Abstract
Mdm2 can mediate p53 ubiquitylation and degradation either in the form of the Mdm2 homodimer or Mdm2/MdmX heterodimer. The ubiquitin ligase activity of these complexes resides mainly in their respective RING finger domains and also requires adjacent C-terminal tails. So far, structural studies have failed to show significant differences between Mdm2 RING homodimers and Mdm2/MdmX RING heterodimers. Here, we report that not only the primary amino acid sequence, but also the length of the C-terminal tail of Mdm2 is highly conserved through evolution and plays an important role in Mdm2 activity toward p53. Mdm2 mutants with extended C termini do not ubiquitylate p53 despite being capable of forming Mdm2 homodimers through both RING-acidic domain and RING-RING interactions. All extended mutants also retained the ability to interact with MdmX, and this interaction led to reactivation of their E3 ubiquitin ligase activity. In contrast, only a subset of extended Mdm2 mutants was activated by the interaction with Mdm2 RING domain, suggesting that Mdm2 homodimers and Mdm2/MdmX heterodimers may not be structurally and functionally fully equivalent.
Key words: p53, Mdm2, RING domain, ubiquitylation, ubiquitin ligase, E3
Introduction
Tumor suppressor p53 maintains genomic integrity of normal cells through transcription-dependent and -independent regulation of cellular responses to genotoxic stress and other types of damage.1 This activity of p53 is lost in the majority of human cancers, most commonly by mutations disrupting the ability of p53 to transactivate its target genes, which are involved in the regulation of cell cycle arrest, apoptosis, DNA repair or epithelial-mesenchymal transition.2–6 In a significant proportion of tumors retaining wild-type p53, overexpression of oncogenes Mdm2 (Hdm2) and MdmX (Mdm4) can lead to p53 inactivation via inhibition of p53-dependent transcription and increased protein degradation and reactivation of p53 by drugs inhibiting Mdm2 or MdmX could have therapeutic potential for this group of cancers. For example, clinical testing is currently underway for a small-molecule inhibitor nutlin-3, which blocks the binding of Mdm2 to p53, activates p53 and induces cell cycle arrest and apoptosis in cancer cells overexpressing Mdm2.7 Deeper understanding of functional interactions between Mdm2 and MdmX and their regulation could help the development of other efficient targeted therapeutic strategies for tumors exhibiting high levels of Mdm2 or MdmX proteins.
Mdm2 serves as a RING finger ubiquitin ligase (E3) with activity toward a number of substrates, including p53, MdmX and Mdm2 itself,8 and Mdm2-mediated ubiquitylation can lead to protein degradation in 26S proteasome. In most tissues, both Mdm2 and MdmX are critically important for keeping p53 levels and activity in check in order to allow for cell proliferation and survival of normal unstressed cells. Targeted deletion of genes coding for Mdm2 or MdmX in mice leads to embryonic lethality, which can be rescued by concomitant deletion of the p53 gene, indicating a critical role for both Mdm2 and MdmX in controlling p53 activity during embryonic development.9,10 Interestingly, while the expression of the p53 inhibitor Mdm2 is stimulated by transcriptionally active p53, thereby forming an autoregulatory feedback loop controlling the cellular levels of both p53 and Mdm2, the MdmX expression is not directly controlled by p53, although Mdm2 expressed in response to p53 activation can also regulate MdmX levels.10,11
Full-length Mdm2 protein consists of 491 amino acid residues and contains several regions with a high degree of evolutionary conservation, of which the non-canonical C-terminal RING domain mediates the interaction with ubiquitin-conjugating enzymes (E2) and is indispensable for the E3 activity of Mdm2.12–14 Despite a strong structural similarity to Mdm2 RING, the MdmX RING domain does not appear to have appreciable E3 ubiquitin ligase activity. On the other hand, we and others have shown that RING-mediated Mdm2 homodimerization or Mdm2/MdmX heterodimerization is required for the ubiquitin ligase activity, and MdmX RING is able to contribute to Mdm2 E3 activity by forming stable heterodimers with Mdm2 RING domain.13,15–17 Depending on the relative ratio between cellular levels of MdmX and Mdm2 proteins, MdmX can either stabilize Mdm2 and enhance its E3 activity and p53 degradation or, when highly overexpressed, inhibit Mdm2-mediated p53 degradation by competing with Mdm2 for p53 binding.18–22 Even though the RING domain structures of both Mdm2 homodimer and Mdm2/MdmX heterodimer have been solved, the molecular mechanisms by which these complexes promote p53 ubiquitylation and functional differences between them are not fully understood.14,23 The structural studies have so far failed to identify any significant structural difference between Mdm2 homodimers and Mdm2/MdmX heterodimers, and there seems to be only a small difference in the ability of isolated Mdm2 and MdmX RING domains to bind the E2 enzyme. However, other studies suggest that MdmX/Mdm2 heterodimer might be thermodynamically more stable than Mdm2 homodimer, and MdmX/Mdm2 heterodimer might be the predominant complex in vivo.13,24,25
In previous studies, we and others have determined that dimerization and ubiquitin ligase activity of the atypical RING domains of Mdm2, MdmX and inhibitor of apoptosis proteins (IAPs) require C-terminal tails, and a single point mutation in this region can cause a complete loss of E3 activity.15,16,26 Analyses of Mdm2 and MdmX RING structures revealed that the C-terminal residues are involved in the formation of the interface between two interacting RINGs and are buried as a consequence of the dimer formation.23 Here, we report that not only the primary sequence but also the length of the C-terminal tail is highly conserved through evolution and is important for the E3 activity of Mdm2 toward tumor suppressor p53. E3 activity of some of the extended Mdm2 mutants can be reactivated by dimerization with Mdm2 or MdmX RING domains containing C-terminal tail of normal length. Surprising differences in the extent of activation of some mutants by dimerization with Mdm2 or MdmX suggest that Mdm2 homodimers and Mdm2/MdmX heterodimers may not be structurally and functionally fully equivalent.
Results
The length of C-terminal tail is evolutionary conserved in Mdm2, MdmX and IAP proteins.
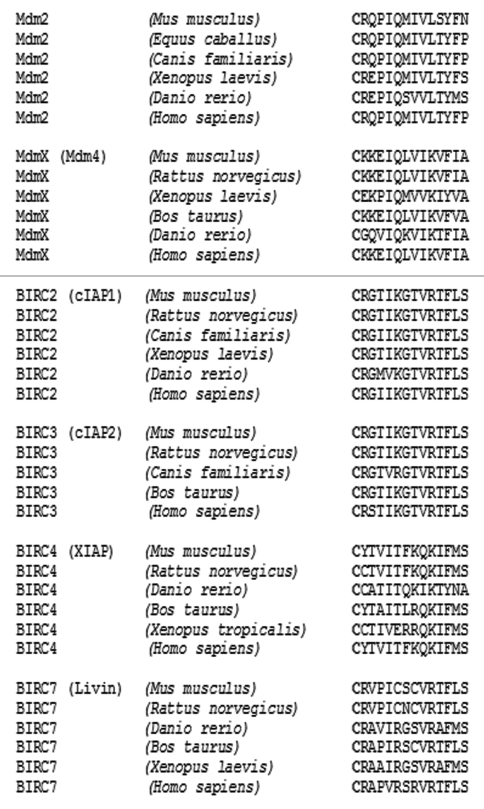
The RING finger domain of human Mdm2 is located at the C terminus of the protein with the last cysteine residue of the RING domain and is followed by only 13 amino acids. Analysis of the C-terminal tail sequences of Mdm2 proteins of various Vertebrates showed that its length is highly conserved through evolution (Fig. 1). Moreover, sequence analysis of other proteins containing C-terminal RING domains, including a close Mdm2 relative, MdmX, or members of an unrelated baculoviral IAP repeat-containing proteins (BIRC) family of proteins (also known as IAPs) revealed conserved length of the C-terminal tail in all proteins. This result suggests that not only the primary amino acid sequence, but also a certain set length of the C-terminal tail might be a prerequisite for the activity of E3 ubiquitin ligases containing a RING domain at the C terminus.
Figure 1.
The length of the C-terminal tail is conserved in ubiquitin ligases containing C-terminal RING domains. Alignments of the C-terminal sequences in Mdm2, MdmX and the IAP (BIRC) family proteins, starting with the last cysteine residue of their RING domain.
Mutations extending the length of Mdm2 C-terminal tail can inhibit Mdm2 activity toward p53.
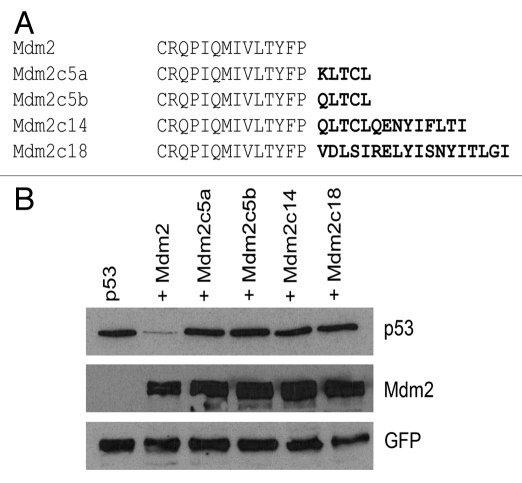
To determine a possible role of the length of the C-terminal tail in Mdm2 activity, we decided to manipulate the length of Mdm2 tail. We created several Mdm2 mutants with a different number (5, 14 and 18) of extra amino acids added to the C terminus of the protein (Fig. 2A) and tested their ability to degrade p53 in a cell-based p53 degradation assay. The mutants were made by introducing missense or frameshift mutations into sites corresponding to STOP codons in the wild-type Mdm2 mRNA sequence using site-directed mutagenesis. In order to account for a possible inhibitory effect of a particular amino acid sequence, we created two variants of the shortest extension differing in the first amino acid and, more importantly, two longer mutants with completely unrelated sequences in the extensions. All four mutants lost the ability to degrade p53 (Fig. 2B), indicating that the conserved length of Mdm2 C terminus may be important for Mdm2 activity toward p53.
Figure 2.
Mutations extending the C-terminal tail can inhibit Mdm2 activity toward p53. (A) Mdm2 mutants containing different number of additional amino acids at the C terminus created in this study. The extra amino acids, which are not present in wild-type Mdm2, are depicted in bold letters. (B) Activity of extended Mdm2 mutants was tested in the p53 degradation assay. U2OS cells were transiently transfected with combinations of plasmid vectors coding for FLAG-tagged p53, GFP and wild-type Mdm2 or the extended Mdm2 mutants. Lysates of transfected cells were analyzed by western blotting.
RING domains of C-terminally extended Mdm2 mutants retain the ability to dimerize with Mdm2 acidic and RING domains.
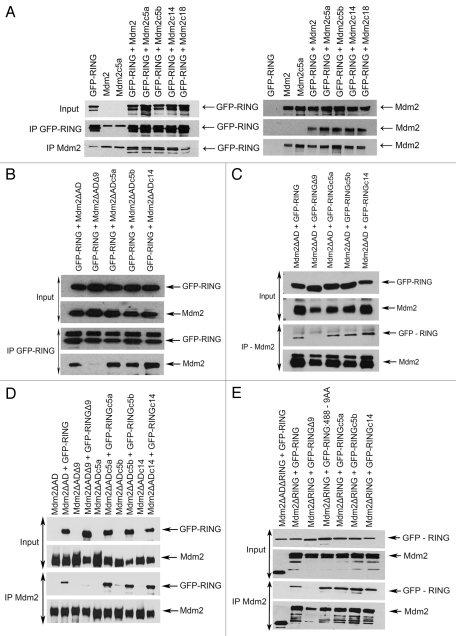
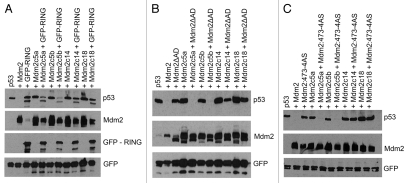
We and others have shown that the E3 ligase activity of Mdm2 depends on the ability of the protein to dimerize,13,16 while some earlier studies indicated that the isolated RING domain of Mdm2 is capable of binding both the C-terminal RING and the central acidic domain (AD) of full-length Mdm2.27 We therefore tested whether the observed drop in the activity of our extended Mdm2 mutants toward p53 could be the result of a loss of their ability to homodimerize through either RING-RING or RING-AD interactions. The initial immunoprecipitation experiments (IPs) with C-terminally extended mutants in the context of full length Mdm2 showed no obvious defect in the binding to isolated GFP-tagged RING domain of Mdm2 (GFP-RING) (Fig. 3A). However, some defects in binding of the extended RING could be masked by the presence of intact acidic domain in the mutant protein, and we therefore decided to test each of the two known modes of binding separately. In order to study only interactions mediated by the RING domains, we created the corresponding C-terminal extensions in the context of Mdm2ΔAD construct, i.e., in Mdm2 lacking the acidic domain, and found that mutant proteins are capable of interacting with wild-type GFP-RING in IPs. As a negative control in our experiments, we used Mdm2 mutant lacking the last nine amino acids of the tail (Mdm2Δ9), which exhibited a clear defect in the RING/RING interaction in our previous studies in reference 16. As shown in Figure 3B, the extended Mdm2 mutants lacking AD, but not the negative control Mdm2ΔADΔ9 double deletion mutant, were able to bind GFP-tagged Mdm2 RING. To confirm these results, we mutated also the isolated GFP-RING to introduce the same extra amino acids at its C terminus, and found that these mutants were still capable of binding to the Mdm2ΔAD protein (Fig. 3C). Moreover, results presented in Figure 3D confirm the ability to dimerize of two mutated Mdm2 RING domains, both containing the same C-terminal extension, one introduced into the GFP-RING protein and the other created in the context of Mdm2ΔAD protein. Finally, we used a similar approach to find out whether the length of the C terminus of Mdm2 could influence the interaction between the Mdm2 RING domain and the acidic domain of another Mdm2 molecule. We used the extended GFP-RING mutants in combination with Mdm2 protein lacking the RING domain (Mdm2ΔRING) and therefore unable to interact through the RING-RING mode. Again, we found no difference between wild-type GFP-tagged RING and extended GFP-RING proteins in their capacity to bind to the Mdm2ΔRING protein (Fig. 3E).
Figure 3.
Extended MDM2 mutants retain the ability to homo-dimerize through RING-RING and RING-acidic domain interactions. C-terminally extended mutants in the context of full-length Mdm2 (A), Mdm2 lacking the acidic domain (Mdm2ΔAD) (B and D) or GFP-RING (A and C–E) were transiently transfected into HEK293 cells together with GFP-RING (A and B), Mdm2ΔAD (C) or Mdm2 (Mdm2ΔRING) (E). Cells were treated 24 h post-transfection with proteasome inhibitor MG132 for 4 h, lysed and immunoprecipitated with anti-Mdm2 (N-20) and anti-GFP (7.1/13.1) antibodies then analyzed by western blotting.
C-terminally extended Mdm2 RING domain is capable of binding MdmX.
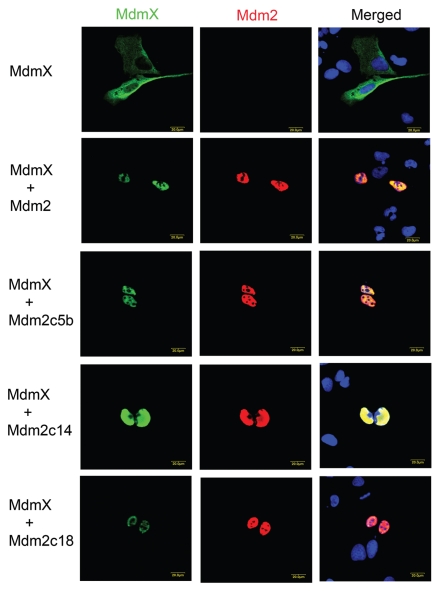
It has been shown that the Mdm2/MdmX heterodimer rather than the Mdm2 homodimer is the predominant intracellular form of Mdm2. Furthermore, the Mdm2/MdmX heterodimer exhibited higher E3 ligase activity than Mdm2 homodimer.13 As our extended Mdm2 mutants retained the capability to form homodimers, we also assessed their ability to physically interact with MdmX. We used an immunofluorescence relocalization assay based on the observation that exogenous wild-type MdmX protein, which does not contain a nuclear localization signal sequence (NLS), is predominantly localized in the cytoplasm of transfected U2OS cells. However, in the presence of an excess of exogenous Mdm2 protein and upon binding to it, MdmX is transported into the cell nucleus in the majority of cells. Using this assay, we detected changes in subcellular localization of MdmX in the presence of all extended Mdm2 mutants, indicating that the extension of Mdm2 C terminus did not prevent the interaction between Mdm2 and MdmX RING domains (Fig. 4).
Figure 4.
The extended Mdm2 mutants retain the ability to interact with MdmX. U2OS cells were transfected with Myc-tagged MdmX together with wild-type Mdm2 or the extended mutants then proteins detected by immunofluorescence using a mixture of anti-Mdm2 mouse monoclonal IF2 (Ab-1, Calbiochem) and anti-Myc rabbit polyclonal (A14, Santa Cruz Biotechnology) antibodies.
Reactivation of C-terminally extended Mdm2 mutants by interaction with MdmX.
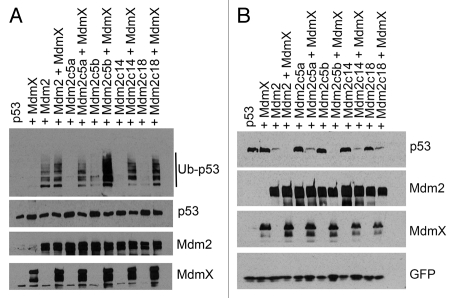
We have shown previously that MdmX is able to restore the activity of Mdm2 proteins containing point mutations in their C-terminal tail. Here, we tested whether binding to MdmX could also reactivate the extended Mdm2 mutants. U2OS cells were transfected with constructs coding for p53 and wild-type Mdm2 or extended Mdm2 mutants in the presence or absence of MdmX, and we studied changes in the levels of p53 ubiquitylation. To our surprise, all four extended Mdm2 mutants were reactivated by MdmX, regardless of their amino acid sequence or the length of the C-terminal extension (Fig. 5A). Moreover, restoration of the E3 activity of the extended mutants lead to a full restoration of their ability to degrade p53 (Fig. 5B).
Figure 5.
MdmX restores the activity of extended Mdm2 mutants in vivo. (A) Co-expression with MdmX restores the E3 activity of the extended Mdm2 mutants, regardless of the length of the C-terminal extension. U2OS cells were transiently cotransfected with FLAG-tagged p53, wild-type Mdm2 or the extended mutants, Myc-tagged MdmX and HA-tagged ubiquitin. Cells were treated 24 h post-transfection with proteasome inhibitor MG132 for 3 h and lysed into 0.5% SDS. Lysates were diluted and p53 was immunoprecipitated with DO-1 antibody. Ubiquitin-conjugated p53 was detected by western blotting using anti-HA mouse monoclonal antibody and p53 levels in the input using anti-FLAG antibody M2 (Sigma-Aldrich). (B) MdmX restores p53 degradation by the C-terminally extended Mdm2 mutants. U2OS cells were transiently co-transfected with FLAG-tagged p53, wild-type Mdm2 or the extended mutants and Myc-tagged MdmX. Cells were lysed 24 h post-transfection, and lysates were analyzed by western blotting.
Reactivation of the extended Mdm2 mutants by interactions with wild-type Mdm2 RING.
Results presented in Figure 5 indicated that the extended C-terminal tails do not significantly interfere with the structure and function of the Mdm2/MdmX heterodimer. As our extended Mdm2 mutants retained the ability to form homodimers, we tested whether they could be activated by the interaction with Mdm2 constructs containing wild-type RING domain. In stark contrast to the full reactivation of all extended mutants by MdmX, only Mdm2c5 a and b mutants were able to degrade p53 as part of complex with GFP-RING (Fig. 6A) and Mdm2ΔAD (Fig. 6B) proteins, both containing wild-type RING domain with C-terminal tail of normal length, but are inactive on their own due to the lack of other important functional domains, such as the p53-binding domain at the C terminus and the central acidic domain. The longer mutants Mdm2c14 and Mdm2c18 extended by 14 and 18 amino acids, respectively, could not be reactivated by these constructs. Interestingly, essentially the same result was obtained when we co-expressed the extended mutants with full-length Mdm2 containing inactivating point mutation directly within the RING domain itself (Mdm2:473-4AS) (Fig. 6C).
Figure 6.
Wild-type Mdm2 RING domain restores the activity of only a subset of extended Mdm2 mutants. U2OS cells were transiently co-transfected with FLAG-p53 and extended MDM2 mutants together with GFP-RING (A), Mdm2ΔAD (B) or inactive Mdm2 RING domain mutant Mdm2:473-4AS (C), and lysates of transfected cells were analyzed by western blotting.
The role of the length of the C-terminal tail in MdmX function.
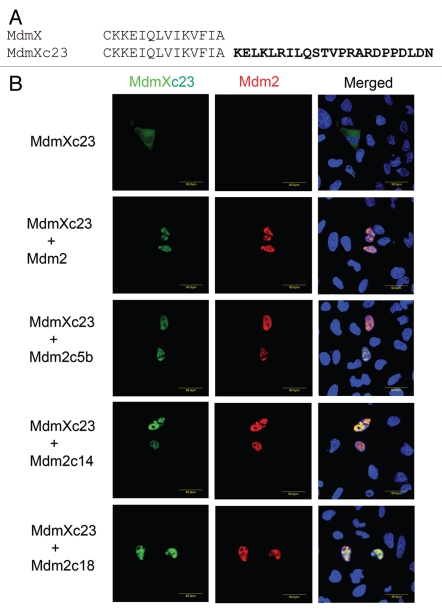
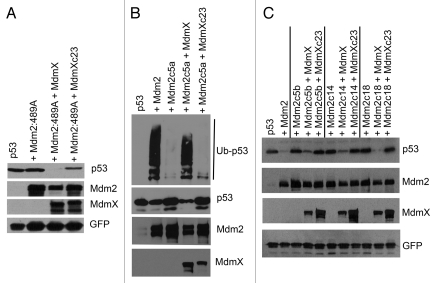
Despite the fact that MdmX protein does not seem to possess a significant ubiquitin ligase activity, it shares the same overall domain architecture with Mdm2, and its RING finger domain exhibits strong structural homology to Mdm2 RING. And just like Mdm2 RING, it contains an adjacent C-terminal tail, the length of which was also conserved in evolution, suggesting that it might be required for MdmX function. To find out whether the length of the MdmX tail might play a significant role in the activity of Mdm2/MdmX heterodimer, using site-directed mutagenesis, we created MdmX mutant containing 23 extra amino acids at the C terminus (MdmXc23; Fig. 7A) and tested its ability to interact with Mdm2 in the above described re-localization assay. Results presented in Figure 7B suggest that the extended MdmX protein is able to physically interact in living cells with wild-type Mdm2 as well as with our extended Mdm2 mutants. However, the activity toward p53 of Mdm2/MdmX heterodimer containing the extended MdmX mutant was markedly impaired. While binding of wild-type MdmX reactivated the C-terminal Mdm2 Y489A point mutant and induced p53 degradation, the MdmXc23 mutant was not capable of such restoration of Mdm2 function (Fig. 8A). In other experiments, wild-type MdmX, but not the extended MdmXc23 mutant, was able to restore the ubiquitin ligase activity toward p53 and p53 degradation mediated by Mdm2/MdmX heterodimers containing C-terminally extended Mdm2 mutants (Fig. 8B and C), indicating that the conserved length of the C-terminal tail of MdmX might be important for the activity of Mdm2/MdmX dimer toward p53 tumor suppressor.
Figure 7.
C-terminal extension of MdmX does not prevent heterodimerization with Mdm2. (A) Using site-directed mutagenesis, we have created an extended MdmX mutant with 23 amino acids added at the C terminus (MdmXc23). The extra amino acids, which are not present in wild-type MdmX, are depicted in bold letters. (B) We examined the ability of wild-type Mdm2 and the extended Mdm2 mutants to bind MdmXc23 and translocate it from the cytoplasm to the nucleus. U2OS cells were transiently co-transfected with plasmids coding for GFP-MdmXc23 and wt Mdm2 or extended Mdm2 mutants. Subcellular localization of MdmX was detected in fluorescence microscope using the fluorescence of the N-terminal GFP tag. Mdm2 was localized in the cells by immunofluorescence using anti-Mdm2 mouse monoclonal antibody IF2 (Ab-1).
Figure 8.
C-terminal extension in MdmX inhibits reactivation of C-terminal Mdm2 mutants by Mdm2/MdmX heterodimerization. (A) Extended MdmX is not able to restore p53 degradation by Mdm2 C-terminal tail mutant Y489A. U2OS cells were transiently co-transfected with FLAG-p53 and Mdm2:489A mutant together with GFP-MdmX or GFP-MdmXc23, and lysates of transfected cells were analyzed by western blotting. (B) Extended MdmX is not able to reactivate p53 ubiquitylation by C-terminally extended Mdm2. U2OS cells were transiently co-transfected with plasmids coding for FLAG-p53, the extended Mdm2c5a mutant and HA-ubiquitin together with GFP-MdmX or GFP-MdmXc23. Cells were treated 24 h post-transfection with proteasome inhibitor MG132 for 3 h and lysed into 0.5% SDS. Lysates were diluted and p53 was immunoprecipitated with DO-1 antibody. Ubiquitin-conjugated p53 was detected by western blotting using anti-HA mouse monoclonal antibody and p53 levels in the input using anti-FLAG antibody M2. (C) Extended MdmX is not able to restore p53 degradation by the extended Mdm2 mutants. U2OS cells were transiently co-transfected with FLAG-p53 and the extended Mdm2 mutants c5, c14 or c18, together with GFP-MdmX or GFP-MdmXc23, and lysates of transfected cells were analyzed by western blotting.
Discussion
Mdm2 and MdmX cooperate in tightly controlling the activity of tumor suppressor p53 during embryonic development and in normal, unstressed tissues. On the other hand, Mdm2 and MdmX overexpression is often found in human cancers and can be responsible for attenuation of the tumor-suppressive functions of p53 and tumor development. Mdm2 dimer serves as a RING finger ubiquitin ligase for p53 and targets p53 for proteasomal degradation, which is something that MdmX on its own cannot accomplish, despite its high degree of homology to Mdm2. However, MdmX can participate on p53 ubiquitylation and degradation as a part of the Mdm2/MdmX complex. Both Mdm2 homodimerization and Mdm2/MdmX heterodimerization is mediated by the RING domains and requires adjacent C-terminal tails. The critical importance of the extreme C terminus for Mdm2 function has been confirmed in studies showing that deletion of the extreme C terminus prevents the formation of RING-RING dimers and inactivates Mdm2.15,16,28 NMR analysis of the isolated Mdm2 RING missing seven C-terminal residues revealed that this mutant is conformationally disrupted and appears to be partially unfolded, suggesting that the extreme C-terminal tail is required for the maintenance of the structural integrity of Mdm2 RING.28 Independent of its role in RING dimerization, the C-terminal tail of Mdm2 is also directly involved in the ubiquitin ligase activity of Mdm2. We and others have shown that point mutations within this region can inhibit Mdm2 activity toward p53 without disrupting RING domain dimerization.15,16,28
In this study, we show that the last cysteine of the RING domain is followed by exactly 13 amino acid residues in Mdm2, MdmX and IAP proteins of vertebrate origin, suggesting that not only the primary amino acid sequence of the C-terminal tail but also its length might be important for the biological function of proteins containing C-terminal RING domain. Interestingly, Mdm2-like and p53-like genes have recently been identified in the genomes of some invertebrates as well, including Arachnid Ixodes scapularis (deer tick) and a very simple multicellular animal Trichoplax adhaerens. Detailed sequence analyses have shown that Mdm2 and p53 genes in Ixodes and Trichoplax retain key features required for p53-Mdm2 interaction and for Mdm2-mediated p53 regulation.29,30 To our surprise, Mdm2-like genes in these organisms also exhibit the same length of the C-terminal tail, indicating that this property of Mdm2 C terminus might have been evolutionary conserved for over one billion years, further highlighting the possibility of its importance for Mdm2 function.
In order to analyze the possible role of the length of Mdm2 C terminus, we created a series of Mdm2 mutants extended by 5, 14 or 18 amino acids. All mutants were inactive toward p53—unable to ubiquitylate p53 in cells and target it for proteasomal degradation. However, the mutants retained the ability to heterodimerize with MdmX, and their activity toward p53 was fully restored in the presence of MdmX in a manner similar to C-terminal tail point mutants Y489A and Y489D, which were described in our previous work in reference 16. Our results also showed that the extended RING domain of Mdm2 is perfectly capable of binding the RING domain or the acidic domain of another Mdm2 molecule, regardless whether wild-type or extended, indicating that the extensions of the C-terminal tail did not pose a sterical hindrance for Mdm2 RING domain dimerization. In contrast to the effect of deletions of the tail, the extensions did not seem to have any significant impact on the overall structure of Mdm2 RING, as this would certainly have a strong negative impact on RING-mediated dimerization.
The Mdm2/MdmX RING domain heterodimer is a highly symmetrical structure in which MdmX RING replaces one of the RINGs of the Mdm2 homodimer, and the extreme C termini of both Mdm2 and MdmX play an important role in the formation of the dimer.14 The C-terminal residues are involved in 13 of 15 hydrogen bonds on the interface between the two RINGs, and six residues of each C terminus are buried as a consequence of dimer formation.23 Importantly, structural studies have so far failed to identify significant differences in the structure of the RINGs of the two proteins. Therefore, if MdmX was able to restore the activity of all extended Mdm2 mutants toward p53, we expected that wild-type Mdm2 RING domain would be also capable of reactivating the extended Mdm2 RINGs. Quite unexpectedly, only Mdm2 RINGs carrying the shortest, i.e., five amino acids, tail extension were fully reactivated by binding to GFP-RING or Mdm2ΔAD, protein constructs containing wild-type Mdm2 RING but not active toward p53 due to the lack of other important functional domains, such as the N-terminal p53-binding domain or the central acidic domain. To our surprise, the shortest extensions were also activated in the presence of otherwise inactive RING domain point mutant Mdm2:473-4AS, showing that only one RING domain of Mdm2 homodimer needs to be functional for the complex to be fully active toward p53 and suggesting a similarity in the activity toward p53 of Mdm2 homodimers and Mdm2/MdmX heterodimers. However, the results were very different when we tested the activity of Mdm2 homodimers containing one wild-type RING domain and the other extended by 14 or 18 amino acids. The homodimers were inactive, even though the Mdm2/MdmX heterodimers containing these extensions were targeting p53 for degradation normally. Recent studies have shown some differences between the Mdm2/MdmX complex and the Mdm2 dimer in their specificity for Mdm2 as a substrate22 and suggested that the heterodimer might be thermodynamically more stable than Mdm2 homodimers.24 Nevertheless, we believe that the clear difference in the reactivation of various Mdm2 extended mutants described here is the first report strongly suggesting that Mdm2 homodimers and Mdm2/MdmX heterodimers might not be fully structurally and functionally equivalent, even though we cannot, at this stage, completely exclude the possibility that the long extensions can interfere with other parts of the Mdm2 protein, thereby preventing reactivation seen with shorter extensions.
We have shown previously that MdmX C-terminal tail participates in the formation of Mdm2/MdmX heterodimer, and it is required for the restoration of the activity the Mdm2 C-terminal tail mutant Y489A.16 In this work, we have created also a C-terminally extended version of MdmX, and show that it can physically interact with both wild-type Mdm2 and the C-terminally extended Mdm2 mutants. However, unlike wild-type MdmX, it is not able to restore the activity of the Y489A Mdm2 point mutant or Mdm2 extension any more.
Taken together, results presented in this study suggest that the length of the C-terminal regions of Mdm2 and MdmX proteins has been conserved in evolution, because it can influence their activity toward tumor suppressor p53. Moreover, the observation that Mdm2 mutants containing C-terminal extensions do not ubiquitylate p53, despite being capable of forming dimers, confirms our previous results indicating that the role of Mdm2 C-terminal tail in RING dimerization can be separated from its function in protein ubiquitylation.16 In addition to that, data presented here indicate that despite their apparent structural similarity, the Mdm2 homodimers and Mdm2/MdmX heterodimers may not be fully functionally equivalent and may, therefore, be differentially regulated, possibly adding an additional level of complexity to the cellular regulation of p53 tumor suppressor.
Materials and Methods
Cell culture.
Human U2OS and HEK293T cells were cultured at 37°C under 5% CO2 in a high-humidity atmosphere in Dulbecco's modified Eangle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin sulfate.
Plasmids and mutagenesis.
Plasmids coding for FLAG-tagged human wild-type p53 (pcDNA3-FLAG-p53), wild-type Mdm2 (pCHDM1A), Mdm2 mutant 1–440 (Mdm2ΔRING) and Myc-tagged MdmX have been described previously in references 8, 20 and 31. Mdm2ΔAD mutant of pCHDM1A, coding for Mdm2 lacking amino acids 245–295, was kindly provided by Mary Hanlon. GFP-MdmX expression plasmid coding for N-terminally tagged full-length human MdmX was kindly provided by Robert Ludwig. Plasmid GFP-RING coding for GFP-tagged human Mdm2 C terminus (amino acids 384–491) was generated by cloning HindIII/AvrI fragment from pCHDM1A into HindIII/XbaI sites of pEGFP-C1 (Clontech). All point and deletion mutations within the C-terminal tail of Mdm2, Mdm2ΔAD, GFP-RING and MdmX were generated by site-directed mutagenesis using Pfu DNA polymerase (Stratagene) and verified by DNA sequencing.
MDM2-mediated p53 degradation.
U2OS cells in 60-mm dishes were transfected with 0.5 µg of FLAG-p53 and 4 µg of pCH-DM1A or extended Mdm2 mutants (5 µg of extended MDM2 mutants and Mdm2:473-4AS, Mdm2ΔAD or GFP-RING transfected either separately or in combinations in a 1:1 ratio in experiments presented in Fig. 6H) using Lipofectamine 2000 reagent according to guidelines of the manufacturer (Invitrogen). For experiments studying the role of MdmX in Mdm2-mediated p53 degradation, the total amount of pCHDM1A (or extended mutants) transfected separately was 2.5 µg, and the total amount of pCHDM1A (or extended mutants) and Myc-MdmX transfected in combination was 5 µg in a 1:1 ratio. Each transfection mixture also contained 0.5 µg of pEGFP-N1 (Clontech) to control for transfection efficiency, and empty plasmid pcDNA3.1 (Invitrogen) was used to bring the total amount of transfected DNA to 6 µg. Cell were cultured for an additional 24 h, washed with PBS and lysed with 0.25 ml of SDS sample buffer. Proteins were resolved by SDS-PAGE and detected using anti-Mdm2 Ab-1 (Calbiochem), anti-Myc 9E10 or anti-GFP 7.1/13.1 (Roche).
Immunoprecipitation.
HEK293T cells were transfected in 100-mm plates with 6 µg of DNA using Lipofectamine 2000 reagent according to guidelines of the manufacturer (Invitrogen). Cells were treated with proteasome inhibitor (15 µM MG132 in cell culture medium) 24 h post-transfection. Four h later, cells were washed with ice-cold PBS and lysed in Triton X-100 lysis buffer (1% Triton X-100, 150 mM NaCl and 50 mM Tris pH 8.0) containing protease inhibitors (Complete Mini EDTA-free, Roche) by incubating on ice for 30 min, and the extracts were centrifuged at 13,000 rpm for 30 min at 4°C. Immunoprecipitation was performed at 4°C overnight with 1 µg of antibody. After incubation, 20 µl of protein G-Sepharose (Sigma-Aldrich) were added for another 45 min on a rotating wheel. Mdm2 was immunoprecipitated with anti Mdm2 rabbit polyclonal antibody (N-20, Santa Cruz Biotechnology) and GFP-RING with anti-GFP 7.1/13.1 (Roche). Immunoprecipitated proteins were washed three times with lysis buffer and resuspended in 2x SDS sample buffer. Proteins from whole-cell extracts and immunoprecipitations were resolved by SDS-PAGE and analyzed by western blotting with anti-Mdm2 Ab-1 (Calbiochem) and anti-GFP 7.1/13.1 (Roche).
Immunofluorescence.
U2OS cells grown on coverslips were transfected with 4.5 µg MDM2 or extended MDM2 mutants and 1.5 µg Myc-tagged MdmX or GFP-tagged MdmX using Lipofectamine 2000 reagent according to guidelines of the manufacturer (Invitrogen), and DMEM was changed 4 h later. Twenty-four hours after transfection, cells were treated with 15 µM MG132 in DMEM for 3 h, washed with PBS and fixed in 3% paraformaldehyde for 20 min at room temperature. After fixation, cell were washed with PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min and blocked with PBS containing 0.5% bovine serum albumin for 30 min at room temperature. The cells were then incubated with the anti-MDM2 monoclonal IF2 (Ab-1, Calbiochem) and anti-c-Myc polyclonal (A14, Santa Cruz Biotechnology) antibodies for 2 h in blocking solution and, following three washes in PBS, were incubated with a mixture of fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit and DyLight™ 594 conjugated AffiniPure donkey anti-mouse antibodies (H + L; Jackson ImmunoResearch Laboratories) in blocking solution containing 1 µg/ml DAPI (Sigma-Aldrich) for visualization of nuclei. Cells were washed with PBS and mounted with Vectashield mounting medium (Vector Laboratories). Images were taken with an Olympus FluoView™ 500 confocal laser scanning fluorescence microscope (Olympus).
Mdm2-mediated p53 ubiquitylation in vivo.
U2OS cells grown in 60-mm dishes were transiently transfected with FLAG-p53 (0.5 µg), hemagglutinin (HA)-ubiquitin (0.5 µg), MDM2 or extended MDM2 mutants (2.5 µg) and Myc-MDMX (2.5 µg) using Lipofectamine 2000 reagent according to guidelines of the manufacturer (Invitrogen). Empty plasmid pcDNA3.1 (Invitrogen) was used to bring the total amount of transfected DNA to 6 µg. Cells were treated 24 h post-transfection with 15 µM MG132 in DMEM for 3 h and lysed in 300 µl 0.5% SDS. Lysates were boiled for 5 min, vortexed, cooled down to room temperature and diluted with 1 ml Triton X-100 lysis buffer. Immunoprecipitation of p53 was performed for 1 h using 1 µg of anti-p53 monoclonal antibody DO-1 at 4°C on a rotating wheel. After incubation, 20 µl of protein G-Sepharose (Sigma-Aldrich) were added for another 45 min. Subsequently, the immunoprecipitates were washed three times with lysis buffer and analyzed by western blotting. Ubiquitylated p53 was detected using anti-HA-tagged mouse monoclonal antibody; FLAG-p53, MDM2 and MDMX levels in the input were detected using anti-FLAG antibody M2 (Sigma-Aldrich), anti MDM2 Ab-1 (Calbiochem) and anti-Myc 9E10 antibody.
SDS-PAGE and western blot analysis.
Total cells lysates of degradation, immunoprecipitation and ubiquitylation assays were resolved by SDS-PAGE. After transfer, the polyvinylidene difluoride membrane (PVDF; Millipore) was blocked in 5% milk in TBS (10 mM TRIS-HCl, 100 nM NaCl and 0.05% Tween 20; pH = 7.4) for 1 h at room temperature, then incubated overnight at 4°C with specific antibody (using dilutions recommended by the manufacturer). After three washes in 1% milk in TBS, sheep anti-mouse (GE Healthcare) secondary horseradish peroxidase conjugated antibody was applied for 1 h at room temperature. Visualization was done on blue sensitive Medical X-ray film (Agfa) using an enhanced chemiluminescence kit (ECL, Amersham Life Science).
Acknowledgements
We would like to thank Borivoj Vojtesek for the anti-p53 antibody and Arnie Levine, Donna George, Mary Hanlon, Robert Ludwig and Allan Weissman for the Mdm2, MdmX and ubiquitin expression plasmids. We also thank Zuzana Prokopova for technical assistance. This work was funded by the Czech Science Foundation (Grant No. 301/09/1324) and Cancer Research UK.
Abbreviations
- AD
acidic domain
- GFP
green fluorescent protein
- RING
really interesting new gene
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 4.Shiota M, Izumi H, Onitsuka T, Miyamoto N, Kashiwagi E, Kidani A, et al. Twist and p53 reciprocally regulate target genes via direct interaction. Oncogene. 2008;27:5543–5553. doi: 10.1038/onc.2008.176. [DOI] [PubMed] [Google Scholar]
- 5.Smit MA, Peeper DS. Deregulating EMT and senescence: double impact by a single twist. Cancer Cell. 2008;14:5–7. doi: 10.1016/j.ccr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 8.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 9.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 10.Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 11.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 12.Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 13.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2's E3 ligase activity. Cancer Res. 2007;67:6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- 14.Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol. 2006;363:433–450. doi: 10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 15.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, et al. The Mdm2 RING domain C terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh RK, Iyappan S, Scheffner M. Heterooligomerization with MdmX rescues the ubiquitin/Nedd8 ligase activity of RING finger mutants of Mdm2. J Biol Chem. 2007;282:10901–10907. doi: 10.1074/jbc.M610879200. [DOI] [PubMed] [Google Scholar]
- 18.Stad R, Little NA, Xirodimas DP, Frenk R, van der Eb AJ, Lane DP, et al. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2001;2:1029–1034. doi: 10.1093/embo-reports/kve227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710–2714. doi: 10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 24.Iyappan S, Wollscheid HP, Rojas-Fernandez A, Marquardt A, Tang HC, Singh RK, et al. Turning the RING domain protein MdmX into an active ubiquitin-protein ligase. J Biol Chem. 2010;285:33065–33072. doi: 10.1074/jbc.M110.115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/S0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 26.Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, et al. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci USA. 2005;102:16182–16187. doi: 10.1073/pnas.0502828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr CJ, Roussel MF. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002;62:1222–1230. [PubMed] [Google Scholar]
- 28.Shloush J, Vlassov JE, Engson I, Duan S, Saridakis V, Dhe-Paganon S, et al. Structural and functional comparison of the RING domains of two p53 E3 ligases, Mdm2 and Pirh2. J Biol Chem. 2011;286:4796–4808. doi: 10.1074/jbc.M110.157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane DP, Cheok CF, Brown CJ, Madhumalar A, Ghadessy FJ, Verma C. The Mdm2 and p53 genes are conserved in the Arachnids. Cell Cycle. 2010;9:748–754. doi: 10.4161/cc.9.4.10616. [DOI] [PubMed] [Google Scholar]
- 30.Lane DP, Cheok CF, Brown C, Madhumalar A, Ghadessy FJ, Verma C. Mdm2 and p53 are highly conserved from placozoans to man. Cell Cycle. 2010;9:540–547. doi: 10.4161/cc.9.3.10516. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Lin J, Levine AJ. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol Med. 1995;1:142–152. [PMC free article] [PubMed] [Google Scholar]