Abstract
Objectives
The subgingival microbiota in Down syndrome and non-Down syndrome (non-DS) adults receiving periodic dental care was examined for 40 bacterial species with checkerboard DNA-DNA hybridization, and related to clinical periodontal attachment loss (AL).
Methods
A total of 44 Down syndrome, 66 non-DS mentally retarded, and 83 mentally normal adults were clinically evaluated, with subgingival specimens removed and pooled per subject from three interproximal sites on different teeth, and assessed for the presence and levels of 40 bacterial species using species-specific whole genomic DNA probes and checkerboard DNA-DNA hybridization. Significant group differences in species proportions averaged across subjects were evaluated using the Kruskal-Wallis test, and associations between subgingival species and mean subject AL within Down syndrome and non-DS subject groups were quantified with Pearson correlation and multiple linear regression analysis.
Results
Down syndrome subjects exhibited greater AL than non-DS subjects (P = 0.05). Down syndrome subjects yielded most microbial species at levels similar to non-DS subjects, except for higher proportions of Selenomonas noxia, Propionibacterium acnes, Streptococcus gordonii, Streptococcus mitis, and Streptococcus oralis as compared to non-DS study subjects, higher Treponema socranskii than non-DS mentally retarded subjects, and higher Streptococcus constellatus relative to mentally normal subjects. Down syndrome adults classified with periodontitis revealed higher subgingival levels of T. socranskii than non-periodontitis Down syndrome subjects (P = 0.02). Higher subgingival proportions of S. constellatus, Fusobacterium nucleatum subsp. nucleatum, S. noxia and Prevotella nigrescens showed significant positive correlations (r = 0.35 to 0.42), and Actinomyces naeslundii II and Actinomyces odontolyticus negative correlations (r = −0.36 to −0.40), with increasing mean subject AL in Down syndrome adults.
Conclusions
Individuals with DS show higher levels of some subgingival bacterial species and specific associations between certain subgingival bacterial species and loss of periodontal attachment. These findings are consistent with the notion that certain subgingival bacteria may contribute to the increased level of periodontal disease seen in DS individuals, and raise the question as to the reason for increased colonization in DS.
Keywords: subgingival microbiota, Down syndrome, periodontitis, periodontal
Introduction
Down syndrome is the most common chromosomal abnormality in live-born infants, and is estimated to occur in approximately 1 in 732 infants in the United States (1). The syndrome is caused by trisomy of chromosome 21 (2). Its most common manifestations include a characteristic physical appearance, and a variety of medical disorders including mental retardation, congenital heart disease, thyroid dysfunction, Alzheimer’s disease and abnormalities of the immune system (2).
It is well documented that individuals with Down syndrome are also at increased risk for developing destructive forms of periodontal disease (3, 4), with the majority affected early in life with extensive gingival tissue inflammation, bleeding on probing, deepened probing depths, loss of periodontal attachment, and crestal alveolar bone loss (3, 4). Since dentogingival microbial biofilm infections are considered essential for the initiation and progression of human periodontal diseases (5), the relationship between subgingival microorganisms and periodontal disease in individuals with Down syndrome has been the focus of several investigations (6–9). However, only mixed and inconclusive findings on this association are presently available, likely due to the limited number of study subjects examined, a primary focus on children and younger-aged adults, and the relatively small range of evaluated periodontopathic taxa.
The main objectives of the present study were to 1.) determine the presence and levels of 40 subgingival bacterial species in adult-aged individuals with Down syndrome using species-specific whole genomic DNA probes and checkerboard DNA-DNA hybridization, 2.) investigate the subgingival microbial profile of Down syndrome adults matched with non-Down syndrome (non-DS) mentally retarded subjects and mentally normal subjects, and 3.) assess the association between 40 subgingival bacterial species and loss of clinical periodontal attachment in individuals with and without Down syndrome.
Materials and Methods
Study sites
This study was done in cooperation with the Georgia Department of Human Resources/Georgia Regional Hospitals (GRH). The study included three subject groups, which were comprised of Down syndrome, non-DS mentally retarded, and mentally normal study subjects, respectively. Down syndrome and non-DS mentally retarded study subjects were recruited from GRH hospital centers in Atlanta, Savannah and Augusta, Georgia. All Down syndrome and non-DS mentally retarded subjects were patients of record and receiving periodic dental care at one of the three hospital locations. Some of these individuals were institutionalized, while others were outpatients living in group-homes or with their families.
Subject inclusion criteria
Study subject inclusion criteria and recruitment were as previously described. In brief, Down syndrome study subjects had a confirmed diagnosis of Trisomy 21, a minimum of 10 teeth present, no other medical conditions known to affect periodontal status (i.e., diabetes mellitus), no systemic antibiotic therapy in the 3 months prior to entry into the study, no history of cigarette smoking, were adults 18 years or older, and were able to cooperate with the study examiners. The same inclusion criteria were used to select the other study group subjects, except for the need of a confirmed diagnosis of mental retardation without Trisomy 21 for non-DS mentally retarded subjects, and an absence of mental retardation for mentally normal subjects. All study subjects needed to be able to communicate with and understand spoken English.
Subject recruitment
The attending dentist-in-charge of the dental clinic at each of the three GRH hospital centers reviewed available records and identified dentate Down syndrome patients whom met the study criteria and would be able to participate in a dental examination. Non-DS mentally retarded subjects, where their mental retardation was secondary to head trauma at birth, were matched to the Down syndrome patients on age, race, and gender using the same hospital records, and selected on the basis of their ability to cooperate and sit for a dental examination without need for sedation. Mentally normal subjects, similarly matched on gender, race and age to the Down syndrome subjects, were recruited from the general population living in the vicinity of the three GRH hospital centers, and were under the care of private practice dentists.
A total of 289 subjects were initially screened for the study, of which 26 were disqualified for medical reasons, 46 completed only portions of the study evaluations, but were not able to return to complete remaining portions, and 217 completed most of the study evaluations. Demographic and clinical data for the entire subject populations are reported elsewhere. For the present study, clinical and microbiological data were obtained from 193 subjects, including 44 Down syndrome, 66 non-DS mentally retarded, and 83 mentally normal control subjects.
Informed consent
The study protocol and informed consent process were approved by the Georgia Regional Hospital Institutional Review Board. Prior to commencing the study, informed consent was obtained and documented for all subjects. The GRH hospital center dentist-in-charge contacted the family or legal guardian/caretaker of each potential Down syndrome or non-DS mentally retarded subject, explained the study protocol, and obtained their consent to enroll the subject in the study. In addition to obtaining family or legal guardian/caretaker written informed consent, the study protocol was also explained directly to each Down syndrome or non-DS mentally retarded subject prior to commencing the study examination procedures, and their personal consent was obtained and witnessed. Mentally normal study subjects consented on their own behalf.
Clinical evaluations
As previously described (10), two experienced dental hygienists blinded to the objectives of the study performed all exams. The examiners were calibrated and standardized in the use of the clinical evaluation measures employed in the study (10). The examiners enumerated missing teeth and teeth with carious lesions then recorded the Loe and Silness gingival index (GI) (11), the Quigley Hein plaque index (12), probing depth (PD) and clinical periodontal attachment levels (AL) on six sites per tooth. Third molars were excluded. Periodontitis was defined as present when 5% or higher of scored teeth exhibited attachment loss =>5 mm (13).
Microbiological evaluations
Three sites per subject were sampled for microbial analysis after completion of the clinical evaluations. In periodontitis subjects, three interproximal sites with PD > 5 mm were randomly chosen from three different teeth in separate quadrants. In non-periodontitis subjects, three interproximal sites with PD < 4 mm were randomly chosen from three different teeth in separate quadrants. After removal of supragingival plaque and isolation to prevent saliva contamination, subgingival plaque samples were collected with a sterile Gracey curette from the selected sites. All subgingival samples per subject were pooled into a single Eppendorf tube containing 450 µl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.6), followed by the addition of 300 µl of 0.5 M NaOH. The samples were immediately frozen at −70°C, and delivered within a three-month period to the Forsyth Institute (Boston, MA, USA) for microbiological analysis.
Upon arrival at the Forsyth Institute, the presence and levels of 40 bacterial species were determined in each pooled subgingival plaque sample using species-specific whole genomic DNA probes and a modification (14) of the checkerboard DNA–DNA hybridization technique (15). In brief, each sample was lysed by boiling for 10 minutes, followed by neutralization with 800 µl of ammonium acetate, with denatured DNA placed in lanes on a nylon membrane using a Minislot device (Immunetics, Cambridge, MA, USA). After DNA fixation to the membrane, the membrane was placed in a Miniblotter 45 (Immunetics), with the lanes of DNA at 90° to the lanes of the device. Digoxigenin-labeled whole genomic DNA probes constructed to each of the 40 bacterial test species, as previously described (16), were hybridized in individual lanes of the Miniblotter. After hybridization, the membranes were washed at high stringency and the DNA probes detected using antibody to digoxigenin conjugated with alkaline phosphatase. Chemifluorescence detection of signals was performed using AttoPhos substrate (Amersham Life Science, Arlington Heights, IL, USA) and a computer-linked Storm Fluorimager (Molecular Dynamics, Sunnyvale, CA, USA) to read the intensity of fluorescence signals resulting from DNA probe-target hybridization. Two lanes in each run contained standards at concentrations of 105 and 106 cells of each of the test bacterial species. The concentration of each DNA probe was adjusted to permit detection of 104 cells of a given test bacterial species. The sensitivity and specificity of the species-specific DNA probes was as previously reported (16). Storm Fluorimager signals were converted to absolute counts by comparison with the standards on the same membrane. Failure to detect a signal was recorded as a zero. A total of 193 pooled subgingival plaque samples were evaluated.
Data analysis
Three-way analysis of variance (ANOVA), followed by Tukey’s HSD test, were used on mean values, and chi-square tests on proportional data, to examine for statistically significant between-group differences in demographic and clinical parameters. The percentage of DNA probe counts comprised by each of the tested bacterial species in pooled subgingival patient samples was determined for each study subject, with species proportions then averaged across subjects in each of the three study subject groups. Significant differences in test species proportions among study subject groups were evaluated using the Kruskal-Wallis test, adjusted for 40 comparisons (17). The association between subgingival species and the mean subject AL within Down syndrome and non-DS subject groups was quantified with Pearson correlation coefficients adjusted for subject age, race, gender, and supragingival plaque levels. In instances where a significant association (P ≤ 0.05) was found in either group, differences in partial associations between groups were tested with multiple linear regression analysis, with interaction terms (group, species, and group by species) modeled in addition to previous adjustment variables. Significance testing of between-group, species-specific interaction terms on mean subject AL were used to identify microbial species, at a critical P-value of ≤ 0.01, that were unique correlates of mean subject AL in individuals with Down syndrome.
Results
Clinical findings
Table 1 summarizes demographic and clinical data of the 193 study subjects. All three subject groups exhibited similar race and gender distributions. ANOVA showed differences between the three subject groups on age (P = 0.0001) and institutional living (P = 0.001). Follow-up analysis with Tukey’s HSD test showed that the non-DS mentally retarded subjects were older than Down syndrome and mentally normal subjects (P = 0.05), and the number of institutionalized subjects in the non-DS mentally retarded group was higher than the Down syndrome (P = 0.05). Clinically, the percentage of subjects with periodontitis (as defined above) was similarly distributed among the three subject groups. ANOVA showed significant differences between the three subject groups on GI, PI, AL (both mean whole-mouth and percentage of sites with AL ≥ 5 mm), and the number of missing teeth (P < 0.05). Follow-up analysis with Tukey’s HSD showed that subjects in both the Down syndrome and non-DS mentally retarded groups had higher values for GI (P = 0.05), PI (P = 0.05), and number of missing teeth (P = 0.05) than mentally normal subjects. Even though similar proportions of subjects in each of the three study groups were classified with periodontitis, Down syndrome subjects exhibited greater loss of clinical periodontal attachment (both mean whole-mouth and percentage of sites with AL ≥ 5 mm) than non-DS subjects (P = 0.05).
Table 1.
Summary of demographic and clinical features of 193 subjects with microbiological data.
| Variable | Down N = 44 |
Non-DS Mentally Retarded N = 66 |
Mentally Normal N = 83 |
p-value |
|---|---|---|---|---|
| Demographics: | ||||
| Age Age range (years) |
35.81 (1.83) 18–56 |
46.15 (1.40) 22–84 |
40.93 (1.33) 18–73 |
0.0001* |
| % Caucasian | 81.82 | 80.30 | 68.67 | NS |
| % Male gender | 47.73 | 57.58 | 44.58 | NS |
| % Institutionalized | 40.91 | 84.85 | NA | 0.001** |
| Clinical: | ||||
| Number of missing teeth | 4.38 (0.58) | 4.60 (0.48) | 1.80 (0.42) | 0.0001# |
| Plaque Index | 1.71 (0.11) | 1.85 (0.09) | 1.28 (0.08) | 0.001# |
| Gingival Index | 0.94 (0.04) | 1.01 (0.04) | 0.71 (0.03) | 0.0001# |
| Probing Depth (mm) | 2.58 (0.08) | 2.34 (0.06) | 2.41 (0.06) | 0.07 |
| Attachment Level (mm) | 2.67 (0.11) | 2.20 (0.09) | 2.24 (0.08) | 0.0006¶ |
| % of sites with AL=>5mm | 11.30 (1.78) | 3.84 (1.45) | 4.46 (1.29) | 0.002¶ |
| % of subjects classified with periodontitis | 72.7 | 71.1 | 78.8 | 0.55 |
Data presented as mean (SE) or percentage. P-value gives the probability that the groups differ in either an ANOVA test or chi-square test for means or proportions respectively.
Follow-up analysis with Tukey’s HSD test showed:
Mental retardation > Down and mentally normal (p = 0.05);
Mental retardation > Down (p = 0.05);
mentally healthy < mental retardation and Down (p = 0.05);
Down > mental retardation and mentally normal (p = 0.05).
Microbiological findings
Figure 1 displays the mean proportions of DNA probe counts of the 40 test bacterial species among the three subject groups, irrespective of the clinical status of subjects within each group. Species were ordered and grouped according to the microbial complexes described by Socransky et al. (18). All of the bacterial species were detected in the subgingival plaque samples of each group, and most species were present at similar levels in each group. No between-group differences in subgingival proportions were found among all evaluated species (N = 9) in the red, purple, and Actinomyces microbial complexes, as well as for 19 (61.3%) of the other 31 evaluated organisms.
Figure 1.
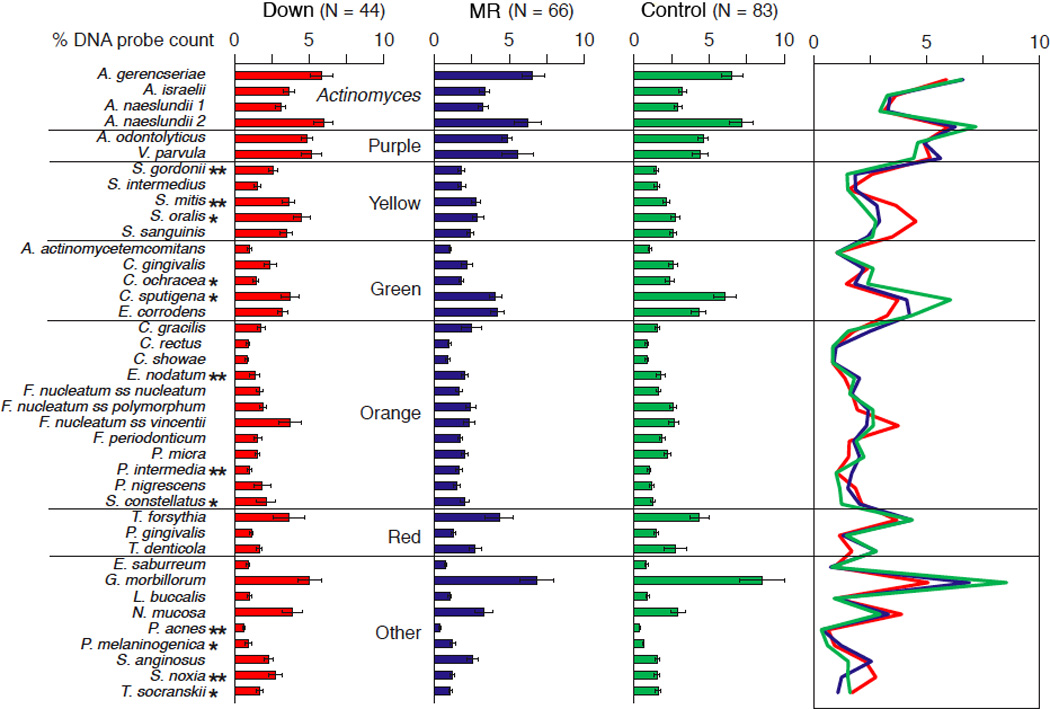
Bar chart of mean % of the DNA probe count of individual species in subgingival plaque samples taken from Down syndrome, non-DS mentally retarded (MR), and mentally normal (control) subjects. The bars represent the mean percentages and the whiskers the standard error of the mean. The % of the DNA probe count for each species was computed per subject and then averaged across subjects in each group.
* P < 0.05, ** P < 0.01.
However, after adjusting for 40 comparisons, Down syndrome subjects yielded significantly higher subgingival levels of S. noxia, P. acnes, S. gordonii, S. mitis, and S. oralis relative to non-DS study subjects; higher T. socranskii than non-DS mentally retarded subjects, and higher S. constellatus as compared to mentally normal subjects. Non-DS mentally retarded subjects had significantly higher subgingival proportions of P. intermedia, P. melaninogenica, and E. nodatum relative to the other two study groups, and higher S. constellatus as compared to mentally normal subjects. Mentally normal subjects revealed significantly higher subgingival levels of T. socranskii than non-DS mentally retarded subjects, higher C. ochracea than Down syndrome subjects, and higher C. sputigena than the other two subject groups.
Comparison of microbial profiles between groups by periodontal status (periodontitis and non-periodontitis) confirmed lack of major differences between the three subject groups (data not shown). However, within the Down syndrome group, periodontitis subjects exhibited higher levels of T. socranskii than non-periodontitis subjects (P = 0.02). Within non-DS groups, periodontitis subjects showed higher proportions of A. actinomycetemcomitans (P = 0.017) and T. denticola (P = 0.005) than non-periodontitis subjects.
Table 2 summarizes the significant correlations (p =< 0.1) noted between mean subject AL and subgingival levels of each test bacterial species, for Down syndrome and non-DS groups, with correlations adjusted for age, race, gender and supragingival plaque levels. Because the non-DS mentally retarded and mentally normal groups showed similar levels of association (data not shown), they were combined into a joint non-DS comparison group. The association between microbial species and mean subject AL markedly differed between Down syndrome and non-DS subjects. In Down syndrome subjects, increasing subgingival proportions of S. constellatus, F. nucleatum subsp. nucleatum, S. noxia, and P. nigrescens were positively associated, and A. naeslundii II and A. odontolyticus were significantly negatively correlated, with increasing mean AL in subjects. In comparison, non-DS subjects showed increasing levels of A. actinomycetemcomitans, E. corrodens, S. mitis, S. oralis, and N. mucosa to be positively associated, and increasing levels of A. gerencseriae to be negatively associated, with increasing mean subject AL.
Table 2.
Correlations relating periodontal attachment loss with levels of each microbial species within Non-Down and Down groups, after adjusting for age, gender, race, and plaque index.
| Microbial Species (Percent DNA) |
Strain* | Non-DS N= 149 |
DS N = 44 |
Interaction significance |
|---|---|---|---|---|
| A. gerencseriae | ATCC 23840 | −0.24 (.005) | > 0.15 | |
| A. israelii | ATCC 12102 | |||
| A. naeslundii 1 | ATCC 12104 | −0.16 (0.06) | ||
| A. naeslundii 2 | ATCC 43146 | −0.40 (.01) | 0.001 | |
| A. odontolyticus | ATCC 17929 | −0.36 (.03) | 0.001 | |
| V. parvula | ATCC 10790 | |||
| S. gordonii | ATCC 10558 | |||
| S. intermedius | ATCC 27335 | |||
| S. mitis | ATCC 49456 | 0.27 (0.001) | 0.03 | |
| S. oralis | ATCC 35037 | 0.20 (0.02) | 0.09 | |
| S. sanguinis | ATCC 10556 | 0.15 (.07) | −0.27 (.10) | 0.006 |
| A. actinomycetemcomitans | ATCC 43718 and 29523 | 0.29 (<0.001) | > 0.15 | |
| C. gingivalis | ATCC 33624 | |||
| C. ochracea | ATCC 33596 | |||
| C. sputigena | ATCC 33612 | |||
| E. corrodens | ATCC 23834 | 0.24 (0.004) | 0.06 | |
| C. gracilis | ATCC 33236 | −0.14 (0.09) | ||
| C. rectus | ATCC 33238 | |||
| C. showae | ATCC 51146 | |||
| E. nodatum | ATCC 33099 | |||
| F. nucleatum ss nucleatum | ATCC 25586 | 0.42 (0.009) | 0.002 | |
| F. nucleatum ss polymorphum | ATCC 10953 | |||
| F. nucleatum ss vincentii | ATCC 49256 | |||
| F. periodonticum | ATCC 33693 | |||
| P. micra | ATCC 33270 | |||
| P. intermedia | ATCC 25611 | |||
| P. nigrescens | ATCC 33563 | 0.35 (0.04) | 0.10 | |
| S. constellatus | ATCC 27823 | 0.42 (0.008) | 0.01 | |
| T. forsythia | ATCC 43037 | |||
| P. gingivalis | ATCC 33277 | 0.15 (0.07) | ||
| T. denticola | FI B1 | |||
| E. saburreum | ATCC 33271 | |||
| G. morbillorum | ATCC 27824 | −0.30 (0.07) | ||
| L. buccalis | ATCC 14201 | |||
| N. mucosa | ATCC 19696 | 0.24 (0.003) | 0.11 | |
| P. acnes | ATCC 11827 and 11828 | |||
| P. melaninogenica | ATCC 25845 | |||
| S. anginosus | ATCC 33397 | |||
| S. noxia | ATCC 43541 | 0.37 (.02) | 0.06 | |
| T. socranskii | FI S1 | 0.31 (.06) |
Data presented as correlation coefficient (p-value). Values with p-levels above 0.10 are not shown.
ATCC = American Type Culture Collection (Manassas, VA), FI = Forsyth Institute (Boston, MA)
Further significance testing of between-group, species-specific interaction terms on mean subject AL showed S. constellatus, F. nucleatum subsp. nucleatum, A. naeslundii II, A. odontolyticus, and S. sanguinis to each demonstrate a non-zero association with mean subject AL within the Down syndrome group that was statistically different from the association found in non-DS subjects, indicating that these bacterial species were related to AL in Down syndrome subjects, but not in non-DS subjects. Interestingly, none of the non-zero microbial species associations with mean subject AL obtained in the non-DS group were significantly different from those found in Down syndrome subjects.
Discussion
The present study findings demonstrated that adult Down syndrome periodontitis subjects, while exhibiting greater periodontal pathology than non-DS subjects (10), yielded subgingival microbiological profiles similar to those found in chronic periodontitis patients. Most of the 40 subgingival bacterial species evaluated with checkerboard DNA-DNA hybridization were present at similar levels in adult Down syndrome subjects as compared to matched non-DS mentally retarded and mentally normal persons, including the major periodontal pathogens A. actinomycetemcomitans, P. gingivalis, T. forsythia, P. intermedia, P. micra, F. nucleatum and C. rectus, consistent with previous culture-based studies (19).
Among the few bacterial species found to exhibit significant between-group differences, Down syndrome adults showed higher subgingival proportions of S. noxia, P. acnes, S. gordonii, S. mitis, and S. oralis as compared to non-DS study subjects, higher T. socranskii levels than non-DS mentally retarded subjects, and higher S. constellatus relative to mentally normal subjects. S. gordonii, S. mitis, and S. oralis are pioneer organisms which initiate early microbial colonization and assist in the further development of microbial plaque by creating a foothold for other bacteria to attach to. Furthermore, S. gordonii contains genes that facilitate the attachment of free floating P. gingivalis to adherent plaque biofilm (20). P. acnes is an opportunistic pathogen normally found on skin and has been associated with persistent apical periodontal infections (21). The presence of P. acnes at elevated levels in the subgingival microbiota of DS subjects may be related to a thumb sucking and/or a finger biting habit (22). S. noxia is a periodontal pathogen associated with periodontal disease activity in interproximal sites (23). T. socranskii has been associated with severity of periodontal tissue destruction (24, 25). S. constellatus is an important periodontal pathogen associated with refractory forms of periodontitis (26–28). The presence of these organisms at elevated levels in DS individuals may contribute to their increased susceptibility to periodontitis.
It is important to note that all subjects in this study received dental care on a regular basis, including periodic periodontal dental prophylaxis/scaling. These repeated periodontal treatment procedures most likely affected the subgingival microbial colonization patterns detected in the study subjects, particularly in regard to suppression of red complex P. gingivalis, T. denticola and T. forsythia species (29), and potentially minimized harmful effects of elevated supragingival plaque levels in Down syndrome and non-DS mentally retarded subjects (30). The positive associations found between various streptococci and mean subject AL in non-DS study subjects may indeed reflect more an outcome of successful periodontal therapy rendered than any potential etiologic association (31).
Among Down syndrome group subjects in the present study, subgingival proportions of S. constellatus, F. nucleatum subsp. nucleatum, S. noxia and P. nigrescens showed statistically significant positive correlations, and A. naeslundii II and A. odontolyticus significantly negative correlations, with increasing mean subject AL, although all of these correlations were relatively modest in magnitude (r = 0.35–0.42). Subgingival proportions of suspected periodontal pathogen T. socranskii also tended to associate (albeit non-significantly) with mean subject AL in Down syndrome subjects. S. constellatus, F. nucleatum subsp. nucleatum, S. noxia, P. nigrescens, and T. socranskii are all frequently associated with the subgingival microbiota of chronic periodontitis lesions in mentally normal adults, whereas A. naeslundii II and A. odontolyticus are strongly associated with stable healthy periodontal conditions (18, 32). S. constellatus and F. nucleatum are difficult to eliminate with scaling and root-palning alone (29). The association of these two organisms with loss of periodontal attachment in DS individuals suggests that organisms resistant to conventional mechanical debridement may contribute to the severity of periodontitis in this vulnerable population.
In previous studies of Down syndrome in children, where little or no periodontal therapy was performed prior to microbiological evaluations, Meskin et al. (7) found no differences in the prevalence of Bacteroides melaninogenicus (now Prevotella melaninogenica) in subgingival plaque from institutionalized children with Down syndrome versus normal or cerebral palsy-affected children. In contrast, increased levels of A. actinomycetemcomitans have been reported in Down syndrome children (6, 33). Similarly, Amano et al. (34) reported ten periodontal pathogens to exhibit an increased PCR-positive prevalence in children with Down syndrome, and Sakellari et al. (8) found various periodontal pathogens to colonize children, adolescents and young adults with Down syndrome earlier and at higher levels than healthy individuals and subjects with cerebral palsy.
In Down syndrome adults most comparable to our study population, Reuland-Bosma et al. (19) compared 17 Down syndrome adults with moderate periodontitis and daily caretaker-performed oral hygiene with a well-matched control group, and found no statistically significant differences in the subgingival prevalence and cultivable proportions of seven periodontal bacterial pathogens. Our findings with 40 subgingival bacterial species confirm and extend their observations by examining a larger number of Down syndrome subjects with varied ethnicities, and evaluating a wider range of disease and health-associated bacterial species. Additionally, this study better controlled for mental challenge and its associated behavioral sequelae as factors that may influence the association between AL and certain bacterial species in Down syndrome subjects. In this regard, we have previously reported that while Down syndrome adults have a higher prevalence of periodontal disease than non-DS mentally retarded adults, they have similar levels of mental challenge (10). Limitations of the present study include the limited number of sampled sites (N = 3) per subject, pooling of subgingival specimens within subjects prior to microbiological evaluation, no inclusion of children or adolescents in the study populations, and no assessments of herpesviruses implicated Down syndrome periodontitis (35).
Nevertheless, the occurrence of more severe periodontal pathology in Down syndrome adults, with subgingival bacterial species typical of chronic periodontitis patients, suggests that altered host-microbial interactions are particularly important in the pathogenesis of Down syndrome periodontitis. Indeed, the presence of immune deficiencies, altered cell functions, and oxidative stress alterations in individuals with Down syndrome support this notion (36–38)). Interestingly, we recently reported that positive carriage of several IL-1 genetic polymorphisms (IL-1A +4845, IL-1B +3954, and IL-1RN +2018) was inversely related to periodontal attachment loss in Down syndrome subjects (39), in contrast to their positive associations in some chronic periodontitis patients (40). Further research to delineate the nature of host immune responses to both bacterial and viral challenges in the subgingival environment of Down syndrome subjects is warranted.
In conclusion, despite the similarities between the subgingival microbial composition of individuals with and without DS, individuals with DS show higher levels of some bacterial species and specific associations between certain bacterial species and loss of periodontal attachment. These findings are consistent with the notion that certain subgingival bacteria may contribute to the increased level of periodontal disease seen in DS individuals, and raise the question as to the reason for such increased colonization in DS.
Acknowledgement
This study was supported by grant R21 DE015012-02 from the National Institute of Dental and Craniofacial Research, Bethesda, Maryland.
References
- 1.Sherman SL, Allen EG, Bean LH, et al. Epidemiology of Down syndrome. Ment Retard Dev Disabil Res Rev. 2007;13:221–227. doi: 10.1002/mrdd.20157. [DOI] [PubMed] [Google Scholar]
- 2.Patterson D, Patterson D. Genetic mechanisms involved in the phenotype of Down syndrome. Ment Retard Dev Disabil Res Rev. 2007;13:199–206. doi: 10.1002/mrdd.20162. [DOI] [PubMed] [Google Scholar]
- 3.Agholme MB, Dahllof G, Modeer T. Changes of periodontal status in patients with Down syndrome during a 7-year period. Eur J Oral Sci. 1999;107:82–88. doi: 10.1046/j.0909-8836.1999.eos107202.x. [DOI] [PubMed] [Google Scholar]
- 4.Reuland-Bosma W, van Dijk J. Periodontal disease in Down's syndrome: a review. J Clin Periodontol. 1986;13:64–73. doi: 10.1111/j.1600-051x.1986.tb01416.x. [DOI] [PubMed] [Google Scholar]
- 5.Kornman KS, Kornman KS. Mapping the pathogenesis of periodontitis: a new look. J Periodontol. 2008;79:1560–1568. doi: 10.1902/jop.2008.080213. [DOI] [PubMed] [Google Scholar]
- 6.Barr-Agholme M, Dahllof G, Linder L, Modeer T. Actinobacillus actinomycetemcomitans, Capnocytophaga and Porphyromonas gingivalis in subgingival plaque of adolescents with Down's syndrome. Oral Microbiol Immunol. 1992;7:244–248. doi: 10.1111/j.1399-302x.1992.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 7.Meskin LH, Farsht EM, Anderson DL. Prevalence of Bacteroides melaninogenicus in the gingival crevice area of institutionalized trisomy 21 and cerebral palsy patients and normal children. J Periodontol. 1968;39:326–328. doi: 10.1902/jop.1968.39.6.326. [DOI] [PubMed] [Google Scholar]
- 8.Sakellari D, Arapostathis KN, Konstantinidis A. Periodontal conditions and subgingival microflora in Down syndrome patients. A case-control study.[see comment] J Clin Periodontol. 2005;32:684–690. doi: 10.1111/j.1600-051X.2005.00737.x. [DOI] [PubMed] [Google Scholar]
- 9.Sakellari D, Belibasakis G, Chadjipadelis T, Arapostathis K, Konstantinidis A. Supragingival and subgingival microbiota of adult patients with Down's syndrome. Changes after periodontal treatment. Oral Microbiol Immunol. 2001;16:376–382. doi: 10.1034/j.1399-302x.2001.160610.x. [DOI] [PubMed] [Google Scholar]
- 10.Khocht A, Janal M, Turner B. Periodontal health in Down syndrome: contributions of mental challenge, personal and professional dental care. Spec Care Dentist. 2010;30:118–123. doi: 10.1111/j.1754-4505.2010.00134.x. [DOI] [PubMed] [Google Scholar]
- 11.Loe H. The Gingival Index, the Plaque Index and the Retention Index Systems. J Periodontol. 1967;38(Suppl):610–616. doi: 10.1902/jop.1967.38.6.610. [DOI] [PubMed] [Google Scholar]
- 12.Quigley GA, Hein JW. Comparative cleansing efficiency of manual and power brushing. J Am Dent Assoc. 1962;65:26–29. doi: 10.14219/jada.archive.1962.0184. [DOI] [PubMed] [Google Scholar]
- 13.Teles FRF, Teles RP, Siegelin Y, Paster B, Haffajee AD, Socransky SS. RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms. Mol Oral Microbiol. 2011;26:127–139. doi: 10.1111/j.2041-1014.2010.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haffajee AD, Cugini MA, Dibart S, Smith C, Kent RL, Jr, Socransky SS. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J Clin Periodontol. 1997;24:324–334. doi: 10.1111/j.1600-051x.1997.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 15.Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. "Checkerboard" DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- 16.Socransky SS, Haffajee AD, Smith C, et al. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol. 2004;19:352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 17.Socransky SS, Haffajee AD, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol. 1991;18:766–775. doi: 10.1111/j.1600-051x.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 18.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 19.Reuland-Bosma W, van der Reijden WA, van Winkelhoff AJ. Absence of a specific subgingival microflora in adults with Down's syndrome. J Clin Periodontol. 2001;28:1004–1009. doi: 10.1034/j.1600-051x.2001.281103.x. [DOI] [PubMed] [Google Scholar]
- 20.Kuboniwa M, Tribble GD, James CE, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
- 21.Fujii R, Saito Y, Tokura Y, Nakagawa KI, Okuda K, Ishihara K. Characterization of bacterial flora in persistent apical periodontitis lesions. Oral Microbiol Immunol. 2009;24:502–505. doi: 10.1111/j.1399-302X.2009.00534.x. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira ACB, Paiva SM, Campos MR, Czeresnia D. Factors associated with malocclusions in children and adolescents with Down syndrome. Am J Orthod Dentofacial Orthop. 2008;133:489.e481–489.e488. doi: 10.1016/j.ajodo.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Tanner ACR, Paster BJ, Lu SC, et al. Subgingival and tongue microbiota during early periodontitis. J Dent Res. 2006;85:318–323. doi: 10.1177/154405910608500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ledder RG, Gilbert P, Huws SA, et al. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007;73:516–523. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi Y, Umeda M, Sakamoto M, Benno Y, Huang Y, Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J Periodontol. 2001;72:1354–1363. doi: 10.1902/jop.2001.72.10.1354. [DOI] [PubMed] [Google Scholar]
- 26.Colombo AP, Haffajee AD, Dewhirst FE, et al. Clinical and microbiological features of refractory periodontitis subjects. J Clin Periodontol. 1998;25:169–180. doi: 10.1111/j.1600-051x.1998.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 27.Colombo APV, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magnusson I, Walker CB. Refractory periodontitis or recurrence of disease. J Clin Periodontol. 1996;23:289–292. doi: 10.1111/j.1600-051x.1996.tb02091.x. [DOI] [PubMed] [Google Scholar]
- 29.Cugini MA, Haffajee AD, Smith C, Kent RL, Jr, Socransky SS. The effect of scaling and root planing on the clinical and microbiological parameters of periodontal diseases: 12-month results. J Clin Periodontol. 2000;27:30–36. doi: 10.1034/j.1600-051x.2000.027001030.x. [DOI] [PubMed] [Google Scholar]
- 30.Ramfjord SP. Maintenance care for treated periodontitis patients. J Clin Periodontol. 1987;14:433–437. doi: 10.1111/j.1600-051x.1987.tb02247.x. [DOI] [PubMed] [Google Scholar]
- 31.Teles RP, Haffajee AD, Socransky SS. Microbiological goals of periodontal therapy. Periodontol 2000. 2006;42:180–218. doi: 10.1111/j.1600-0757.2006.00192.x. [DOI] [PubMed] [Google Scholar]
- 32.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 33.Sreedevi H, Munshi AK. Neutrophil chemotaxis in Down syndrome and normal children to Actinobacillus actinomycetemcomitans. J Clin Pediatr Dent. 1998;22:141–146. [PubMed] [Google Scholar]
- 34.Amano A, Kishima T, Akiyama S, Nakagawa I, Hamada S, Morisaki I. Relationship of periodontopathic bacteria with early-onset periodontitis in Down's syndrome. J Periodontol. 2001;72:368–373. doi: 10.1902/jop.2001.72.3.368. [DOI] [PubMed] [Google Scholar]
- 35.Hanookai D, Nowzari H, Contreras A, Morrison JL, Slots J. Herpesviruses and periodontopathic bacteria in Trisomy 21 periodontitis. J Periodontol. 2000;71:376–384. doi: 10.1902/jop.2000.71.3.376. [DOI] [PubMed] [Google Scholar]
- 36.Komatsu T, Kubota E, Sakai N. Enhancement of matrix metalloproteinase (MMP)-2 activity in gingival tissue and cultured fibroblasts from Down's syndrome patients. Oral Dis. 2001;7:47–55. [PubMed] [Google Scholar]
- 37.Komatsu T, Lee MC, Miyagi A, et al. Reactive oxygen species generation in gingival fibroblasts of Down syndrome patients detected by electron spin resonance spectroscopy. Redox Rep. 2006;11:71–77. doi: 10.1179/135100006X101039. [DOI] [PubMed] [Google Scholar]
- 38.Zaldivar-Chiapa RM, Arce-Mendoza AY, De La Rosa-Ramirez M, Caffesse RG, Solis-Soto JM. Evaluation of surgical and non-surgical periodontal therapies, and immunological status, of young Down's syndrome patients. J Periodontol. 2005;76:1061–1065. doi: 10.1902/jop.2005.76.7.1061. [DOI] [PubMed] [Google Scholar]
- 39.Khocht A, Heaney K, Janal M, Turner B. Association of interleukin-1 polymorphisms with periodontitis in Down syndrome. J Oral Sci. 2011;53:193–202. doi: 10.2334/josnusd.53.193. [DOI] [PubMed] [Google Scholar]
- 40.Grigoriadou ME, Koutayas S-O, Madianos PN, Strub J-R. Interleukin-1 as a genetic marker for periodontitis: review of the literature. Quintessence Int. 2010;41:517–525. [PubMed] [Google Scholar]


