Ezh2, a PRC2 core component that is associated with DNA methyltransferases, has been implicated as having both tumor suppressor and a tumor-promoting activity in various human cancers. In this study, Sauvageau and colleagues demonstrate that Ezh2 suppresses T-cell leukemia development in mice and show a similar role for Ezh2 in human adult acute T-cell leukemia (T-ALL). This study provides novel insights into the mechanistic link between T-ALL and alterations in chromatin regulation.
Keywords: T-ALL, EZH2, PRC2, leukemia, JARID2, DNMT3A
Abstract
In this study, we show the high frequency of spontaneous γδ T-cell leukemia (T-ALL) occurrence in mice with biallelic deletion of enhancer of zeste homolog 2 (Ezh2). Tumor cells show little residual H3K27 trimethylation marks compared with controls. EZH2 is a component of the PRC2 Polycomb group protein complex, which is associated with DNA methyltransferases. Using next-generation sequencing, we identify alteration in gene expression levels of EZH2 and acquired mutations in PRC2-associated genes (DNMT3A and JARID2) in human adult T-ALL. Together, these studies document that deregulation of EZH2 and associated genes leads to the development of mouse, and likely human, T-ALL.
Enhancer of zeste homolog 2 (EZH2) catalyzes di- and trimethylation of Lys 27 on histone H3 (H3K27me2/3) and establishes chromatin marks associated with gene silencing (Surface et al. 2010). The enzymatic activity of EZH2 depends on the formation of a PRC2 complex comprising EZH2, embryonic ectoderm development (EED), and suppressor of zeste 12 homolog (SUZ12) (Pasini et al. 2004; Montgomery et al. 2005). Variations in the expression levels of individual components could therefore affect the activity of the PRC2 holoenzyme. We (Lessard et al. 1999) and others (Richie et al. 2002; Majewski et al. 2008, 2010) have shown that PRC2 restricts the activity of hemopoietic progenitor/stem cells and that heterozygosity for mutant Eed alleles accelerates lymphomagenesis (Sauvageau et al. 2008). EZH2 mutations representing loss-of-function alleles have recently been identified in myelodysplastic syndromes (MDSs) and myeloproliferative neoplasms (MPNs) (Ernst et al. 2010; Nikoloski et al. 2010; Jankowska et al. 2011; Score et al. 2012). Conversely, overexpression of Ezh2 has also been implicated in the progression of various types of human cancers (Sauvageau and Sauvageau 2010; Margueron and Reinberg 2011), and a recurrent EZH2 mutation identified in B-cell lymphomas (Morin et al. 2010) was proposed to act as a dominant, cancer-promoting EZH2 allele (Yap et al. 2011). Genetic data therefore suggest that gene dosage could be deterministic for the apparently contradictory oncogenic and tumor-suppressing activities of EZH2, but no functional data supporting these possibilities have so far been presented.
In this study, we exploit an inducible gene inactivation approach to show that Ezh2, as a PRC2 core component, suppresses T-cell leukemia (T-ALL) development in mice and provide evidence indicating a similar function for this complex in human adult T-ALL. More broadly, we provide key observations linking this disease with alterations in chromatin regulation.
Results and Discussion
For functional studies presented in this study, we exploited the conditional Ezh2F allele carrying loxP sites flanking exons 14 and 15, which code for the SET domain (Shen et al. 2008). The Cre-mediated deletion generates a mutated Ezh2Δ allele and abrogates production of EZH2 protein (Shen et al. 2008). Ezh2F/Δ mice breed normally, are healthy, and have a lifespan comparable with wild-type and Ezh2F/+ mice.
To ablate Ezh2 function in adult bone marrow cells, we crossed the conditional Ezh2F/F and Ezh2F/Δ mice with mice carrying the polyinosine–polycytidine (pIpC)-inducible Mx-Cre transgene and induced Cre expression with pIpC in 7- to 8-wk-old animals (Fig. 1A). At 12 d after pIpC induction, a PCR-based genotyping approach (Fig. 1B) demonstrated a partial Ezh2 deletion in the spleens of Ezh2F/F Mx-Cre+ and Ezh2F/Δ Mx-Cre mice (hereafter, Ezh2Δ/Δ mice) and nearly complete recombination of the floxed allele in the bone marrow and thymus. In the bone marrow cells of Ezh2Δ/Δ mice, no EZH2 protein could be detected by Western blot analysis (Fig. 1C). Phenotypical analyses revealed a sevenfold to 10-fold decrease in the numbers of Ezh2Δ/Δ thymocytes compared with Ezh2+/+ Mx-Cre+ controls (hereafter, wild-type controls) (Supplemental Fig. 1A) associated with a loss of Ezh2Δ/Δ CD4+CD8+ cells and accumulation of the immature CD4−CD8− cell population (Supplemental Fig. 1B), confirming that EZH2 activity is essential for normal thymocyte development (Su et al. 2005; Mochizuki-Kashio et al. 2011).
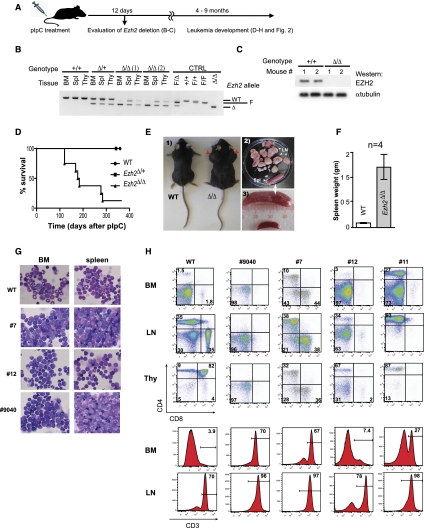
Figure 1.
Leukemia development in Ezh2-deficient mice. (A) Outline of the experimental approach. (B) PCR-based analysis of Ezh2 deletion in hemopoietic tissues at 12 d after pIpC treatment. Primer pairs (sequence available on request) amplify a 314-base-pair (bp) wild-type (WT) Ezh2 fragment, a 280-bp fragment identifying the conditional Ezh2F allele, and a 200-bp fragment of the excised Ezh2Δ allele. Tail clippings were used as a source of control DNA. (C) Western blot analysis of EZH2 levels in bone marrow cells of wild-type and Ezh2F/Δ mice 12 d after pIpC treatment. α-Tubulin levels are shown as a loading control. (D) Kaplan-Meier survival plot of pIpC-treated wild-type, Ezh2F/+, and Ezh2F/Δ mice. The day of the last pIpC injection was designated as day 0 of the experiment. (E, panel 1) Photograph of a representative sick Ezh2F/Δ mouse showing enlarged lymph nodes (red arrows). Note also a bloated appearance, suggesting the presence of an abdominal mass. An age-matched mouse from the control wild-type cohort is shown for comparison. (Panel 2) Lymph nodes and spleen of an Ezh2F/Δ mouse shown in panel 1. A wild-type spleen is shown for comparison in panel 3. (F) Splenomegaly of the sick Ezh2F/Δ mice. Result shown represents mean spleen weight ± SD; n = 4. (G) Wright stain-dyed cytospin preparations of bone marrow and spleen cells from a sick Ezh2F/Δ mouse; 40-fold magnification. Mouse identification numbers are shown at the left. (H) Phenotypical characterization of Ezh2F/Δ leukemias. (Top panels) Proportions of CD4+ and CD8+ cells. (Bottom panels) Proportions of CD3+ cells in the bone marrow and lymph nodes. Mouse identification numbers are shown at the top, and tissues analyzed are indicated at the left. (BM) Bone marrow; (LN) lymph node; (Thy) thymus.
During a 10-mo observation period, all pIpC-treated wild-type and Ezh2Δ/+ mice remained healthy, while complete inactivation of Ezh2 resulted in the development of spontaneous T-ALL. In the first cohort of Ezh2Δ/Δ mice (three out of four mice), the shortest latency of leukemia development was 152 d (152–281 d) (Fig 1D), and in the second cohort (n = 10), they started to succumb at 122 d after deletion. Diseased animals presented with grossly enlarged lymph nodes (Fig. 1E, panels 1,2) and spleens (Fig. 1E [panels 2,3], F). Leukemic cells infiltrated the bone marrow, spleen (Fig. 1G, right panel), liver, and kidney (Supplemental Fig. 2A). Infiltrates comprised a mixture of relatively mature lymphoid cells with open chromatin and cells with blastic appearance and prominent nucleoli (Fig. 1G). Leukemias appeared to be phenotypically heterogeneous (Fig. 1H, top panel): In mice #9040 and #12, the majority of cells were CD4−CD8−; leukemia #7 comprised CD4+, CD8+, and CD4−CD8− cells; and leukemia #11 was predominantly CD4+. However, all leukemias expressed CD3 (Fig. 1H, bottom panel) and TCRγδ (Supplemental Fig. 2B, right panel) and were negative for the cell surface TCRαβ (Supplemental Fig. 2B, left panel).
The CD4+ and CD8+ cells detected in sick Ezh2Δ/Δ mice could represent the progeny of the nondeleted Ezh2F/Δ progenitors. Arguing against this possibility, the PCR-based analyses of Ezh2 (Fig. 2A) showed that the nondeleted Ezh2F/Δ cells represented only a minor fraction of leukemic cell populations and could not contribute to the production of CD4+ and CD8+ cells as seen in leukemias #7 and #11.
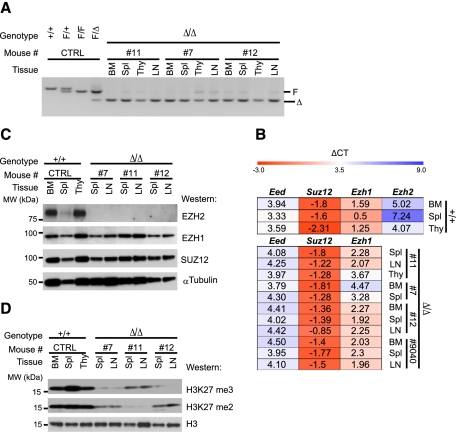
Figure 2.
Genetic and biochemical characterizations of Ezh2Δ/Δ leukemias. (A) PCR-based analysis of Ezh2 deletion in leukemic cell populations. Mouse identification numbers are shown at the top, and the positions of the conditional (F) and deleted (Δ) Ezh2 alleles are shown at the right. Controls were as described for Figure 1A. (B) Heat map showing expression of PRC2 transcripts in hemopoietic tissues of wild-type (WT) controls. Expression values of genes were normalized relative to endogenous HPRT (hypoxanthine-guanine phosphoribosyltransferase) controls. (Top panel) Wild-type controls. (Bottom panel) Ezh2Δ/Δ leukemias. Transcripts are shown at the top, and mouse identification numbers and tissues analyzed are shown at the right. Red and blue represent high and low expression levels, respectively. (C) Western blot analysis of PRC2 protein levels in hemopoietic tissues of sick Ezh2Δ/Δ mice. α-Tubulin levels are shown as a loading control. Mouse identification numbers and tissues analyzed are shown at the top, the blotting antibodies are identified at the right, and the positions of molecular weight markers are indicated at the left. (D) Western blot analysis of global H3K27me2/3 levels. Histone H3 levels are shown as loading controls. Blot labels are as described for C.
Hemopoietic tissues in wild-type controls expressed detectable mRNA levels of the core PRC2 members Eed, Suz12, Ezh1, and Ezh2 (Fig. 2B, top panel), and Ezh2 deletion had no major impact on the mRNA levels of other PRC2 genes (Fig. 2B, bottom panel). In Ezh2Δ/Δ leukemias, no protein recognized by an antibody directed against the region upstream of SET domain of EZH2 (Fig. 2C) could be detected, while EZH1 and SUZ12 proteins were present. The absence of EZH2 correlated with a noticeable decrease in the global levels of H3K27me2/3 (Fig. 2D). Together with our previous work showing T-cell and B-cell tumor development in Eed mutant mice (Sauvageau et al. 2008), these observations suggested that PRC2 represents a suppressor complex with nonredundant functions in mouse T-ALL.
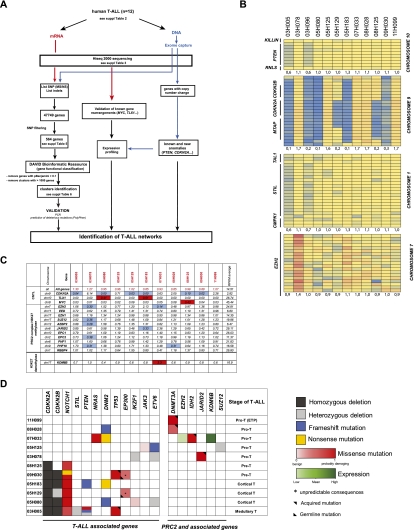
Very little is known about the integrity of PRC2 and the associated genes in human T-ALL. To address this, we analyzed a series of 12 human adult T-ALL specimens (including one paired relapsed sample: 03H005 and 03H096) (Supplemental Table 2) for nonsynonymous mutations and large and small insertions and/or deletions (indels) in PRC2 and other associated genes. Using the Illumina HiSeq2000 platform, we sequenced transcriptomes and exomes using a strategy described in Figure 3A. Sequencing and mapping statistics are shown in Supplemental Table 3. Exome coverage was determined at >45× (mean, 54.8) with 97.2 ± 0.8% of all exons captured. The density of exon sequence reads was first used for in silico reconstitution of chromosomes, which revealed a good correlation with structural and numerical changes determined by G-banding cytogenetic analyses (Fig. 3B; Supplemental Table 2). The density of the sequence reads for the selected exons also allowed for a more accurate assessment of deletions in PRC2 and other T-ALL-associated genes (n > 30; CDKN2A/CDKN2B, PTEN, etc.) (Fig. 3B). This approach was validated by fluorescent in situ hybridization (FISH) (Supplemental Table 2) and analysis of expression data from transcriptomes (Fig. 3C,D). Using this approach, we found that CDKN2A/CDKN2B are deleted in about half of our specimens (Fig. 3B). In these cases, CDKN2A expression was extremely low to undetectable (see blue in the second row of Fig. 3C). We also found deletions in STIL, PTEN (Fig. 3B), IKZF1, and ETV6 (Fig. 3D), the last two being recently associated with T-ALL (Van Vlierberghe et al. 2011; Zhang et al. 2012). Using this approach, we could not identify deletions in EED or EZH1 but found a 1.47-Mb deletion that included SUZ12 (chromosome 17: position 29059504–30538257, hg19, patient 05H125) and one patient (03H078) with an extra copy of EZH2 (Fig. 3B; Supplemental Fig. 3). Transcriptome analyses revealed that EZH2 expression levels were reduced to 30% of the mean values in this specimen (Fig. 3C; Supplemental Fig. 3). Of interest, EZH2 neighbor genes, also in more than two copies in this leukemia, were expressed at levels ∼50% higher than those detected in other leukemias (Supplemental Fig. 3B). We also found one specimen (07H033) that expressed EZH2 at only 14% of mean values. Interestingly, this leukemia expressed much higher levels of the H3K27 demethylase gene KDM6B (Fig. 3C). We observed, in other ongoing studies, that human leukemia with low EZH2 and high KDM6B levels show deregulation of HOXA gene expression (data not shown). Similar findings were observed for this 07H033 leukemia in which 5′ HOXA genes are highly expressed (Supplemental Fig. 3C).
Figure 3.
Characterization of genetic anomalies in human adult T-ALL. (A) Flow chart for the identification of genetic alterations in human T-ALL. Transcriptome and exome sequencing allowed the identification of 47,749 nonsynonymous SNPs, indels, and other anomalies (copy number variants and deletions). Following filtering (see the Materials and Methods), 564 genes were retained (listed in Supplemental Table 6). Gene annotation enrichment analysis with DAVID Bioinformatics Resources (Huang et al. 2009) identified chromatin regulators (shown in C,D; Supplemental Fig. 4) and the ubl conjugation pathway as significantly perturbed in these specimens (see Supplemental Table 7). All genetic anomalies were validated using exon sequencing, and acquired mutations were validated as described in the Materials and Methods and are shown in Supplemental Table 8. (B) Copy number variants in selected genes using exon capture sequencing (see the Materials and Methods for details). All chromosomes were manually inspected: Selected genes that appeared relevant (e.g., EZH2) or redundantly deleted (CDKN2A/B) are shown. The correlation between expression level and copy number variation is presented in Supplemental Figure 3B for EZH2. The color code refers to change from mean RPKM values (yellow) per exon. Blue and red indicate lower and higher RPKM values, respectively. (C) Expression of relevant control (CDKN2A, TLX1, and MYC) and EZH2-associated genes in all T-ALL patients studied herein. Data are expressed in relative RPKM values, with mean absolute values per gene is shown in the last column. Note high levels of MYC and TLX1 restricted to leukemia with rearrangements in these genes (see Supplemental Table 2) and low EZH2 expression in two leukemias, including one (07H033) with high KDM6B expression. (D) Representation of mutations, indels, and deletions found in T-ALL genes (left panel) and PRC2 and associated genes (right panel). The functional effects of nonsynonymous substitutions (color gradient) were predicted by the PolyPhen-2 program (http://genetics.bwh.harvard.edu/pph/data).
We next exploited transcriptome sequencing (average, 151 million ± 24 million mapped reads) (Supplemental Table 3) to identify the nonsense and missense mutations in our T-ALL specimens. More than 47,000 alleles were identified, a vast majority of which represented missense mutations of already known variants (single-nucleotide polymorphisms [SNPs]). Validated indels (not in the context of poly[A] or poly[T] sequences and likely underestimated) comprised only six genes (listed in Fig. 3D as a frameshift mutation [n = 4]; Supplemental Table 5). SNP filtering, detailed in Figure 3A and the Materials and Methods, reduced the number of SNPs to 564 (see Supplemental Table 5 for SNP position and patient attribution). Several of the newly identified SNPs occurred in DNA or chromatin-modifying genes that are functionally and/or physically linked to PRC2, such as DNMT3A (n = 2, including one with a homozygous R882 mutation) (Fig. 3D shows only acquired mutations for PRC2-associated genes), DNMT1 (n = 1) (Supplemental Fig. 4), JARID2 (core PRC2 gene, n = 1) (Fig. 3D), and IDH2 (n = 1) (Fig. 3D). DNMT3A, JARID2, and IDH2 mutations were confirmed as newly acquired, since they were not present in patient-matched normal (nonleukemic) DNA (Supplemental Table 8). Mutant IDH2 interfere with activity of jumonji histone demethylases such as KDM6B (Xu et al. 2011) and thus were included in this functional category.
Western blot analysis indicated that H3K27me1/2/3 levels were comparable between our T-ALL samples (Supplemental Fig. 5) and were much higher than those detected in our homozygous mouse Ezh2Δ/Δ tumors (cf. results in Fig. 2D [mouse] and those in Supplemental Fig. 5B [human]). Given that within a 1-year observation period no heterozygous Ezh2 mutant mice developed tumors, these results may suggest that human T cells are more sensitive to PRC2 dosage than their mouse counterparts.
Several other mutations, likely not functionally related to PRC2, were identified in the previously characterized T-ALL-associated genes: NOTCH1 (n = 6), PTEN (n = 2), and NRAS (n = 1). This group also included TP53 (n = 2) and recently reported T-ALL mutations or deletions such as DNM2 (n = 4), JAK3 (n = 1), EP300 (n = 2), IKZF1 (n = 2), and ETV6 (n = 3). A summary of the known and novel T-ALL-associated gene anomalies detected in our patient cohort is provided in the left panel of Figure 3D. Interestingly, in this patient population, NOTCH1 mutations were associated with CDKN2A/B deletions and were inversely correlated with the pro-T-cell phenotype. PRC2 and associated gene anomalies, on the other hand, were not found in cortical and medullary T-ALL (Fig. 3D). Mutations found in the genes shown in Figure 3D and Supplemental Figure 4 are presented in Supplemental Table 4.
To identify possible genetic networks affected in these leukemias, we submitted the list of modified alleles (Supplemental Table 6) to the DAVID bioinformatic resource (Huang et al. 2009; http://david.abcc.ncifcrf.gov/home.jsp) for functional classification. To rule out methodological or population biases (e.g., strong founder effect in the Quebec population), we compared functional gene clusters identified in T-ALL with those determined for normal karyotype acute myeloid leukemia (AML; 19 specimens) and found that nine of the top 15 gene clusters are distinct between the two diseases. Twenty-two functional gene clusters were found affected in T-ALL (Supplemental Table 7). Of interest, and consistent with PRC2 involvement in this disease, chromatin regulation was the category most significantly affected in this cohort (Benjamini-corrected −P-value [Huang et al. 2009] at 5 × 10−6). String analysis (Jensen et al. 2009; http://string-db.org) of this gene cluster suggested that several of the identified genes functionally interact with PRC2 (Supplemental Fig. 6).
Three of the other most significant functional clusters (ubl conjugation pathway, ligase, and ubl conjugation) are involved in the ubiquitin conjugation pathway. Most interestingly, string analysis of these genes revealed TP53 and CDKN2A/CDKN2B, also mutated or deleted in our tumors (Fig. 3D), as common targets for several of the mutated E2 or E3 ubiquitin ligase genes.
Our studies provide the first in vivo validation of the proposed tumor-suppressive activity of Ezh2 in mice. These results are in line with our previous findings indicating that a loss of Eed function sensitizes mice for the development of T-ALL and B-cell leukemia (B-ALL). Moreover, the high incidence of other PRC2 genetic alterations observed in human T-ALL, whether in the adult population (mutations: DNMT3A and JARID2; expression changes: EZH2 and KDM6B) (this study) or the childhood population (EZH2, EED, and SUZ12) (Zhang et al. 2012), argues for a more generalized deregulation of this Polycomb protein complex and associated genes in T-ALL. More globally, our results also suggest that several other regulators of the chromatin state, not necessarily linked with PRC2, may be involved in the pathogenesis of T-ALL.
Material and methods
Mice and genotyping
C57BL/6 Ezh2F/+ mice were obtained from Stuart H. Orkin (Harvard Stem Cell Institute, Boston, MA), transgenic B6.Cg-Tg(Mx1-Cre)1Cgn/J were obtained from The Jackson Laboratory, and MeuCre40 mice [EM:01372, C57BL/6-Tg(rtetR-tetO-cre) 40Mhz/Orl (MeuCre40)] were obtained from the European Mouse Mutant Archive. To generate the Ezh2F/Δ mouse strain, we crossed Ezh2F/+ mice with transgenic MeuCre40 mice and backcrossed the Ezh2Δ/+ progeny with Ezh2F/F mice. Mice were bred and manipulated in a specific pathogen-free animal facility. Animal handling followed the guidelines of the Canadian Council on Animal Care, and the experimental procedures were approved by the University of Montreal Deontology Committee on Animal Experimentation. Primers and PCR conditions used for genotyping of mice are available on request.
Human normal and leukemic samples
All of the samples used in these studies were collected by the Quebec Leukemia Cell Bank with an informed consent and approval of the project by the Research Ethics Board of the Maisonneuve-Rosemont Hospital and Université de Montréal. Seventy-eight blood or bone marrow samples used to assess the mRNA expression levels of the core PRC2 components (Supplemental Fig. 5A) comprised acute biphenotypic leukemias (n = 10), B-ALLs (n = 18), T-ALLs (n = 5), plasmocytic leukemias (n = 3), multiple myeloma (n = 5), and chronic lymphoproliferative disorders of different morphological types (n = 37). Normal peripheral blood cells were obtained from healthy volunteers and sorted to isolate the CD4+, CD8+, and CD19+ control cell populations.
Twelve T-ALL samples were analyzed in this study (nine diagnostic and three relapsed samples, including one paired relapsed sample). Morphologic, immunophenotypic, and cytogenetic characteristics are shown in Supplemental Table 2. Normal DNA paired with each of these T-ALL specimens was obtained from buccal swabs or from saliva (sample 11-H099). DNA was extracted as recommended by the manufacturers (Isohelix buccal DNA Isolation kit, Cell Projects; Oragene•DNA, DNA Genotech).
Data analysis
Sequence data were mapped to the reference genome using the Illumina Casava 1.8.1 package and Elandv2 mapping software. Seventy-five percent and 76% of the RNA-seq and exon captured sequences, respectively, were unambiguously mapped to the genome (Supplemental Table 3). On average, 97.2% of the regions targeted by the TruSeq capture kit were sequenced, with a mean fold coverage of 54.8. RNA-seq yielding 15 Gb of mapped reads per sample, with an average exon expression of 15.2 reads per kilobase per million (RPKM). Variants (SNPs and small indels) were identified using Casava 1.8.1 and were categorized as homozygous or heterozygous. We used custom-designed software to categorize each variant as occurring in introns, exons, or untranslated regions (UTRs) and identified missense and nonsense mutations (Supplemental Table 5). Specific regions were further examined across all samples for the presence of large single-allele deletions or uniparental disomy (UPD) by graphically assessing the heterozygocity of known SNPs (based on dbSNP 1.3.2). Exome data were then used to identify copy number variants at these locations by comparing local coverage between samples. Specifically, we used RPKM values computed (local scripts available on request) for all exons as a proxy for assessing local copy numbers at the exon level. To avoid exon length and sequence composition biases, we normalized these values by transforming them to Z-scores—subtracting the average and dividing by the standard deviation of the RPKM measured for all samples. However, the values obtained cannot be directly interpreted as a copy number, since they represent deviations from the mean.
SNP filtering
SNP filtering represented the critical step in our analysis, since SNPs were by far (>99.9%) the most abundant genetic anomalies detected in this study (Fig. 3A). For this, we focused only on SNPs resulting in amino acid substitution (nonsynonymous) and then excluded those present in the dbSNP Human Build 1.3.2 database (http://www.ncbi.nlm.nih.gov/projects/SNP) unless they occurred in leukemia-associated genes such as DNMT3A. Next, we focused on SNPs that occurred in expressed genes, defined here as genes with RPKM values >10 (see Wilhelm et al. 2011 for a description of expressed genes using Next-Gen Sequencing). We further filtered this data set by eliminating SNPs in which the allelic ratio was <0.39 (considered not significant; e.g., late occurrence in disease progression or early occurrence not selected for) and those for which <40 sequences were available (see mean variant depth ALL in Supplemental Table 5). Finally, SNPs common to AML (n = 19 specimens, to be reported elsewhere) and ALL (>75%) were excluded because they most likely represented misaligned sequences (pseudogenes, etc.) or previously unappreciated common variants. The final list of genetic anomalies (>560) (Supplemental Table 5) was manually validated in the sequenced exome using the Integrative Genomics Viewer version 2.0.31 (Robinson et al. 2011). Suspicious sequences (e.g., mutations following stretch of polyadenine), were eliminated from the final list. Subsets of SNPs in genes selected for this study (see Fig. 3D; Supplemental Fig. 4) were verified by genomic PCR–Sanger sequencing to determine whether the mutation was acquired in the leukemic cells or was constitutive.
Acknowledgments
We thank Stuart H. Orkin (Harvard Stem Cell Institute, Boston) for providing us with Ezh2F/+ mice. We also acknowledge Mélanie Fréchette and Andrea Evelyn Mejia Alfaro for their assistance with animal care and transplantation experiments, Julie Hinsinger from the Histology Core Facility of IRIC for preparation and staining of tissue sections, and Nadine Mayotte and Simon Girard for their excellent technical assistance. This work was supported by grants from Genome Quebec and the Canadian Cancer Society to G.S., and from the Cancer Research Network of the FRSQ, the Cole Foundation, and Genome Quebec to J.H. G.S. holds a Canada Research Chair in the Molecular Genetics of Stem Cells, and J.H. holds a Research Chair in Leukemia supported by Industrielle-Alliance (Université de Montréal).
Note added in proof
While this work was under review, two other studies describing mutations for EZH2 and other PRC2 genes in T-ALL were published (Ntziachristos et al. 2012; Zhang et al. 2012).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.186411.111.
References
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. 2010. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42: 722–726 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, Visconte V, Sugimoto Y, Prince C, O'Keefe C, et al. 2011. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 118: 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416 doi: 10.1093/nar/gkn760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G 1999. Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev 13: 2691–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Blewitt ME, de Graaf CA, McManus EJ, Bahlo M, Hilton AA, Hyland CD, Smyth GK, Corbin JE, Metcalf D, et al. 2008. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol 6: e93 doi: 10.1371/journal.pbio.0060093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, Busslinger M, Koseki H, Hu Y, Smyth GK, et al. 2010. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 116: 731–739 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, Shinga J, Koseki H, Iwama A 2011. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 118: 6553–6561 [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T 2005. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 15: 942–947 [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. 2010. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 42: 665–667 [DOI] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Vlierberghe PV, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. 2012. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie ER, Schumacher A, Angel JM, Holloway M, Rinchik EM, Magnuson T 2002. The Polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene 21: 299–306 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G 2010. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Miller M, Lemieux S, Lessard J, Hebert J, Sauvageau G 2008. Quantitative expression profiling guided by common retroviral insertion sites reveals novel and cell type specific cancer genes in leukemia. Blood 111: 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Score J, Hidalgo-Curtis C, Jones AV, Winkelmann N, Skinner A, Ward D, Zoi K, Ernst T, Stegelmann F, Dohner K, et al. 2012. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood 119: 1208–1213 [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A 2005. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 121: 425–436 [DOI] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA 2010. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7: 288–298 [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M, Tosello V, Della Gatta G, Paietta E, Racevskis J, et al. 2011. ETV6 mutations in early immature human T cell leukemias. J Exp Med 208: 2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Briau M, Austin P, Faubert A, Boucher G, Chagnon P, Hope K, Girard S, Mayotte N, Landry JR, et al. 2011. RNA-seq analysis of 2 closely related leukemia clones that differ in their self-renewal capacity. Blood 117: e27–e38 doi: 10.1182/blood-2010-07-293332 [DOI] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, et al. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117: 2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]