Abstract
Expansion of trinucleotide repeat (TNR) sequences in human DNA is considered to be a key factor in the pathogenesis of more than 40 neurodegenerative diseases. TNR expansion occurs during DNA replication and also, as suggested by recent studies, during the repair of DNA lesions produced by oxidative stress. In particular, the oxidized guanine base, 8-oxoG, within sequences containing CAG repeats may induce formation of pro-expansion intermediates through strand slippage during DNA base excision repair (BER). In this article, we describe how oxidized DNA lesions are repaired by BER and discuss the importance of the coordinated activities of the key repair enzymes, such as DNA polymerase β, flap endonuclease 1 (FEN1) and DNA ligase, in preventing strand slippage and TNR expansion.
Trinucleotide repeat expansion and DNA metabolism
Trinucleotide repeat (TNR) expansions are associated with more than 40 neurodegenerative diseases; for example, the genomes of patients suffering from Huntington’s disease or myotonic dystrophy often have expansions of CAG or CTG repeats [1–3]. TNR expansion occurs in germ cells, as well as in non-dividing and dividing somatic cells [2, 4, 5]. Somatic TNR expansion is particularly significant in brain striatal neurons and exhibits an aging dependency in mouse models and humans [4, 5], and it has been suggested that somatic CAG repeat expansion plays a role in the onset and progression of Huntington’s disease [5]. TNR expansion was initially identified as an intergenerational event occurring during DNA replication. However, later studies using bacteria, yeast and mice as model systems indicated that TNR expansion occurs not only during DNA replication [6–8] but also during DNA repair [9–13], transcription [14, 15] or recombination [16, 17]. Although it has been proposed that formation of non-B form DNA structures by TNRs is required for TNR expansion [18, 19], the molecular mechanisms of expansion remain poorly understood.
Several DNA repair mechanisms, such as mismatch repair [10] and transcription-coupled repair (also known as nucleotide excision repair) [2, 20], have been proposed to be involved in TNR somatic expansion. For example, in mice, the mismatch repair protein complex MSH2-MSH3 is essential for CAG repeat expansion in both germ cells [10] and somatic cells [12]. In addition, the MSH2-MSH3 complex binds to and stabilizes TNR hairpin structures [21], thereby suggesting a mechanism of promoting TNR expansion. However, age-dependent somatic TNR expansion also appears to be mediated through an alternate DNA repair pathway. Since aging is considered to be associated with increased oxidative stress [22, 23], it is likely that age-dependent neuronal CAG repeat expansion involves oxidative DNA damage and repair.
Oxidative base damage is a type of genomic DNA lesion that commonly results from the action of reactive oxygen species (ROS; see Glossary). ROS can be endogenously generated either by normal cellular metabolism (through electron transport “leaks” from the inner membrane of mitochondria), or during inflammatory processes. In addition, exogenously-generated ROS are induced by environmental oxidative stressors such as bromate, chromate, arsenate, sunlight and ionizing radiation [24]. In the human genome, oxidative DNA base lesions can occur in gene coding, regulatory and other regions. These lesions, if not repaired, can lead to mutations, abnormal gene transcription and epigenetic instability, which ultimately can cause adverse effects [24]. To combat the adverse effects of oxidative DNA damage, mammalian cells maintain robust DNA repair mechanisms that remove oxidative lesions. In particular, the DNA repair system known as base excision repair (BER) plays a role in repairing various types of oxidative lesions, among which 8-oxoguanine (8-oxoG) is a frequently produced form in mammals. In this review, we focus on the repair of 8-oxoG lesions within TNR sequences via the BER pathway. We discuss molecular mechanisms of this repair pathway and its linkage with trinucleotide repeat expansion, based upon recent results.
Repair of 8-oxoG and trinucleotide repeat expansion
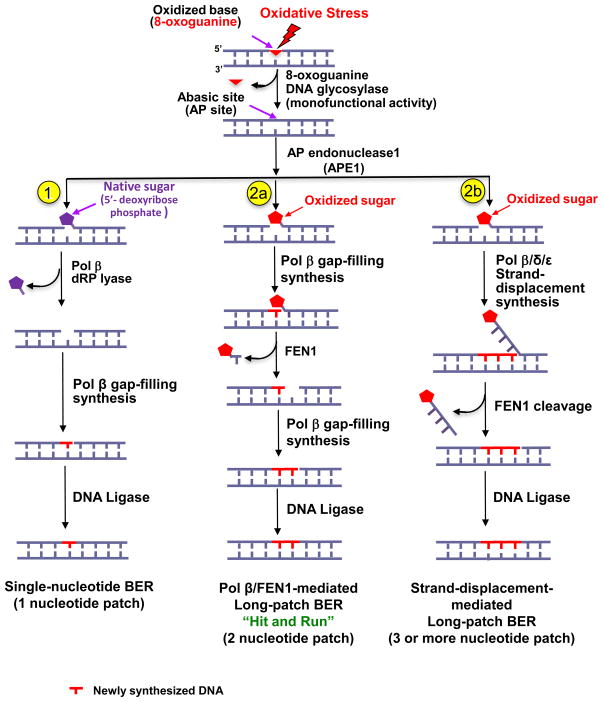
The repair of 8-oxoG lesions via the BER pathway is initiated by a damage-specific DNA glycosylase, i.e., 8-oxoguanine DNA glycosylase (OGG1), that removes the 8-oxoguanine lesion (Figure 1). OGG1 is a bifunctional enzyme (DNA glycosylase and AP lyase) that releases the damaged base and may incise the resulting AP site using its AP lyase activity, leaving a single-strand DNA break intermediate with a 3′-end that blocks DNA polymerase synthesis [25–27]. However, the OGG1 AP lyase activity is about 500-fold less efficient than the glycosylase activity [28], which suggests that OGG1 often acts as a monofunctional glycosylase to create an intact AP site-containing repair intermediate. Cleavage on this AP site by AP endonuclease 1 (APE1) leaves a single-stranded break intermediate with 3′-hydroxyl and 5′-deoxyribose phosphate (dRP) groups at the margins of a single-nucleotide gap [29] (Figure 1, sub-pathway 1). A 5′-dRP-containing intermediate with a native (non-oxidized) 5′-sugar phosphate group is repaired by the single-nucleotide BER (SN-BER) sub-pathway. SN-BER is mediated by DNA polymerase β (pol β), which has 5′-dRP lyase and polymerase gap-filling activities (Figure 1, sub-pathway 1) [30]. By contrast, an oxidized AP site or an oxidized 5′-sugar phosphate group cannot be processed through SN-BER because the 5′-sugar phosphate group cannot be removed by pol β. This intermediate is repaired through the long-patch BER (LP-BER) sub-pathway (Figure 1, sub-pathway 2a or 2b) [31]. In the most efficient form of LP-BER, pol β cooperates with flap endonuclease 1 (FEN1) to remove the oxidized sugar through coordinated single-nucleotide gap-filling synthesis and excision of a one-nucleotide flap along with the oxidized sugar. This LP-BER process, mediated by pol β and FEN1 and termed the “hit and run” mechanism [32] (Figure 1), consists of stepwise and distributive gap-filling by pol β and single-nucleotide gap formation by FEN1. This process yields repair patches of two nucleotides or more. Thus, this LP-BER mechanism is mediated by functional coordination between pol β and FEN1 and does not generate a long 5′-flap created by strand-displacement DNA synthesis. The notion of pol β-FEN1 functional coordination is consistent with structural analyses of FEN1 binding to a pol β-DNA complex, which suggest that FEN1 dislodges pol β and binds to a downstream 5′-flap along with an upstream one-nucleotide 3′-flap, within the polymerase and DNA complex [33]. However, in some cases, a presumably less efficient LP-BER process may be accomplished through strand-displacement DNA synthesis (see Glossary) by pol β or by replicative DNA polymerases pol δ or pol ε, which would create a long 5′-flap with several nucleotides [31] (Figure 1, sub-pathway 2b). The strand-displacement flap is excised by FEN1 and the resulting nick is sealed by DNA ligase.
Figure 1. Base excision repair (BER) of oxidized DNA base lesions.
Oxidative stress may result in oxidized DNA base lesions, i.e., 8-oxoG and/or oxidized abasic site. The DNA glycosylase OGG1 removes 8-oxoG leaving an abasic site (AP site). Then, APE1 incises at the 5′-side of the AP site sugar, leaving either a native (non-oxidized) 5′-sugar phosphate or an oxidized 5′-sugar phosphate. A native 5′-sugar phosphate group can be repaired by the single-nucleotide BER (SN-BER) sub-pathway (1), whereas an oxidized sugar phosphate group can be repaired by the long-patch BER (LP-BER) sub-pathway (2a and 2b). (1) Incision of a native AP site by APE1 results in formation of an intermediate containing a single-nucleotide gap, and the pol β dRP lyase activity removes this native 5′-sugar phosphate. Subsequently, pol β gap-filling synthesis fills the gap, leaving a nicked DNA intermediate for ligation. In this scenario, the pol β-mediated repair involves one-nucleotide replacement only (i.e., SN-BER). (2a) For the scenario where a 5′-sugar phosphate group is oxidized and resistant to the pol β dRP lyase activity, removal of this sugar phosphate will occur by the “hit and run” mechanism of LP-BER, which is mediated by pol β gap-filling synthesis and FEN1 flap excision of the nucleotide linked to the 5′-sugar phosphate group. This is an efficient LP-BER process that generally involves the replacement of only two nucleotides. (2b) Removal of the oxidized sugar phosphate also can occur by an alternate LP-BER sub-pathway that is mediated by DNA strand-displacement synthesis by pol β or pol δ/ε, followed by FEN1 flap cleavage. This LP-BER process involves the replacement of three or more nucleotides.
FEN1 may cooperate with pol β through partnerships with additional BER cofactors, such as proliferating cell nuclear antigen (PCNA). PCNA can interact directly with both FEN1 [34] and pol β [35]. PCNA can also stimulate FEN1 cleavage activity on a 5′-flap [34], thereby facilitating FEN1 function in processing Okazaki fragments during DNA lagging strand synthesis and in removing an oxidized 5′-sugar group during LP-BER. Functional coordination between FEN1 and pol β in LP-BER is stimulated by poly(ADP)ribose polymerase-1 (PARP-1). In the presence of FEN1, PARP-1 increases pol β synthesis activity, and the effect is dependent on FEN1 cleavage activity. This suggests that PARP-1 may accelerate the “hit and run” mechanism by promoting FEN removal of a damaged sugar along with one nucleotide. Furthermore, FEN1-pol β functional coordination can be stimulated by the high mobility group box 1 (HMGB1) protein, in that HMGB1 increases pol β synthesis and FEN1 incision [36]. Although the detailed molecular mechanisms by which PARP-1 and HMGB1 stimulate FEN1 cleavage activity remains to be elucidated, protein-protein interaction and functional coordination between FEN1 and pol β, as well as other BER cofactors, seems to be crucial for efficient LP-BER.
Results from several studies support a linkage between age-dependent somatic CAG repeat expansion and oxidative DNA damage [37, 38]. BER may be directly involved in somatic TNR expansion, because knockout of the Ogg1 gene abolished age-dependent neuronal CAG repeat expansion in a Huntington’s disease model in mice [4]. In addition, pol β, FEN1 and HMGB1 facilitate CAG repeat expansion in vitro during LP-BER of oxidative DNA lesions. This TNR expansion involves DNA lesion-containing strand slippage, hairpin formation and disruption of pol β-FEN1 functional coordination [39]. Although the in vitro BER of the oxidative base lesion results in only 1 to 5 repeat unit expansions with one round of repair, this could be sufficient to generate single-strand DNA breaks for multiple rounds of BER and eventually cause large TNR expansions, because thousands of oxidative DNA base lesions are estimated to occur in a cell every day [40]. Thus, multiple oxidative DNA base lesions and repair events could lead to a “toxic oxidation cycle” [2]. Furthermore, 8-oxoG lesions are unevenly distributed in the genome and concentrate at certain gene promoters [41], which indicates that oxidative base lesions can cluster and form lesion hotspots in the genome. Such clustering of lesions could trigger multiple rounds of localized damage, repair and expansion.
Dual functions of FEN1 in modulating TNR stability
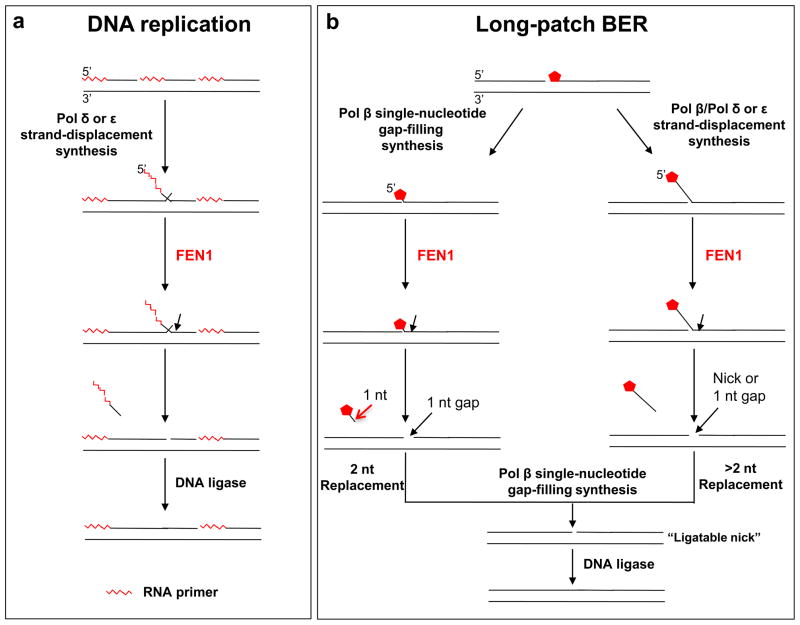
FEN1 can remove RNA primers in Okazaki fragment maturation during lagging strand DNA replication [42], and is required for removing damaged sugar residues during LP-BER [43]. In addition, FEN1 is crucial for maintaining genome stability by removing flap structures during DNA replication, because a deficiency in FEN1 function results in increased TNR sequence variation [44]. This role has been emphasized in studies showing that FEN1 is involved in preventing TNR expansion [45–49]. During DNA lagging-strand synthesis (Figure 2a), FEN1 binds its preferred substrate, a “double-flap” intermediate that has a 3′-one-nucleotide flap and a longer 5′-flap. FEN1 then removes the 5′-flap, along with the RNA primer, that was generated by pol δ or pol ε strand-displacement synthesis. This results in a nicked DNA suitable for ligation of the Okazaki fragments [42, 50, 51]. During LP-BER (Figure 2b), FEN1 coordinates with pol β to remove an oxidized sugar using the “hit and run” mechanism [32]. This results in efficient LP-BER with a two-nucleotide repair patch (Figure 2b). Alternatively, FEN1 can cleave a multi-nucleotide flap generated by strand-displacement gap-filling DNA synthesis. This leads to a presumably less efficient LP-BER process and multi-nucleotide repair patch (Figure 2b). Should FEN1 cleavage result in a one nucleotide gap instead of a ligatable nick, the gap can be readily filled by pol β.
Figure 2. The multifaceted functions of FEN1 in DNA replication and long-patch BER.
(a) FEN1 plays a crucial role in Okazaki fragment maturation during DNA lagging strand synthesis by removing flaps that contain the RNA primer (
 ) of Okazaki fragments. The flaps are created by pol δ or pol ε strand-displacement synthesis. FEN1 usually captures a dual flap intermediate with a one-nucleotide 3′-flap along with a 5′-flap and removes the 5′-flap, leaving a nicked DNA that is a substrate for ligation, a “ligatable nick.” (b) During long-patch BER, FEN1 removes a modified (reduced or oxidized) sugar (
) of Okazaki fragments. The flaps are created by pol δ or pol ε strand-displacement synthesis. FEN1 usually captures a dual flap intermediate with a one-nucleotide 3′-flap along with a 5′-flap and removes the 5′-flap, leaving a nicked DNA that is a substrate for ligation, a “ligatable nick.” (b) During long-patch BER, FEN1 removes a modified (reduced or oxidized) sugar (
 ) by either coordinating with pol β through the “Hit and Run” mechanism (the sub-pathway on the left) or capturing a 5′-flap associated with a modified sugar (the sub-pathway on the right) created by pol β or pol δ/ε. This latter process results in multi-nucleotide replacement. In the case where FEN1 cleavage results in a one-nucleotide gap, pol β will fill the gap readily leaving a ligatable nick.
) by either coordinating with pol β through the “Hit and Run” mechanism (the sub-pathway on the left) or capturing a 5′-flap associated with a modified sugar (the sub-pathway on the right) created by pol β or pol δ/ε. This latter process results in multi-nucleotide replacement. In the case where FEN1 cleavage results in a one-nucleotide gap, pol β will fill the gap readily leaving a ligatable nick.
FEN1 and its homologues are well known for their function in preventing repeat sequence expansion in bacteria [52, 53], yeast [54–57] and mice [48]. In yeast, deletion of the FEN1-encoding gene (RAD27) increased the frequency of expansion of certain TNR repeats, consisting of 40 and 70 CTG units, by about 30-fold and 60-fold, respectively [56]. In addition, CTG and CGG repeat expansions exhibited a DNA replication orientation-dependency in RAD27-null yeast strains [57, 58], with the expansion occurring in a newly synthesized lagging strand [57, 58]. The prevention of TNR expansions by FEN1 is significant, because one copy of FEN1 (haploinsufficiency) is sufficient for maintaining the stability of CAG or CTG repeats containing 110 to 130 repeat units in both yeast and mammals [48, 59, 60]. It has been proposed that different FEN1 concentrations are required for maintaining TNR stability in different cell types, with a high concentration required for removing long TNRs in rapidly dividing cells and germ cells, and low concentrations required for removing short TNRs in cells that do not divide rapidly [59]. In a Huntington’s disease mouse model, low expression of FEN1 is associated with an increase in somatic CAG repeat expansion in the striatum [61]; this suggests that weaker FEN1 cleavage activity results in accumulation of CAG repeat hairpins in this tissue, thereby leading to CAG repeat expansion.
Results from several studies involving Drosophila, animal models, human cells, and human populations appear to contradict a role of FEN1 in preventing TNR expansion. However, we argue that these results may still be consistent with a role of FEN1 in preventing TNR expansion. For example, haploinsufficiency of FEN1 did not affect the stability of (CTG)84 repeats in a myotonic dystrophy mouse model [62]. Similarly, (CGG)130 repeats were unaffected by FEN1 haploinsufficency in a fragile × syndrome mouse model [63]. We suggest that the data from these mouse models could indicate that the (CTG)84 and (CGG)130 TNR sequences are associated with stable hairpin or tetraplex structures that block FEN1 cleavage activity. Such inhibitory effects have been observed for CTG hairpins and CGG tetraplexes in biochemical studies [45, 49, 64], with the CTG repeat having a stronger inhibitory effect on FEN1 incision than the CGG repeat [64]. Moreover, different trinucleotide repeats can adopt distinct secondary structures that may result in different consequences for the roles of FEN1 in modulating repeat stability [64]. Other factors that could affect FEN1 are the genomic location of the CAG repeat-containing gene and its transcriptional activity. By contrast, the Huntington’s disease mouse model carrying shorter CAG repeats may have an advantage in studying FEN1 function, as the shorter CAG repeat is considered to form relatively unstable hairpins. Thus, it is possible that FEN1 maintains TNR stability in vivo by removing unstable – but not stable – secondary structures.
The fact that a human cell line with a 10-fold FEN1 knockdown can still maintain the stability of short CAG repeats (13–27 repeat units) in the huntingtin gene locus [65] may indicate that a residual level of FEN1 expression is sufficient to remove unstable hairpin structures formed by these repeats. FEN1 mutations and polymorphic variants were not identified in a study of Huntington’s disease parent-child pairs with ten or more CAG repeat units of expansion in the huntingtin gene locus [66]. However, this population study involved only 15 parent-child pairs, and future studies with larger sample sizes and better characterized FEN1 structure-function deficiencies will be needed to exclude a FEN1 role in preventing CAG repeat expansion. Finally, the fact that (CAG)90 in the Drosophila model of cerebellar ataxia 7 (SCA7) did not exhibit instability under deficiency of FEN1, PCNA and MutS, suggests (in our opinion) that Drosophila may not be an appropriate model for studying the molecular mechanisms of CAG repeat instability in mammalian systems [67].
Mechanism of FEN1 incision in modulating TNR stability
The role played by FEN1 in maintaining TNR stability may be a consequence of its ability to cleave DNA flap structures containing TNRs. However, the efficiency of FEN1 cleavage on TNR sequences is determined by multiple factors, such as repeat length, the dynamics of alternate secondary structures within repeat flaps [45–47, 49], as well as protein-protein interactions and functional coordination between FEN1 and other replication and repair proteins [50]. During DNA replication, long TRNs induce the formation of stable hairpin structures within a flap, which can inhibit FEN1 loading and cleavage of the structured flap [45, 49]. By contrast, short TNRs can be cleaved by FEN1 [50] because they form flaps with unstable hairpin structures on which FEN1 can load by capturing the 5′-end of a transiently formed single-strand molecule. The loading occurs before the length of the 5′-flap is significantly increased by DNA polymerase strand-displacement synthesis; at this point, FEN1 could track along the 5′-repeat flap to the base of a double-flap substrate (now with a one-nucleotide 3′-flap and a 5′-flap) and excise it [50] (Figure 3a). A structural study of FEN1 has highlighted the crucial role of the double-flap structure in facilitating the most efficient FEN1 flap cleavage activity [33]. Thus, in DNA replication, FEN1 can make use of transiently formed short repeat flaps created by DNA strand-displacement synthesis and circumvent the inhibitory effects of the longer stable hairpin or tetraplex structured TNR flaps.
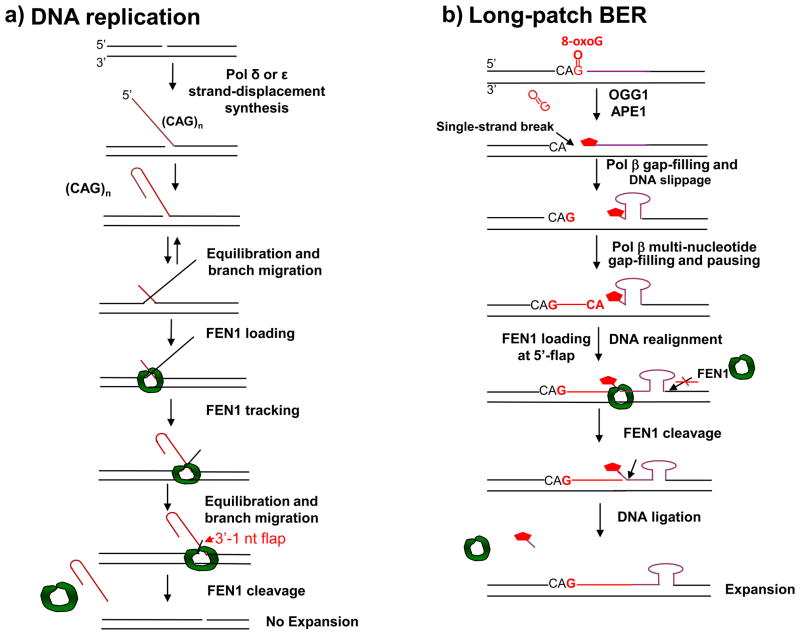
Figure 3. Dual functions of FEN1 in regulating trinucleotide repeat stability.
FEN1 modulates trinucleotide repeat stability in different ways during DNA replication and long-patch BER. During DNA replication (a), pol δ/ε strand-displacement DNA synthesis creates a repeat containing 5′-flap that subsequently folds back into a hairpin. The repeat-containing hairpin competes with upstream repeats to anneal to the template strand resulting in a dynamic equilibration between the repeat-containing hairpin configuration and a double-flap configuration with a long 3′-flap and a short 5′-flap. FEN1 subsequently captures the 5′-short flap, loads from the 5′-end of the flap and tracks down through the flap along with DNA branch migration until FEN1 captures an intermediate with a 1 nt-3′-flap. FEN1 then cleaves the repeat-containing 5′-flap or hairpin, and expansion does not occur. However, during BER of a base lesion such as 8-oxoG (b), single-strand DNA breaks in the context of trinucleotide repeats (induced by base removal and APE1 5′-incision of the AP site) often results in DNA slippage. This is especially the case when the 5′-sugar phosphate group (
 ) cannot be removed by pol β’s dRP lyase. Strand slippage results in formation of intermediates with multi-nucleotide gaps and repeat-containing hairpins. Pol β conducts gap-filling synthesis to fill the gaps, but terminates synthesis at the base of the hairpin. Formation of a stable hairpin structure inhibits conventional FEN1 cleavage activity for removing the entire length of the hairpin. Instead, FEN1 “alternate cleavage” of a short flap generated at the 5′-end of the hairpin by DNA realignment is allowed, resulting in removal of the flap with the 5′-end dRP group, a ligatable nick, ligation and repeat expansion.
) cannot be removed by pol β’s dRP lyase. Strand slippage results in formation of intermediates with multi-nucleotide gaps and repeat-containing hairpins. Pol β conducts gap-filling synthesis to fill the gaps, but terminates synthesis at the base of the hairpin. Formation of a stable hairpin structure inhibits conventional FEN1 cleavage activity for removing the entire length of the hairpin. Instead, FEN1 “alternate cleavage” of a short flap generated at the 5′-end of the hairpin by DNA realignment is allowed, resulting in removal of the flap with the 5′-end dRP group, a ligatable nick, ligation and repeat expansion.
During BER, however, the scenario is quite different from that in DNA replication (Figure 3b). During repair of oxidative base damage in the sequence context of CAG repeats, CAG repeat hairpin structures can be formed immediately after a single-strand DNA break is created by OGG1 and APE1 [4, 39]. This leads to hairpin-structured CAG repeat-containing flaps along with multi-nucleotide gaps that are filled in by pol β (Figure 3b). The hairpin-containing flap inhibits “conventional” FEN1 cleavage at the base of the structured flap, and an “alternate cleavage” of a modified sugar at the 5′-end of the hairpin-structured flap can be carried out by FEN1 instead [39] [64]. This cleavage leads to production of a “ligatable nicked” intermediate (as in Figure 2b), facilitating ligation of the CAG repeat-containing hairpin into genomic DNA [39].
In some cases, after a TNR-containing hairpin is ligated into genomic DNA, the hairpin can become a hotspot of oxidative DNA damage. For example, a guanine on the hairpin loop region of a CAG repeat structure is highly susceptible to oxidative DNA damage [68] [69]. Such damage may result in an oxidative base lesion in a TNR hairpin loop or its stem region that may be resistant to BER [69], because these structures can inhibit repair activity. This could result in “a cycle of toxic oxidation” [69], in which multiple rounds of oxidative stress cause DNA damage at various positions in TNR sequences, leading to cycles of TNR expansion, some of which may occur with different efficiencies. However, the BER mechanisms for repair of oxidative DNA damage on hairpins, and their impact on TNR expansion, remain poorly understood.
FEN1-pol β coordination in modulating TNR stability during LP-BER
Functional coordination between FEN1 and pol β plays a role in determining whether CAG repeats are expanded or not during LP-BER [39]. During repair of an 8-oxoG lesion, pol β conducts single-nucleotide gap-filling synthesis in the context of a non-repetitive DNA sequence, whereas it mainly performs multi-nucleotide gap-filling synthesis in the context of CAG repeats [39]. Thus, multi-nucleotide gaps are produced and filled by pol β in the context of CAG repeats but not within a non-repetitive sequence (Figure 4). The pol β multi-nucleotide gap-filling synthesis on CAG repeats exhibits periodic pausing sites, consistent with formation of CAG repeat hairpins with various repeat lengths [39]. These results suggest that single-strand break intermediates resulting from removal of 8-oxoG and incision of abasic sites allow CAG repeat-containing strands to disassociate from the template and form hairpin structures (Figure 4). This strand slippage may subsequently allows pol β multi-nucleotide gap-filling synthesis, and after ligation, newly synthesized extra copies of the CAG repeat units may be sealed into the repaired strand. Because the most efficient LP-BER process is achieved by pol β-FEN1 functional coordination in the “hit and run” mechanism (involving excision and replacement of only two nucleotides), formation of multi-nucleotide gaps and CAG repeat hairpins probably result from disruption of pol β-FEN1 functional coordination. The size of the TNR expansion accompanying this disruption of coordination might be determined by the size of the multi-nucleotide gaps and the stability of hairpin structures.
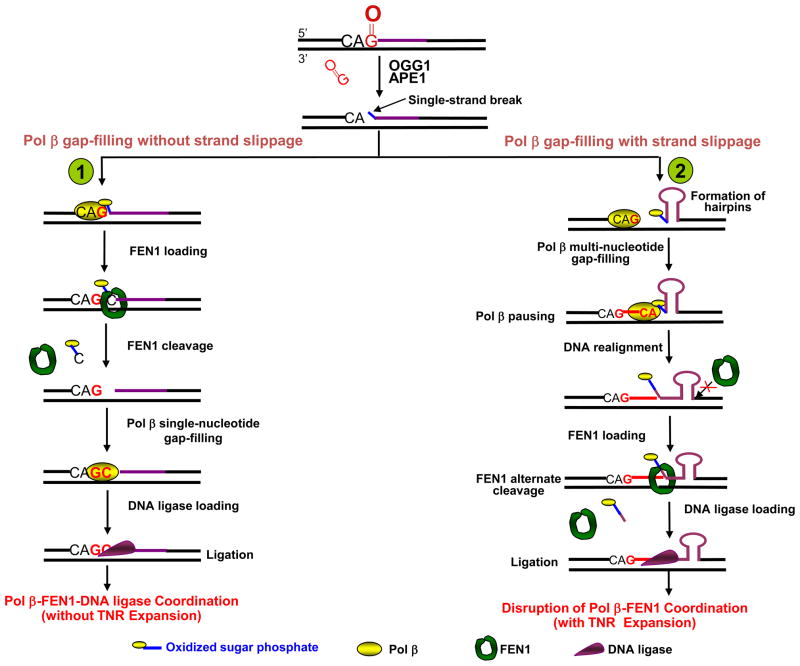
Figure 4. Hypothetical models illustrating CAG repeat stability modulated by coordination among BER enzymes during BER of 8-oxoG.
The scheme illustrates the important role of coordinated handoff of single-stranded BER intermediates between pol β and FEN1 in regulating the stability of CAG repeats during BER of 8-oxoG in the context of CAG repeats (shown in purple). (Pathway 1) The single-stranded repair intermediates are handed off effectively between pol β and FEN1. After pol β gap-filling with dGMP insertion and FEN1 removal of the sugar phosphate (
 ) plus 1 nucleotide, pol β then conducts gap-filling (dCMP insertion) and a nicked DNA intermediate is produced. This process occurs without strand slippage that could result in hairpin structures. The nicked intermediate is subsequently sealed by DNA ligase, and repeat expansion will not occur.
) plus 1 nucleotide, pol β then conducts gap-filling (dCMP insertion) and a nicked DNA intermediate is produced. This process occurs without strand slippage that could result in hairpin structures. The nicked intermediate is subsequently sealed by DNA ligase, and repeat expansion will not occur.
(Pathway 2) If pol β and FEN1 coordination is disrupted by spontaneous strand slippage, CAG repeat hairpin structures will form, the multi-nucleotide gap will be filled by pol β, and pol β terminates synthesis at the base of the hairpin. FEN1 flap cleavage activity also will be inhibited. Thus, stabile hairpin structures inhibit FEN1 conventional cleavage at the 3′-base of the hairpin, as illustrated. Alternatively, FEN1 “alternate cleavage” of a short 5′-flap at the base of the hairpin will result in a ligatable nick, ligation and repeat expansion.
FEN1 can remove a 5′-modified sugar phosphate from a stable CAG repeat hairpin by partially trimming the 5′-end near the base of the hairpin [39]. This trimming leads to production of a “ligatable nick” that, after ligation, leads to TNR expansion (Figure 4, sub-pathway 2). However, filling of multi-nucleotide gaps by pol β will prevent re-annealing of the slipped CAG repeat hairpin strand to its original template strand, thereby stabilizing the hairpin structures [39]. Therefore, we propose that the gap-filled hairpin BER intermediate is ligated into the expansion product only if the base of the hairpin at the 5′-end is rearranged and annealed to the template strand, leaving a substrate for FEN1 and the potential of forming a “ligatable nick” [46] (Figure 4, sub-pathway 2). FEN1 would then remove a flap from the intermediate that is formed after the putative rearrangement and re-annealing. If a damaged sugar at the 5′-end of the flap is present, this will be removed by FEN1 facilitating ligation.
The efficient processing of the LP-BER intermediates by pol β, FEN1 and DNA ligase activities is expected to be the outcome in most cases, leading to expansion-free BER (Figure 4, sub-pathway 1). However, if the LP-BER process in the sequence context of CAG repeats escapes the coordinated action of these enzymes, spontaneous hairpin formation may occur (Figure 4, sub-pathway 2). This will result in both pol β multi-nucleotide gap-filling synthesis and inhibition of FEN1 flap cleavage. After the rearrangement discussed above and FEN1 alternate cleavage, a ligatable nick will be formed either by FEN1 alone or FEN1 and pol β gap-filling synthesis.
BER cofactors such as PCNA, HMGB1 and PARP-1 could affect TNR stability by modulating the coordination between pol β and FEN1 through protein-protein interactions. For example, HMGB1 stimulates FEN1 flap cleavage activity and moderately accelerates pol β strand-displacement synthesis [36]. Furthermore, the presence of HMGB1 increases CAG repeat expansion products formed during BER in vitro [39], which suggests that HMGB1 could facilitate FEN1 alternate cleavage during processing of hairpin structures, enabling ligation. HMGB1 can bind to hairpin structures of CAG repeats [70] and therefore could promote TNR expansion by stabilizing hairpin structures, as well as by modulating BER enzymatic activities. PARP-1 deficiency in mice leads to a smaller size of duplication of a palindromic sequence when the animals are exposed to alkylating DNA damaging agents [71]. Because PARP-1 is a nick sensing protein, duplication or expansion of repeat sequences could result from inefficient repair of single-stranded DNA breakage intermediates, leading to their formation and accumulation as hairpin structures.
In addition, FEN1 may be involved in removing hairpin structures by cooperating with an endonuclease that can specifically incise the middle of a TNR hairpin loop [72] in a proposed incision-dependent repair of the TNR hairpin [73]. This mechanism calls for involvement of mismatch repair proteins in the repair. Indeed, the MSH2-MSH3 complex is essential for CAG repeat expansion in an animal model [10], and the purified MSH2-MSH3 complex can bind to the TNR hairpin loop structure, suggesting that the protein complex can stabilize hairpin structures [21]. The stabilization of hairpin structures during DNA replication and repair by MSH2-MSH3 could prevent removal of hairpins, leading to TNR expansion [74]. However, the effect of the MSH2-MSH3 complex on TNR expansion during BER remains to be further elucidated.
Hypothetical formation of a ligatable nick by hairpin realignment during LP-BER
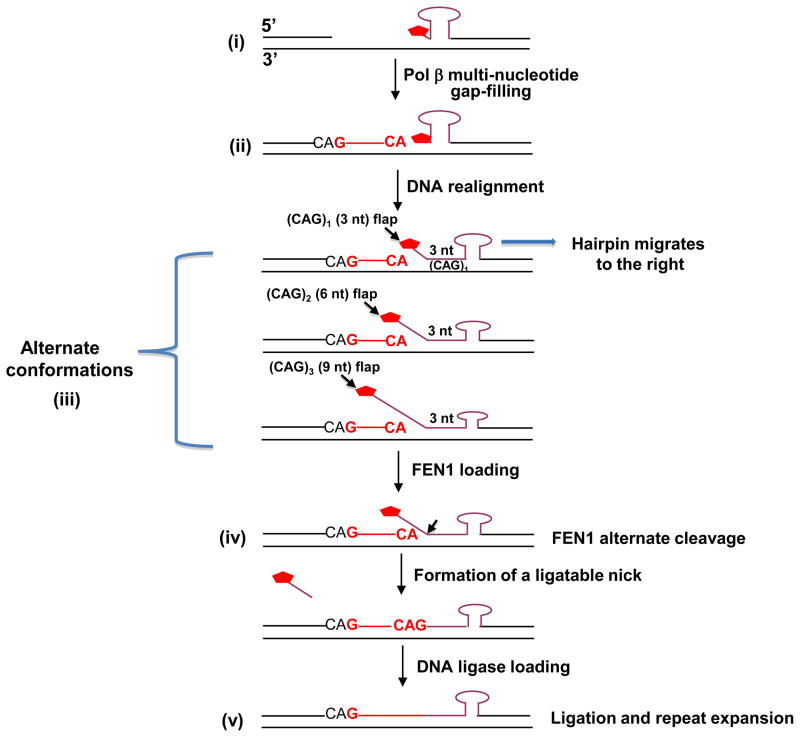
Results from biochemical experiments have indicated that FEN1 cleavage along with pol β synthesis could produce ligatable nicks, even with short 5′-flaps [39]. To inform design of future experiments in this area, we present a hypothetical model (Figure 5), which we consider the simplest among many possibilities. According to this model, after pol β collides with the base of the hairpin stem and pauses or terminates synthesis, intermediate structures containing various sizes of hairpins and multi-nucleotide gaps (Figure 5) are formed. We assume that the newly formed strand remains in position, annealed to the template strand. At this stage, FEN1 cannot use the hairpin as substrate. We propose that the hairpin then rearranges (as indicated by an arrow Figure 5), leaving a mixture of hairpin structural isoforms. These isoforms would have at least one CAG triplet annealed at the 5′-side of the hairpin base along with multiple 5′-end flaps of various sizes (i.e., alternate conformations). After FEN1 cleavage at a point corresponding to one nucleotide into the flap junctions, ligatable nicks would be formed directly or after pol β single-nucleotide gap-filling synthesis. It is noteworthy that the expansion by this mechanism would occur in multiples of three nucleotides.
Figure 5. Hypothetical model for formation of a ligatable nick by hairpin realignment accompanying LP-BER.
In the context of CAG repeats, the single-stranded BER intermediates with a 5′-sugar phosphate (
 ) may undergo spontaneous DNA slippage resulting in formation of multi-nucleotide gaps and a CAG repeat-containing hairpin with 5′-sugar phosphate (i). Pol β performs multi-nucleotide gap-filling synthesis to produce a newly synthesized CAG repeat strand, eventually colliding with the 5′-base of the hairpin. Pol β pauses at the base of the hairpin and dissociates from this product (ii). The CAG repeat hairpin undergoes spontaneous DNA realignment. This leads to formation of intermediates with alternate conformations (iii) of an annealed 5′-region consisting of 3 nucleotides (one CAG repeat) and various lengths of 5′-repeat flap, consisting of 3 nt [(CAG)1], 6 nt [(CAG)2], 9 nt [(CAG)3], etc. FEN1 loading and alternate cleavage (iv) then removes the 5′-CAG repeat-containing flaps, creating the ligatable nick for ligation, resulting in TNR expansion (v). During the spontaneous DNA realignment and formation of alternate conformations proposed, the hairpin migrates to the right, as illustrated by the arrow.
) may undergo spontaneous DNA slippage resulting in formation of multi-nucleotide gaps and a CAG repeat-containing hairpin with 5′-sugar phosphate (i). Pol β performs multi-nucleotide gap-filling synthesis to produce a newly synthesized CAG repeat strand, eventually colliding with the 5′-base of the hairpin. Pol β pauses at the base of the hairpin and dissociates from this product (ii). The CAG repeat hairpin undergoes spontaneous DNA realignment. This leads to formation of intermediates with alternate conformations (iii) of an annealed 5′-region consisting of 3 nucleotides (one CAG repeat) and various lengths of 5′-repeat flap, consisting of 3 nt [(CAG)1], 6 nt [(CAG)2], 9 nt [(CAG)3], etc. FEN1 loading and alternate cleavage (iv) then removes the 5′-CAG repeat-containing flaps, creating the ligatable nick for ligation, resulting in TNR expansion (v). During the spontaneous DNA realignment and formation of alternate conformations proposed, the hairpin migrates to the right, as illustrated by the arrow.
Concluding remarks and future directions
Pol β has poor strand-displacement synthesis activity and strong single-nucleotide gap-filling activity, and therefore LP-BER can take place by successive single-nucleotide gap-filling steps by pol β and FEN1 coordination [32]. As a result, TNR expansion usually does not occur. However, a long 5′-flap could be produced if FEN1 fails to process the shorter flap or if a replicative polymerase along with PCNA conducts processive gap-filling synthesis in the absence of pol β. This second scenario could lead to TNR expansion in the following way. Because pol β can promote CAG repeat expansion in vitro [39], we propose a hypothetical model that emphasizes the importance of coordination between pol β, FEN1 and DNA ligase in maintaining TNR stability, specifically during LP-BER. If an oxidized AP site or an oxidized sugar phosphate is subjected to the most efficient LP-BER sub-pathway (mediated by coordination between pol β, FEN1 and DNA ligase), the repair product will be a two-nucleotide replacement, DNA slippage in the damaged strand will not occur, and TNR stability is maintained (Figure 4, sub-pathway 1). By contrast, if the lesion-containing strand escapes efficient BER, it will be subjected to a less efficient LP-BER sub-pathway, involving DNA strand slippage of the TNR strand and resulting in formation of hairpin structures and multi-nucleotide gaps. DNA synthesis to fill the multi-nucleotide gaps and ligation of the hairpin structures would lead to TNR expansion (Figure 4, sub-pathway 2, and Figure 5) [39].
This proposed series of repair events involving elaborate coordination of BER enzymes and cofactors, either preventing or promoting CAG repeat expansion, warrants further testing in simple in vitro systems with purified BER proteins, including testing in the context of nucleosomes. Knowledge of the coordination between BER enzymes also will be necessary in understanding the processing of the oxidized bases by bifunctional DNA glycosylases. These enzymes leave blocked 3′-margins in gaps, and specialized enzymes (not considered here) are involved in the repair [75]. The possible connections between these additional BER enzymes and TNR expansion should be interesting to consider.
While our current knowledge of molecular mechanisms of TNR expansion during BER is sufficient to proposed working models, many questions remain to be addressed (Box 1). It is anticipated that new research into these questions and a deeper understanding of the molecular mechanisms of TNR expansion may lead to new strategies for prevention and treatment of TNR-related human neurodegenerative diseases.
Box 1. Future research questions.
How do BER enzymes and cofactors cooperate to process hairpin structures and manipulate their stability?
How do differences in BER enzyme and cofactor concentration alter ligation or removal of hairpins for TNR expansion or stabilization?
How does BER cooperate with other DNA repair pathways, such as mismatch repair, to modulate TNR expansion?
How may BER interact with DNA replication machinery to prevent TNR expansion when oxidative DNA damage is encountered during DNA replication?
Acknowledgments
The authors thank Bonnie Mesmer for editorial assistance. The authors also thank Dr. Lawrence Loeb and students at the University of Washington for discussion. This work was supported by National Institutes of Health Projects Z01-ES050158 and Z01-ES050159 from the NIEHS Intramural Research Program and National Institutes of Health grant ES017476 (to Y.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paulson HL, Fischbeck KH. Trinucleotide repeats in neurogenetic disorders. Annu Rev Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- 2.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson CE, et al. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 4.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy L, et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 6.Wells RD. Molecular basis of genetic instability of triplet repeats. J Biol Chem. 1996;271:2875–2878. doi: 10.1074/jbc.271.6.2875. [DOI] [PubMed] [Google Scholar]
- 7.Wilson SH, et al. Studies of DNA polymereases in replication-based repeat expansion. In: Wells RD, Warren ST, editors. Genetic Instabilities and Neurological Disease. 1. 1998. pp. 693–698. [Google Scholar]
- 8.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc Natl Acad Sci U S A. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair. 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 11.Pearson CE, et al. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 12.Manley K, et al. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. java/Propub/genetics/ng1299_1471.fulltext java/Propub/genetics/ng1299_1471.abstract. [DOI] [PubMed] [Google Scholar]
- 13.Parniewski P, et al. Nucleotide excision repair affects the stability of long transcribed (CTG*CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 1999;27:616–623. doi: 10.1093/nar/27.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, et al. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 15.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard GF, et al. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. Embo J. 2000;19:2381–2390. doi: 10.1093/emboj/19.10.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meservy JL, et al. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol Cell Biol. 2003;23:3152–3162. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J Biol Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 19.Wells RD, et al. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salinas-Rios V, et al. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011;39:7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen BA, et al. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 22.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 23.Cardozo-Pelaez F, et al. Oxidative DNA damage in the aging mouse brain. Movement Disorders. 1999;14:972–980. doi: 10.1002/1531-8257(199911)14:6<972::aid-mds1010>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Migliore L, Coppede F. Environmental-induced oxidative stress in neurodegenerative disorders and aging. Mutat Res. 2009;674:73–84. doi: 10.1016/j.mrgentox.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Radicella JP, et al. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCullough AK, et al. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 27.Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair. 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Krishnamurthy N, et al. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008;47:1043–1050. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 30.Prasad R, et al. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson SH, et al. Base excision repair and design of small molecule inhibitors of human DNA polymerase beta. Cell Mol Life Sci. 2010;67:3633–3647. doi: 10.1007/s00018-010-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, et al. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- 33.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tom S, et al. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J Biol Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 35.Kedar PS, et al. Direct interaction between mammalian DNA polymerase beta and proliferating cell nuclear antigen. J Biol Chem. 2002;277:31115–31123. doi: 10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- 36.Prasad R, et al. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell. 2007;27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy L, Shelbourne PF. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum Mol Genet. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- 38.Bogdanov MB, et al. Increased oxidative damage to DNA in a transgenic mouse model of Huntington’s disease. J Neurochem. 2001;79:1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, et al. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J Biol Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 42.Bambara RA, et al. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 43.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tishkoff DX, et al. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 45.Henricksen LA, et al. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J Biol Chem. 2000;275:16420–16427. doi: 10.1074/jbc.M909635199. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Bambara RA. Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J Biol Chem. 2003;278:13728–13739. doi: 10.1074/jbc.M212061200. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, et al. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol Cell Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spiro C, McMurray CT. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol Cell Biol. 2003;23:6063–6074. doi: 10.1128/MCB.23.17.6063-6074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spiro C, et al. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, et al. Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 51.Kao HI, et al. Cleavage Specificity of Saccharomyces cerevisiae Flap Endonuclease 1 Suggests a Double-Flap Structure as the Cellular Substrate. J Biol Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 52.Morel P, Reverdy C, Michel B, Ehrlich SD, Cassuto E. The role of SOS and flap processing in microsatellite instability in Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:10003–10008. doi: 10.1073/pnas.95.17.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagata Y, Mashimo K, Kawata M, Yamamoto K. The roles of Klenow processing and flap processing activities of DNA polymerase I in chromosome instability in Escherichia coli K12 strains. Genetics. 2002;160:13–23. doi: 10.1093/genetics/160.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson RE, et al. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 55.Kokoska RJ, et al. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freudenreich CH, et al. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 57.White PJ, et al. Stability of the human fragile X (CGG)(n) triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol Cell Biol. 1999;19:5675–5684. doi: 10.1128/mcb.19.8.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schweitzer JK, Livingston DM. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 59.Yang J, Freudenreich CH. Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene. 2007;393:110–115. doi: 10.1016/j.gene.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vande Berg BJ, et al. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase beta. Implication for the identity of the rate-limiting conformational change. J Biol Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 61.Goula AV, et al. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van den Broek WJ, et al. Fen1 does not control somatic hypermutability of the (CTG)(n)*(CAG)(n) repeat in a knock-in mouse model for DM1. FEBS Lett. 2006;580:5208–5214. doi: 10.1016/j.febslet.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 63.Entezam A, et al. Potassium bromate, a potent DNA oxidizing agent, exacerbates germline repeat expansion in a fragile × premutation mouse model. Hum Mutat. 2010;31:611–616. doi: 10.1002/humu.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vallur AC, Maizels N. Complementary roles for exonuclease 1 and Flap endonuclease 1 in maintenance of triplet repeats. J Biol Chem. 2010;285:28514–28519. doi: 10.1074/jbc.M110.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moe SE, et al. Huntingtin triplet-repeat locus is stable under long-term Fen1 knockdown in human cells. Journal of Neuroscience Methods. 2008;171:233–238. doi: 10.1016/j.jneumeth.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 66.Otto CJ, et al. The “flap” endonuclease gene FEN1 is excluded as a candidate gene implicated in the CAG repeat expansion underlying Huntington disease. Clin Genet. 2001;59:122–127. doi: 10.1034/j.1399-0004.2001.590210.x. [DOI] [PubMed] [Google Scholar]
- 67.Jackson SM, et al. A SCA7 CAG/CTG repeat expansion is stable in Drosophila melanogaster despite modulation of genomic context and gene dosage. Gene. 2005;347:35–41. doi: 10.1016/j.gene.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 68.Jarem DA, et al. Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry. 2009;48:6655–6663. doi: 10.1021/bi9007403. [DOI] [PubMed] [Google Scholar]
- 69.Jarem DA, et al. Incidence and persistence of 8-oxo-7,8-dihydroguanine within a hairpin intermediate exacerbates a toxic oxidation cycle associated with trinucleotide repeat expansion. DNA Repair. 2011;10:887–896. doi: 10.1016/j.dnarep.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, et al. HMGB1: roles in base excision repair and related function. Biochim Biophys Acta. 2010;1799:119–130. doi: 10.1016/j.bbagrm.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shibata A, et al. Parp-1 deficiency causes an increase of deletion mutations and insertions/rearrangements in vivo after treatment with an alkylating agent. Oncogene. 2005;24:1328–1337. doi: 10.1038/sj.onc.1208289. [DOI] [PubMed] [Google Scholar]
- 72.Hou C, et al. Incision-dependent and error-free repair of (CAG)(n)/(CTG)(n) hairpins in human cell extracts. Nat Struct Mol Biol. 2009;16:869–875. doi: 10.1038/nsmb.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Panigrahi GB, et al. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 74.Mirkin SM. Toward a unified theory for repeat expansions. Nat Struct Mol Biol. 2005;12:635–637. doi: 10.1038/nsmb0805-635. [DOI] [PubMed] [Google Scholar]
- 75.Hegde ML, et al. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]