:1147–1152. doi: 10.1128/JB.185.4.1147-1152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh T, Bernhardt TG. Using superfolder green fluorescent protein for periplasmic protein localization studies. J Bacteriol. 2011;193:4984–4987. doi: 10.1128/JB.00315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Dziadek J, Madiraju MV, Rutherford SA, Atkinson MA, Rajagopalan M. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology. 2002;148:961–971. doi: 10.1099/00221287-148-4-961. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Molec. Biol. Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh K. Cell polarity and the control of apical growth in Streptomyces. Curr. Opin. Microbiol. 2010;13:758–765. doi: 10.1016/j.mib.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant CV, Daniels C, Leung JM, Ghosh AS, Young KD, Kotra LP, Burrows LL. Common β-lactamases inhibit bacterial biofilm formation. Mol Microbiol. 2005;58:1012–1024. doi: 10.1111/j.1365-2958.2005.04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci. USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Molecular Microbiology. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AS, Young KD. Sequences near the active site in chimeric penicillin binding proteins 5 and 6 affect uniform morphology of Escherichia coli. J. Bacteriol. 2003;185:2178–2186. doi: 10.1128/JB.185.7.2178-2186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullbrand B, Akerlund T, Nordstrom K. On the origin of branches in Escherichia coli. J. Bacteriol. 1999;181:6607–6614. doi: 10.1128/jb.181.21.6607-6614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of Bacteriology. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel AM, Wang SB, Letek M, Gil JA, Flardh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J. Bacteriol. 2008;190:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson TA, Young KD, Denome SA, Elf PK. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. Journal of Bacteriology. 1997;179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Molec. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Li SY, Höltje J-V, Young KD. Comparison of high-performance liquid chromatography and fluorophore-assisted carbohydrate electrophoresis methods for analyzing peptidoglycan composition of Escherichia coli. Anal. Biochem. 2004;326:1–12. doi: 10.1016/j.ab.2003.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Meberg BM, Paulson AL, Priyadarshini R, Young KD. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J Bacteriol. 2004;186:8326–8336. doi: 10.1128/JB.186.24.8326-8336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg BM, Sailer FC, Nelson DE, Young KD. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J Bacteriol. 2001;183:6148–6149. doi: 10.1128/JB.183.20.6148-6149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Ghosh AS, Paulson AL, Young KD. Contribution of membrane-binding and enzymatic domains of penicillin binding protein 5 to maintenance of uniform cellular morphology of Escherichia coli. J Bacteriol. 2002;184:3630–3639. doi: 10.1128/JB.184.13.3630-3639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Young KD. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 2000;182:1714–1721. doi: 10.1128/jb.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Young KD. Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 2001;183:3055–3064. doi: 10.1128/JB.183.10.3055-3064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T, Ghosh AS, Goldberg MB, Young KD. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 2004;52:1045–1054. doi: 10.1111/j.1365-2958.2004.04050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J Bacteriol. 2011;193:4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PC, Migocki MD, Thoni C, Harry EJ. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol Microbiol. 2007;64:487–499. doi: 10.1111/j.1365-2958.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Overview of cell shape: cytoskeletons shape bacterial cells. Curr. Opin. Microbiol. 2007;10:601–605. doi: 10.1016/j.mib.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim S, Kolb-Maurer A, Gentschev I, Goebel W, Kuhn M. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect. Immun. 2003;71:3473–3484. doi: 10.1128/IAI.71.6.3473-3484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potluri L, Karczmarek A, Verheul J, Piette A, Wilkin JM, Werth N, Banzhaf M, Vollmer W, Young KD, Nguyen-Disteche M, den Blaauwen T. Septal and lateral wall localization of PBP5, the major D,D-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol Microbiol. 2010;77:300–323. doi: 10.1111/j.1365-2958.2010.07205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, Popham DL, Young KD. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol. 2006;188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Letek M, Campelo AB, Vaquera J, Mateos LM, Gil JA. Altered morphology produced by ftsZ expression in Corynebacterium glutamicum ATCC 13869. Microbiology. 2005;151:2563–2572. doi: 10.1099/mic.0.28036-0. [DOI] [PubMed] [Google Scholar]
- Rothfield L. New insights into the developmental history of the bacterial cell division site. J. Bacteriol. 2003;185:1125–1127. doi: 10.1128/JB.185.4.1125-1127.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 2005;3:959–968. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- Schmidt JM, Stanier RY. The development of cellular stalks in bacteria. J Cell Biol. 1966;28:423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Dobrinski B, Hakenbeck R. Unusual septum formation in Streptococcus pneumoniae mutants with an alteration in the D,D-carboxypeptidase penicillin-binding protein 3. J. Bacteriol. 1990;172:6499–6505. doi: 10.1128/jb.172.11.6499-6505.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Montgomery BL. Determining cell shape: adaptive regulation of cyanobacterial cellular differentiation and morphology. Trends Microbiol. 2011;19:278–285. doi: 10.1016/j.tim.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Suit JC, Barbee T, Jetton S. Morphological changes in Escherichia coli strain C produced by treatments affecting deoxyribonucleic acid synthesis. J. Gen. Microbiol. 1967;49:165–173. doi: 10.1099/00221287-49-1-165. [DOI] [PubMed] [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30:154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. Embo J. 2010 doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, de Pedro MA, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 2007;189:5692–5704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Huang KC, Young KD. The Min system as a general cell geometry detection mechanism: branch lengths in Y-shaped Escherichia coli cells affect Min oscillation patterns and division dynamics. J. Bacteriol. 2008;190:2106–2117. doi: 10.1128/JB.00720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Young KD. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 2004;186:6768–6774. doi: 10.1128/JB.186.20.6768-6774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Young KD. In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2. J. Bacteriol. 2009;191:3526–3533. doi: 10.1128/JB.01812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Ward JE, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes FB, Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J. Bacteriol. 1989;171:3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL, Zaritsky A, Grover NB. Nucleoid partitioning and the division plane in Escherichia coli. J. Bacteriol. 1994;176:6030–6038. doi: 10.1128/jb.176.19.6030-6038.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. Bacterial shape. Mol. Microbiol. 2003;49:571–580. doi: 10.1046/j.1365-2958.2003.03607.x. [DOI] [PubMed] [Google Scholar]
- Yu XC, Margolin W. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J. Bacteriol. 2000;182:6203–6213. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Vernet T, Pinho MG. The different shapes of cocci. FEMS Microbiol. Rev. 2008;32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- Zaritsky A, Wang P, Vischer NO. Instructive simulation of the bacterial cell division cycle. Microbiol. 2011;157:1876–1885. doi: 10.1099/mic.0.049403-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.