The transcription factor Ebf1 regulates early B lymphopoiesis by acting in a network with E2A and Pax5. However, the function of Ebf1 at later stages of differentiation in unclear. In this study, Grosschedl and colleagues investigate the role of Ebf1 in B lymphopoiesis by using conditional gene inactivation. The authors show that Ebf1 is required for proliferation and survival of pro-B and mature B cells. In addition, the proliferation defect of Ebf1fl/fl pro-B cells can be overcome by transformation with v-Abl, and the survival defect can be rescued by the expression of the Ebf1 targets c-Myb or Bcl-xL. In mature B cells, Ebf1 deficiency interferes with normal BCR/Akt signaling and the generation of germinal center B cells. Thus, these results delineate novel targets and pathways controlled by Ebf1 at different stages of lymphomagenesis.
Keywords: Ebf1, c-Myb, B-cell differentiation, BCR signaling, Ig class switch recombination, germinal center B cells
Abstract
The transcription factor Ebf1 is an important determinant of early B lymphopoiesis. To gain insight into the functions of Ebf1 at distinct stages of differentiation, we conditionally inactivated Ebf1. We found that Ebf1 is required for the proliferation, survival, and signaling of pro-B cells and peripheral B-cell subsets, including B1 cells and marginal zone B cells. The proliferation defect of Ebf1-deficient pro-B cells and the impaired expression of multiple cell cycle regulators are overcome by transformation with v-Abl. The survival defect of transformed Ebf1fl/fl pro-B cells can be rescued by the forced expression of the Ebf1 targets c-Myb or Bcl-xL. In mature B cells, Ebf1 deficiency interferes with signaling via the B-cell-activating factor receptor (BAFF-R)- and B-cell receptor (BCR)-dependent Akt pathways. Moreover, Ebf1 is required for germinal center formation and class switch recombination. Genome-wide analyses of Ebf1-mediated gene expression and chromatin binding indicate that Ebf1 regulates both common and distinct sets of genes in early and late stage B cells. By regulating important components of transcription factor and signaling networks, Ebf1 appears to be involved in the coordination of cell proliferation, survival, and differentiation at multiple stages of B lymphopoiesis.
B lymphopoiesis is a complex process of differentiation that converts lineage-committed progenitors into highly specialized antibody-secreting plasma cells. In the bone marrow, differentiation of lymphoid progenitor cells into pro-B cells involves the rearrangement of the immunoglobulin heavy chain (IgH) locus and proliferation in response to interleukin-7 (IL-7) (Dias et al. 2005; Nutt and Kee 2007; Malin et al. 2010). Successful IgH gene rearrangement generates pre-B cells that express the pre-B-cell receptor (pre-BCR) and undergo rearrangement of the Ig light chain loci to further differentiate into immature B cells. These cells egress into the peripheral lymphoid organs and are competent to respond to antigenic stimulation via the BCR. Survival of circulating mature B cells is dependent on tonic BCR signals via the PI3 kinase (PI3K)/Akt pathway and on B-cell-activating factor receptor (BAFF-R) signals (Kraus et al. 2004; Mecklenbrauker et al. 2004; Srinivasan et al. 2009). Antigenic stimulation of B cells results in the formation of germinal center (GC) B cells that undergo extensive proliferation, somatic hypermutation of Ig genes, and IgH class switch recombination (Klein and Dalla-Favera 2008).
Early specification of the B-cell lineage program and loss of alternate lineage potential are regulated by a network of transcription factors, including E2A, Ebf1, and Pax5 (for review, see Nutt and Kee 2007; Mandel and Grosschedl 2010). Loss-of-function analyses of Ebf1 and E2a (Tcf3) in mice indicated that both transcription factors are necessary for the generation of pro-B cells that undergo rearrangement of the IgH gene locus (Bain et al. 1994; Zhuang et al. 1994; Lin and Grosschedl 1995). Gain-of-function and genetic bypass experiments, in which Ebf1 was ectopically expressed in hematopoietic progenitor cells from wild-type mice or mutant mice lacking transcription factors Ikaros or Pu.1, showed that Ebf1 is involved in specifying the B-cell lineage (Zhang et al. 2003; Medina et al. 2004; Pongubala et al. 2008; Reynaud et al. 2008; Zandi et al. 2008). In addition, Ebf1 was found to repress alternative cell fate determinants (Pongubala et al. 2008; Thal et al. 2009; Lukin et al. 2011). Early B lymphopoiesis also depends on the functions of Myb (c-Myb) and Miz1, which appear to act upstream of Ebf1 and regulate survival and differentiation of pro-B cells (Fahl et al. 2009; Greig et al. 2010). Myb has been shown to regulate the expression of the α chain of the IL-7 receptor and cell survival via an IL-7R-independent pathway (Fahl et al. 2009; Greig et al. 2010). Transcription factors of the Foxo family also regulate the expression of IL-7R, whereby they act in an autoregulatory loop in which IL-7R signaling results in PI3K-dependent phosphorylation and inactivation of Foxo1 and cell cycle progression (Dengler et al. 2008; Kerdiles et al. 2009). Foxo1 has been identified as a direct target of both Ebf1 and E2A and might act downstream from these lineage determinants (Zandi et al. 2008; Lin et al. 2010; Treiber et al. 2010).
Many of the transcription factors that regulate early steps in B lymphopoiesis are also required for peripheral B-cell functions. In particular, Myb has been shown to regulate survival of splenic B cells and the expression of the BAFF-R (Thomas et al. 2005). The survival of splenic B cells is also regulated by the PI3K arm of the BCR pathway, which results in the inactivation of Foxo transcription factors (Dengler et al. 2008; Srinivasan et al. 2009). The proliferative response of splenic B cells to lipopolysaccharide (LPS) involves the function of Pax5, which also represses alternative lineage markers in mature B cells (Wakatsuki et al. 1994; Horcher et al. 2001). E2A has a relatively minor role in the generation and survival of splenic B cells (Kwon et al. 2008). However, E2A is required for the formation of GC B cells, and the E47 isoform of E2A regulates BCR editing upon exposure to self-antigen (Kwon et al. 2008; Beck et al. 2009). Moreover, class switch recombination in GC B cells depends on the regulation of activation-induced deaminase (Aid) by Foxo1 and IRF4, the activation of I promoters located upstream of the IgH constant region gene segments, and the function of enhancers in the locus control region (LCR) at the 3′ end of the IgH locus (Wolniak et al. 2004; Klein and Dalla-Favera 2008; Stavnezer et al. 2008). Despite the extensive analysis of the role of Ebf1 in the specification of the B-cell lineage, the early developmental block in Ebf1-null mice has obscured its functions at later stages of B lymphopoiesis.
Results
Ebf1 regulates early B lymphopoiesis in a network of multiple transcription factors
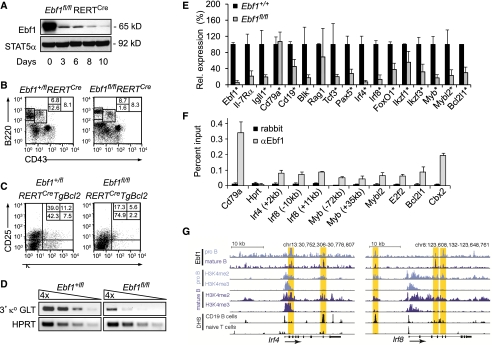
To inactivate Ebf1 conditionally at various stages of B-cell differentiation, we generated mice carrying a targeted allele of Ebf1 in which exons #2 and #3, encoding part of the DNA-binding domain, were flanked by loxP sites (Supplemental Fig. S1A). The Neo cassette of the targeting construct, flanked by Frt sites, was permanently deleted by crossing Ebf1+/fl,Neo mice with Act-FLPe mice that express the Flp recombinase (Rodriguez et al. 2000). Successful targeting of the Ebf1 locus was determined by DNA blot and PCR analysis (Supplemental Fig. S1B,C). To confirm the loss of Ebf1 function upon deletion by Cre-mediated recombination of the loxP sites, we crossed Ebf1fl/fl mice with the mb1Cre strain, which expresses Cre recombinase in early pro-B cells (Hobeika et al. 2006). Analysis of the bone marrow of Ebf1fl/flmb1Cre mice by flow cytometry revealed a block of B-cell differentiation at the pre-pro-B-cell stage, which is identical to that observed in Ebf1Δ/Δ-null mice (Supplemental Fig. S1D; Lin and Grosschedl 1995). This early developmental block in Ebf1fl/flmb1Cre mice was not rescued by the expression of a TgBcl2 transgene (Supplemental Fig. S1D).
To examine the function of Ebf1 at various stages of B-cell differentiation, we crossed Ebf1+/fl mice with RERTCre mice in which Ebf1 can be deleted in a tamoxifen-inducible manner (Guerra et al. 2003). Within 3–6 d after tamoxifen treatment of Ebf1fl/flRERTCre mice or pro-B-cell cultures, the protein and RNA expression of Ebf1 was markedly reduced in the spleen, bone marrow, fetal liver, and pro-B-cell cultures (Fig. 1A; Supplemental Fig. S1E–G). Flow cytometric analysis of Ebf1fl/flRERTCre bone marrow revealed a markedly reduced frequency of B220intCD43− pre-B cells and increased numbers of recirculating B220hiCD43− B cells, as compared with Ebf1+/flRERTCre bone marrow (Fig. 1B). We also observed a developmental block of Ebf1-deficient pre-B cells in vitro. In fetal liver cell cultures from Ebf1fl/flRERTCreTgBcl2 mice that were tamoxifen-treated and sorted for CD19+CD43+c-kit− early stage B cells, we found fewer CD25+κ− pre-B cells and CD25−κ+ immature B cells than in corresponding Ebf1+/flRERTCreTgBcl2 cultures (Fig. 1C). Consistent with a block in the differentiation of pre-B cells, we also detected fewer Igκ germline transcripts (GLTs) in Ebf1-deficient cell cultures (Fig. 1D).
Figure 1.
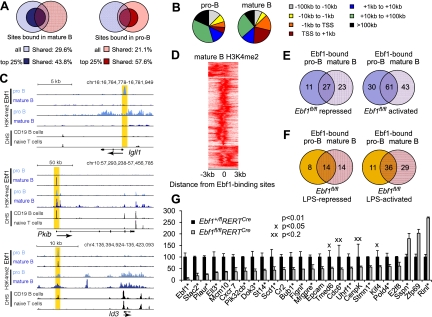
Impaired B lymphopoiesis and expression of regulatory genes in Ebf1fl/flRERTCre bone marrow. (A) Immunoblot analysis to detect Ebf1 and Stat5a in lysates of splenic B cells from mice that were treated with tamoxifen 4-OHT (3 mg) on two consecutive days and sacrificed on various days after first administration. (B) Flow cytometric analysis of bone marrow to detect B220+CD43+ pro-B cells, B220intermediateCD43− pre-B cells, and B220highCD43− recirculating B cells. Numbers in quadrants indicate percentage of cells. (C) Flow cytometric analysis to detect differentiation and generation of CD25+κ− pre-B cells and CD25−κ+ immature B cells from CD19+CD43+c-kit− pro-B cells in fetal liver cell cultures. Cells cultured in the presence of OP9 feeders and IL-7 were treated with 2 μM 4-OHT for 24 h and cultured for an additional 4 d. Differentiation was induced by withdrawal of IL-7. (D) Semiquantitative RT–PCR analysis of κ light chain GLTs in pro-B cells that were cultured in the presence of OP9 feeders and IL-7 for 6 d and treated with 2 μM 4-OHT during the first day of culture. Data are representative of at least four (A) or three (B–D) experiments. (E) Quantitative RT–PCR analysis to examine the expression of B-cell-specific regulatory genes in pro-B-cell cultures. Data represent mean values of three independent biological replicates, and the raw cycle values were normalized to actin expression. Error bars indicate standard deviation (SD). (F) Quantification of Ebf1 binding to target genes in 38B9 cells by ChIP and quantitative PCR analysis. Binding is represented as percentage of input chromatin, and error bars represent SD of duplicate ChIP experiments (see also Supplemental Fig. S1; Supplemental Table S1). (G) Sequence tag profiles in pro-B cells and splenic B cells at the Irf4 and Irf8 loci. Regions of Ebf1 occupancy and H3K4me2 and H3K4me3 modifications in both pro-B cells (Lin et al. 2010) and splenic B cells (this study), as well as DNase I hypersensitivity sites (DHS) in CD19+ B cells and naive T cells (http://genome.ucsc.edu/ENCODE; Sabo et al. 2006), are shown. Tag density profiles are visualized using the University of California at Santa Cruz (UCSC) Genome Browser. Vertical yellow bars highlight genomic positions of Ebf1-binding sites that were identified by ChIP-seq analysis in pro-B cells (Lin et al. 2010; Treiber et al. 2010) and splenic B cells (this study).
Differentiation of pre-B cells is dependent on transcription factors Foxo1, Irf4, and Irf8, whereby Foxo1 regulates IL-7Rα expression in pro-B cells and Igκ rearrangement in pre-B cells (Amin and Schlissel 2008; Dengler et al. 2008; Herzog et al. 2008). Irf4 and Irf8 inhibit cell proliferation and promote κ rearrangement by binding to the κ 3′ enhancer (Lu et al. 2003; Lazorchak et al. 2006; Johnson et al. 2008). Ebf1-deficient pro-B cells showed impaired expression of Foxo1 and additional Ebf1-bound transcription factor genes, including Irf4, Irf8, Tcf3 (E2a), Pax5, Aiolos (Ikzf3), Ikaros (Ikzf1), Myb, and Mybl2 (Fig. 1E; Supplemental Fig. S1H; Lin et al. 2010; Treiber et al. 2010). We also observed reduced expression of IL-7Rα and Bcl2l1. We confirmed direct binding of Ebf1 to Irf4, Irf8, Myb, Mybl2, and Bcl2l1 sequences by chromatin immunoprecipitation (ChIP) analysis (Fig. 1F). Moreover, previous ChIP-seq analysis of pro-B cells indicated that Ebf1 peak regions in the Irf4 and Irf8 loci overlapped with regions of H3K4me2 chromatin modification (Fig. 1G; Lin et al. 2010; Treiber et al. 2010). No Ebf1 binding was detected at the IL-7Rα and Igκ loci (data not shown), suggesting that the decreased IL-7Rα and κ germline transcription in Ebf1-deficient pro-B cells is likely due to the impaired expression of Foxo1 and Irf4/8, respectively. Thus, Ebf1 regulates early B-cell differentiation not only in combination with E2A and Pax5, but in an extended network that includes Foxo and Irf transcription factors.
Ebf1 has been implicated in the repression of alternative lineage determinants. In particular, the Id2 gene has been found to be repressed by Ebf1 (Pongubala et al. 2008; Thal et al. 2009). Moreover, several genes that are expressed in natural killer (NK) cells are up-regulated in pro-B-cell Ebf1+/− pro-B cells (Lukin et al. 2011). Therefore, we examined whether the conditional inactivation of Ebf1 in pro-B cells results in derepression of genes specifically expressed in myeloid cells, T cells, or NK cells. With the exception of Fcer1γ, no significant changes in myeloid gene expression were observed (Supplemental Fig. S2A; data not shown). Among the other genes examined, we only observed deregulation of Embigin and Flt3 (Supplemental Fig. S2A). These data indicate that the suppression of alternative lineage markers is generally maintained immediately after conditional inactivation of Ebf1. Unfortunately, the proliferation defect of Ebf1-deficient pro-B cells obscured an assessment of the maintenance of gene expression after multiple rounds of cell divisions.
Survival and proliferation defects of Ebf1-deficient pro-B cells are rescued by forced Myb expression and A-MuLV transformation, respectively
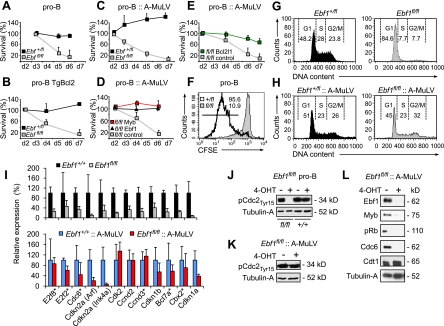
To examine the effects of Ebf1 inactivation on pro-B-cell survival, we determined the numbers of annexin V-positive and annexin V-negative cells between 2 and 6 d after tamoxifen-induced deletion of Ebf1 in vitro. Consistent with the decrease of IL-7Rα expression in Ebf1-deficient pro-B cells, the frequency of annexin/7-AAD-double-negative cells was markedly reduced (Fig. 2A). This defect was not overcome by the transgenic expression of Bcl2 or the transformation of mutant cells with A-MuLV, which overcomes the need of IL-7R signaling (Fig. 2B,C; Danial et al. 1995; Beck et al. 2009). Therefore, the cell death of Ebf1-deficient pro-B cells is not a consequence of only defective IL-7R signaling.
Figure 2.
Ebf1 regulates proliferative expansion and survival of pro-B cells. (A) Analysis of apoptosis of pro-B cells cultured in the presence of feeders and IL-7 after induced deletion of Ebf1. Numbers of viable (annexin V- and PI-negative) cells were determined by flow cytometry at the indicated time points after 4-OHT treatment, and the data were converted to the percentage of viable cells at day 3. Line graphs represent the average survival of three independent cell cultures at various days (d). (B,C) Analysis of survival of primary Ebf1fl/flRERTCreTgBcl2 pro-B cells (n = 3) and transformed Ebf1fl/flRERTCre::A-MuLV pro-B cells (n = 4), as described above. (D,E) Analysis of apoptosis in A-MuLV-transformed Ebf1fl/fl pro-B cells that have been transduced with a GFP-expressing retrovirus or bicistronic retroviruses expressing both GFP and various Ebf1 target genes. Cells sorted for GFP expression were treated with 4-OHT, and the percentages of viable cells were determined at the indicated time points after treatment (n = 4). Full and partial rescue of the survival defect of Ebf1-deficient cells is observed by expression of c-Myb and Bcl2l1, respectively. (F) Flow cytometric analysis of the proliferation of Ebf1+/flRERTCre (thick line) and Ebf1fl/flRERTCre (gray fill) pro-B cells treated with 2 μM 4-OHT for 24 h, cultured for 2 d without 4-OHT, labeled with CFSE, and assessed by CFSE dilution after 3 d (n = 4). Numbers indicate the percentage of cells in the area marked by the thin bar. (G,H) Flow cytometric analysis of cell cycle progression of Ebf1+/flRERTCre (black fill) and Ebf1fl/flRERTCre (gray fill) primary pro-B (G) or A-MuLV-transformed pro-B (H) cell cultures 5 d after 4-OHT administration. Cells were fixed, stained with PI to assess DNA content, and gated for intact cells. Representative plot and quantitative analysis of cell cycle distribution are indicated (n = 3). Numbers indicate percentage of cells. (I) Quantitative RT–PCR analysis to determine the expression of regulatory genes involved in cell cycle or survival and identified as indirect or direct (*) Ebf1 targets. Primary pro-B cells (top panel) and A-MuLV-transformed pro-B cells (bottom panel) were harvested 5 d after 4-OHT treatment. Data represent mean values of three biological replicates, and raw cycle values were normalized to actin. Ebf1+/+ samples were set to 100%. (J,K) Immunoblot analysis to detect Cdc2 phosphorylation on Tyr 15 in primary (J) and A-MuLV-transformed (K) Ebf1fl/flRERTCre pro-B cells. (L) Immunoblot analysis to detect Rb phosphorylation and expression of Ebf1, c-Myb, Cdc6, and Cdt1 in A-MuLV-transformed Ebf1fl/flRERTCre pro-B cells.
To attempt to rescue the survival defect of primary or A-MuLV-transformed Ebf1fl/fl pro-B cells, we transduced mutant cells with a GFP-expressing control retrovirus or bicistronic retroviruses expressing both GFP and various Ebf1-regulated target genes. Expression of IL-7Rα or a constitutive active form of Stat5 failed to overcome the survival defect of primary and transformed Ebf1fl/fl pro-B cells (Supplemental Fig. S2B; data not shown). Myb rescued the survival defect of transformed Ebf1fl/fl pro-B cells as efficiently as Ebf1, although Myb was not sufficient to allow for survival of primary Ebf1fl/fl pro-B cells (Fig. 2D; Supplemental Fig. S2B). We also observed a partial rescue of transformed Ebf1fl/fl pro-B cells with Bcl2l1 (Fig. 2E), consistent with its role in B-cell survival (Grillot et al. 1996; Cheng et al. 2003). Despite its implied role in pro-B-cell survival, the Ebf1 target Foxo1 did not rescue the survival defect of transformed Ebf1fl/fl pro-B cells (Supplemental Fig. S2C). Quantitative RT–PCR analysis indicated that forced Myb expression restored the expression of Bcl2l1, Foxo1, and Ikzf1, but did not rescue the impaired expression of IL-7Rα and Igll1 (λ5) (Supplemental Fig. S2D). Thus, Myb acts downstream from Ebf1 to regulate the survival of pro-B cells and the expression of Bcl2l1, but is not sufficient to overcome the requirement for Ebf1 function in cell cycle progression.
We also determined the effects of Ebf1 deletion on cell proliferation and cell cycle progression by performing flow cytometric analysis of cells labeled with CFSE or PI. Ebf1fl/fl pro-B cells showed marked proliferation and cell cycle defects (Fig. 2F,G). In contrast to the survival defect, the cell cycle defect of primary Ebf1-deficient pro-B cells, which accumulate in the G1 phase, was overcome by transformation with A-MuLV (Fig. 2H). Thus, Ebf1 regulates proliferation and survival of pro-B cells by distinct mechanisms.
Cell cycle progression is regulated by cyclins, cyclin-dependent kinases (CDKs), retinoblastoma proteins, and CDK inhibitors. In particular, D-type and E-type cyclins regulate G1-phase progression and G1/S transition, respectively. D-type cyclins integrate extracellular signals to G1-phase progression, and complexes of Cdk4/5 and D-cyclins enhance Rb phosphorylation, resulting in activation of E2F transcription factors. Several of these genes are directly or indirectly regulated by Ebf1 (Supplemental Table S1; Lin et al. 2010; Treiber et al. 2010). By quantitative RT–PCR analysis of primary Ebf1-deficient pro-B cells, we confirmed marked changes in the expression of direct Ebf1 targets implicated in cell cycle progression (Fig. 2I). These genes include E2f2, E2f8, Cdk2, cyclin D2 (Ccnd2), and cyclin D3 (Ccnd3), as well as Cdc6, an essential regulator of the initiation of DNA replication that is also under the control of E2Fs. In addition, the expression of the prosurvival gene Bcl2l1, but not that of Bcl2a1 and Mcl1, was altered in primary and transformed Ebf1-deficient pro-B cells (Fig. 1E; Supplemental Fig. S2D; data not shown). A-MuLV-mediated transformation targets the expression of both D-type cyclins and E2Fs (Coutts et al. 2000; Parada et al. 2001). In A-MuLV-transformed Ebf1fl/fl pro-B cells, E2f8, Ccnd2, and Ccnd3 were expressed at similar levels in Ebf1fl/fl and Ebf1+/fl pro-B cells (Fig. 2I), suggesting that v-Abl compensates for the function of Ebf1 in the regulation of these genes and allows for cell cycle progression in the absence of Ebf1.
In primary Ebf1-deficient pro-B cells, we also detected a decrease in Ccnd3 RNA and protein expression (Fig. 2I; Supplemental Fig. S2E). However, we could not rescue the cell cycle defect by forced expression of Ccnd3 (Supplemental Fig. S2F,G). Primary Ebf1-deficient pro-B cells also showed impaired expression of Cdk2 and the absence of phosphorylation of Cdc2 (Cdk1) (Fig. 2I,J). Phosphorylation of Cdc2 and expression of Cdk2 were restored in transformed Ebf1-deficient pro-B cells, whereas the phosphorylation of Rb and the expression of Cdc6 remained low (Fig. 2K,L). This observation raises the possibility that an origin activation checkpoint is bypassed in transformed Ebf1-deficient pro-B cells, allowing them to proceed into an abortive S phase, followed by apoptosis (Tudzarova et al. 2010).
Ebf1 regulates generation of B1 and marginal zone (MZ) B cells as well as BAFF-R-mediated B-cell survival
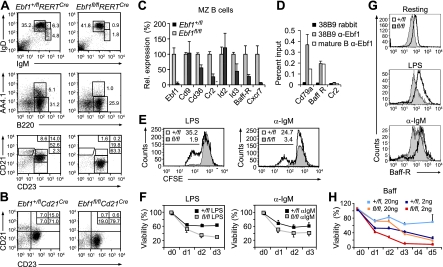
To gain insight into the role of Ebf1 in peripheral B cells, we analyzed splenic B cells in tamoxifen-treated Ebf1fl/flRERTCre mice, as well as in Ebf1fl/flCd21Cre and Ebf1fl/flCd19Cre mice, which delete the floxed genes in transitional and mature B cells (Rickert et al. 1997; Kraus et al. 2004). In tamoxifen-treated Ebf1fl/flRERTCre mice, the frequencies and numbers of CD19+ splenic B cells were similar to those found in Ebf1+/flRERTCre mice (Supplemental Fig. S3A,B). However, homozygous mutant mice had markedly reduced frequencies of transitional B cells (B220+AA4.1+), immature B cells, and MZ B cells (CD21+CD23−) (Fig. 3A). In these mutant mice, follicular (FO) B cells (B220+IgM+IgD+) were found at normal frequencies but showed reduced surface expression of CD21 (Fig. 3A).
Figure 3.
Roles of Ebf1 in peripheral B-cell subsets and BAFF-R-mediated cell survival. (A) Flow cytometric analysis of Ebf1+/flRERTCre and Ebf1fl/flRERTCre splenocytes to detect IgMhiIgD+ immature B cells and IgMhiIgDhi mature B cells (top panels), IgMhiIgDhiB220+AA4.1+ transitional B cells (middle panels), and B220+CD23−Cd21hi MZ B cells and B220+CD23+CD21+ FO B cells (bottom panels). Numbers in quadrants indicate percentage of cells. (B) Flow cytometric analysis of Ebf1+/flCd21Cre and Ebf1fl/flCd21Cre splenocytes to detect MZ B cells and FO B cells. (C) Quantitative RT–PCR analysis of Ebf1-regulated targets in sorted MZ B cells. Fold expression values are relative to heterozygote control cells. (D) ChIP analysis to examine binding of Ebf1 to the Tnfrsf13c (Baff-R) locus in 38B9 pro-B cells or MACS-enriched splenic B cells. Binding is represented as percentage of input chromatin. (E) Flow cytometric analysis of proliferation of splenic B cells that were depleted of non-FO B cells with antibodies directed against CD43, CD4, CD8a, Gr1, AA4.1, and CD9. Cells sorted 10 d after the initial 4-OHT treatment were CFSE-labeled and stimulated with LPS or αIgM F(ab′)2. Proliferation was determined by CFSE dilution 3 d after stimulation. Numbers indicate percentages of cells in the area marked by the thin bar. (F) Analysis of apoptosis of LPS- or α-IgM-stimulated splenic B cells at the specified days (d) (n = 3). (G) Flow cytometric analysis of surface expression of BAFF-R in resting and stimulated splenic B cells. Cells were activated with the indicated stimuli for 40 h. The graph is representative of three experiments. (H) Analysis of the survival of Ebf1+/flRERTCre and Ebf1fl/flRERTCre resting splenic B cells in the presence of optimal (20 ng/mL) and limiting (2 ng/mL) exogenous BAFF (n = 3).
In Ebf1fl/flCd21Cre and Ebf1fl/flCd19Cre mice, MZ B cells were also diminished (Fig. 3B; Supplemental Fig. S3C–E). Moreover, FO B cells showed a reduced surface expression of CD21 and CD19 (Fig. 3B; Supplemental Fig. S3E,F). Thus, the entire CD19/CD21 coreceptor complex is affected by the inactivation of Ebf1. In residual MZ B cells and in FO B cells that express CD21 at a high level, we detected abundant unrecombined Ebf1fl alleles or expression of Ebf1 (Supplemental Fig. S3E,G). In contrast, very few unrecombined Ebf1fl alleles were found in the corresponding Ebf1+/fl cell populations. These findings suggest that MZ B cells with recombined Ebf1fl/fl alleles have been depleted and replaced by cells that have escaped inactivation of Ebf1.
Analysis of peritoneal B cells in Ebf1fl/flRERTCre and Ebf1fl/flCd21Cre mice revealed an almost complete loss of B1 cells (Supplemental Fig. S3H,I; data not shown). Thus, Ebf1 is required for the generation and/or maintenance of transitional and MZ B cells in the spleen and B1 cells in the peritoneum, but is dispensable for the maintenance of resting FO B cells.
By quantitative RT–PCR analysis of Ebf1fl/flRERTCre MZ B cells, sorted 6 d after tamoxifen treatment to minimize the appearance of cells with nondeleted alleles, we found reduced expression of Id3 and Cxcr7, which regulate the generation and maintenance of MZ B cells, respectively (Fig. 3C; Quong et al. 2004; Wang et al. 2012). In Ebf1-deficient MZ B cells, we also observed reduced expression of Baff-R (Tnfrsf13c) and Cr2 (Cd21). ChIP analysis to detect binding of Ebf1 to these gene loci indicated that Baff-R is a direct target of Ebf1 in both pro-B and mature B cells (Fig. 3D).
Stimulation of sorted resting B cells from Ebf1fl/flRERTCre and Ebf1+flRERTCre mice by LPS, CpG, or anti-IgM indicated that proliferation of activated Ebf1fl/fl cells, as determined by flow cytometric tracing of CFSE, was markedly impaired relative to Ebf1+/fl cells (Fig. 3E; Supplemental Fig. S4A). Moreover, the frequencies of cells in S phase of the cell cycle, measured by incorporation of EDU, were lower in Ebf1fl/fl cells as compared with Ebf1+/fl cells (Supplemental Fig. S4B). To determine whether the pronounced proliferation defect of LPS-stimulated Ebf1fl/fl cells includes a signaling defect, we examined the nuclear translocation of NF-κB. The nuclear accumulation of c-Rel and p65 was similar with LPS-stimulated Ebf1fl/fl and Ebf1+/fl cells, suggesting that the defect does not involve impaired signaling (Supplemental Fig. S4C).
We also determined the survival of stimulated Ebf1fl/flRERTCre and Ebf1+/flRERTCre splenic B cells in vitro and found a decrease in the survival of Ebf1fl/fl B cells, irrespective of the mode of stimulation (Fig. 3F; Supplemental Fig. S4D). Survival of mature B cells depends on two signaling determinants, BCR and BAFF-R, which monitor the presence of antigen and neighboring cells, respectively (Mecklenbrauker et al. 2004; Beck et al. 2009; Srinivasan et al. 2009). Analysis of the surface expression of BAFF-R by flow cytometry revealed a reduction in resting and stimulated Ebf1fl/fl B cells, whereby the defect was more pronounced in stimulated cells (Fig. 3G). To further examine the requirement of Ebf1 for BAFF-mediated B-cell survival, we determined the effects of exogenous BAFF ligand in unstimulated in vitro cultures. Addition of BAFF rescued, at least in part, the survival defect of Ebf1fl/fl splenic B cells in a dose-dependent manner (Fig. 3H). However, we did not observe an enhanced rescue of the survival defect by combining either BAFF with LPS or IL10 and IL6, or anti-IgM with anti-CD40 treatment (data not shown).
Impaired BCR and Akt signaling in Ebf1-deficient B cells
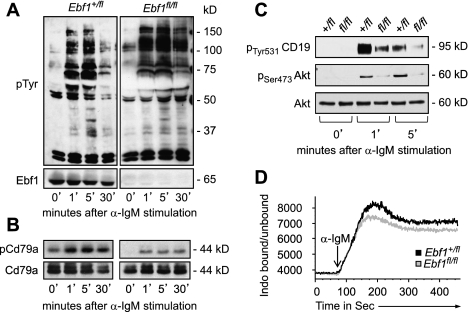
The BCR activates several signaling pathways, whereby the BCR-mediated survival signal is governed by PI3K/Akt signaling (Srinivasan et al. 2009; Okkenhaug and Fruman 2010; Werner et al. 2010). Ebf1 regulates multiple components of BCR signaling pathways, including Cd79a (Igα), Cd79b (Igβ), and Cd19. In anti-IgM-stimulated Ebf1fl/fl B cells, tyrosine phosphorylation of a subset of proteins, including proteins migrating at ∼75 and ∼37 kDa, is impaired (Fig. 4A). In particular, we observed reduced phosphorylation of CD79a, despite normal CD79a protein expression (Fig. 4B). Moreover, phosphorylation of CD19, a BCR-proximal process, was reduced in anti-IgM-stimulated Ebf1fl/fl B cells (Fig. 4C). Consistent with the phosphorylation defects, Ca2+ mobilization in response to anti-IgM stimulation was also impaired in Ebf1fl/fl B cells (Fig. 4D). We also examined the PI3K arm of BCR by analyzing the phosphorylation of Akt/PKB. BCR-stimulated phosphorylation of Akt was markedly reduced (Fig. 4C), suggesting that Ebf1 regulates both canonical and Akt-mediated BCR signaling. In resting Ebf1-deficient B cells, we did not detect an altered expression of genes that are regulated by tonic Akt signaling (Supplemental Fig. S4E; Srinivasan et al. 2009), suggesting that Ebf1 is required for stimulated but not tonic BCR-mediated Akt signaling.
Figure 4.
Role of Ebf1 in BCR and Akt signaling. (A) Immunoblot analysis of tyrosine phosphorylation in MACS-purified and αIgM F(ab′)2-stimulated Ebf1+/flRERTCre and Ebf1fl/flRERTCre splenic B cells in vitro. Efficient deletion of Ebf1 is shown in the bottom panel. (B) Immunoblot analysis of anti-CD79a-immunoprecipitated protein to detect phosphorylated and total CD79a protein. (C) Immunoblot analysis of phosphorylated CD19 (pCD19) (top panel) and Akt (middle panel) in FACS-purified splenic B cells stimulated with αIgM F(ab′)2 for the indicated times. Blots are representative of three independent experiments. (D) Analysis of experimentally induced Ca2+ fluxes in resting mature B cells of Ebf1+/fl and Ebf1fl/flRERTCre mice. Indo-1AM-loaded cells were stimulated with αIgM F(ab′)2, and free intracellular Ca2+ was measured in real time. Data are representative of six independent experiments.
Analysis of Ebf1 targets in mature B cells by RNA expression and ChIP-seq analyses
Microarray analysis of resting splenic B cells of Ebf1+/flRERTCre and Ebf1fl/flRERTCre mice revealed differences in the expression of multiple cell cycle- and signaling-related genes (Supplemental Fig. S5A,B). Of ∼200 genes that showed threefold or higher differences in expression, 80% and 20% were down-regulated and up-regulated in Ebf1fl/fl cells, respectively. Among the Ebf1-activated genes, we identified several genes related to cell cycle control, such as Mcm10, Bub1, and E2f8, as well as genes regulating BCR/Akt signaling, including Cr2, Dok3, and Hck. However, we detected no significant changes in the expression of known Ebf1 target genes, like Cd79a, Pax5, and Blnk (data not shown).
Many putative Ebf1 target genes differed from the target genes previously identified in our microarray and ChIP-seq analyses of pro-B cells (Treiber et al. 2010). To identify Ebf1-bound genes in mature splenic B cells, we performed a ChIP-seq analysis. In total, we identified 7424 significant Ebf1 peaks by comparing Ebf1 and control data sets using the CCAT peak-calling algorithm (Treiber et al. 2010). Assigning the Ebf1-binding sites to genes within 100 kb of the transcription start sites (TSSs), we found 4825 genes that are putatively bound by Ebf1 in mature B cells (Supplemental Table S2). To assess the quality of the ChIP-seq data, we performed quantitative ChIP experiments on 46 randomly chosen ChIP-seq peaks and could validate 15 of the peaks, which correspond to a greater than eightfold enrichment of Ebf1 tags (Supplemental Table S3). For further analysis, we used the top 25% of the peaks (1528 genes), corresponding to the eightfold enrichment. We also compared the data sets with our previous data sets of Ebf1 binding in pro-B cells. We found that ∼30% of genes bound in pro-B cells overlapped with those bound in mature B cells (Fig. 5A, left panel). Using the top 25% of the pro-B-cell library, this fraction of overlapping sites increased to ∼44%. Conversely, ∼58% of genes bound in mature B cells overlapped with those bound in pro-B cells when the top 25% of the mature B-cell library were used for comparison (Fig. 5A, right panel). This analysis also showed that the distribution of Ebf1-binding sites is altered in mature B cells and includes significantly more binding sites in promoter-proximal regions (Fig. 5B). Bioinformatic analysis of Ebf1-binding regions to detect overrepresented transcription factor-binding sites identified different motifs than those found in pro-B cells. In mature B cells, Sp1 and E2F transcription factor-binding motifs were overrepresented in Ebf1-bound regions (Supplemental Table S4).
Figure 5.
Analysis of Ebf1 binding and Ebf1-mediated gene expression in mature B cells. (A) Overlap of genes occupied by Ebf1 in splenic B cells, as determined by ChIP-seq analysis, with Ebf1-bound genes in pro-B cells (Treiber et al. 2010). Shades of blue and red represent genes bound in pro-B cells and mature B cells, respectively. Dark colors represent Ebf1 targets in the top 25% of either library (see Supplemental Table S1). (B) Distribution of Ebf1-bound regions relative to annotated gene loci in mature B cells and pro-B cells. (C) Association of Ebf1-bound regions with H3K4me2 modification and DHS in Igll1, Pkib, and Id3 as representatives for genes bound by Ebf1 at different stages of differentiation. UCSC Genome Browser was used to visualize binding patterns. DHS data were obtained from ENCODE (http://genome.ucsc.edu/ENCODE). Vertical yellow band pattern indicates the position of Ebf1-binding sites. (D) Correlation of Ebf1 occupancy and H3K4me2 patterns in mature B cells. The heat map reflects the enrichment of H3K4 dimethylation in the vicinity of the top 25% Ebf1 peaks in mature B cells. The center of the Ebf1-binding sites in mature B cells is represented by 0. (E,F) Overlap of Ebf1-bound genes in pro-B cells and mature B cells (<100 kb from the TSS) that were also identified as functional targets by loss-of-function microarray analyses in unstimulated B cells (E) and LPS-stimulated B cells (F). The numbers represent Ebf1-bound genes that were identified as genes differentially expressed in Ebf1fl/flRERTCre versus Ebf1+/flRERTCre B cells. The identity of the genes and positions of the binding sites are shown in Supplemental Tables S5–S8. (G) Quantitative RT–PCR analysis of Ebf1-regulated targets that were identified in the microarray analysis. A representative set of 26 genes of the top 200 Ebf1-regulated target genes is shown, and fold expression values are relative to heterozygote control cells. Ebf1-bound genes among these targets are marked by an asterisk (see also Supplemental Fig. S3; Supplemental Tables S5, S6).
Previous analysis of Ebf1 binding in pro-B cells revealed a good correlation of Ebf1 binding and H3K4me2 modifications (Treiber et al. 2010). By ChIP-seq analysis of H3K4me2 and H3K4me3 modifications, we also observed this correlation in mature B cells, although in some genes with Ebf1 binding only in mature B cells, such as Pkib, H3K4me2 modifications can already be detected in pro-B cells (Fig. 5C,D).
Matching the data of the microarray analysis of resting splenic B cells of Ebf1fl/fl mice with the ChIP-seq data set indicated that 104 of the Ebf1-occupied genes were down-regulated and 50 were up-regulated relative to Ebf1+/fl B cells (Fig. 5E; Supplemental Tables S5, S6). We also examined the effects of Ebf1 deletion on gene expression in LPS-stimulated B cells. Microarray analysis indicated that a relatively small subset of LPS-regulated genes is dependent on Ebf1, consistent with our finding that NF-κB-mediated gene induction is not impaired in Ebf1fl/fl B cells (Fig. 5F; Supplemental Fig. S6; Supplemental Tables S7, S8). Virtually all of the functional Ebf1 targets that were confirmed by quantitative RT–PCR were also identified in the ChIP-seq analysis, suggesting that Ebf1 may directly regulate these genes (Fig. 5G; Supplemental Table S2).
Impaired GC formation in the absence of Ebf1
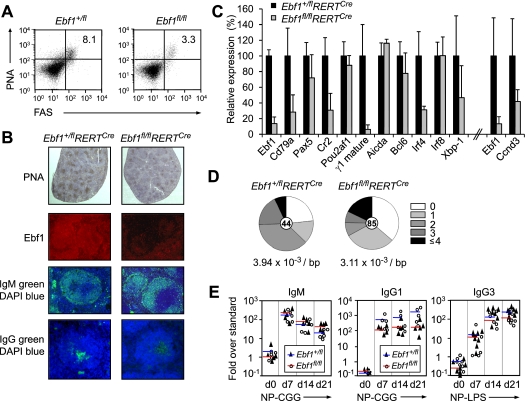
Activation of splenic B cells by antigen encounter results in the generation of GC B cells that undergo class switch recombination and somatic hypermutation (Klein and Dalla-Favera 2008). To assess the role of Ebf1 in GC formation, we treated Ebf1fl/flRERTCre mice with tamoxifen in vivo and subsequently immunized the mice with sheep red blood cells (SRBCs). Seven days after immunization, the number of splenic GC B cells (B220+PNA+FAShi) was not significantly elevated above basal levels in Ebf1fl/flRERTCre mice in contrast to Ebf1+/flRERTCre mice (Fig. 6A). We also detected impaired proliferation of Ebf1-deficient GC B cells, as determined by BrDU incorporation in vivo (Supplemental Fig. S7A). Twelve days after immunization, however, the reduction of the number of GC B cells in Ebf1fl/flRERTCre mice was less severe, which could be accounted for by the accumulation of cells with nonrecombined Ebf1fl/fl alleles (Supplemental Fig. S7B). A progressive loss of Ebf1-deficient GC B cells was also inferred from the analysis of Ebf1fl/flCγ1Cre mice. In these mice, Cre recombinase is induced specifically in GC B cells by transcription of the Igγ1 constant region gene segment 4 d after immunization with T-cell-dependent (TD) antigens (Casola et al. 2006). In immunized Ebf1fl/flCγ1Cre mice, we detected two to three times fewer GC B cells by flow cytometry, and the frequency of nonrecombined Ebf1fl/fl alleles increased between 7 and 12 d after immunization, consistent with a requirement for Ebf1 in the maintenance of GC B cells (Supplemental Fig. S7C).
Figure 6.
Impaired GC development in the spleens of Ebf1fl/flRERTCre mice. (A) Flow cytometric analysis to detect GC B cells in the spleens of SRBC-immunized Ebf1fl/flRERTCre and Ebf1+/flRERTCre mice. Mice were immunized 8 d after the first 4-OHT treatment and were analyzed 6 d after immunization. Samples were gated on living B220+ cells, and the numbers on the FACS plots represent the percentage of PNA+FAShi GC B cells (n = 6). (B) Analysis of immune responses of Ebf1fl/flRERTCre mice. Spleen sections were stained with PNA (top panel) to reveal GC cells, and with antibodies to IgM (green) to reveal B-cell follicles and total IgG (green, bottom panel). Nuclei were stained with DAPI. (C) Gene expression analysis of sorted PNA+FAShi GC B cells of the indicated genotypes by quantitative RT–PCR. Black and gray bars represent the averages of individually sorted samples (n = 3). (D) Normal somatic hypermutation of Ebf1fl/flRERTCre GC B cells upon immunization with SRBCs. Mice were immunized as described in A; genomic DNA was extracted from sorted GC B cells and subjected to nested PCR amplification with two degenerate V and J segment primer pairs, and the amplified products corresponding to JH3 were separated and cloned. The number of clones sequenced is shown in the circles in the middle of the pie diagrams; each diagram is representative of two independent experiments, with at least 40 cloned sequences each. (E) Relative titers of immunoglobulin subtypes in sera of Ebf1+/flRERTCre mice (black triangles and blue lines represent individual samples and averages, respectively) and Ebf1fl/flRERTCre mice (open circles and red lines represent individual samples and averages, respectively) at the specified days (d), determined by ELISA.
To gain insight into the defect of Ebf1-deficient GC B cells, we immunized Ebf1+/fl and Ebf1fl/flRERTCre mice with SRBCs and analyzed the spleen by immunohistochemistry 7 d after immunization. In Ebf1fl/fl spleens, we detected a significant decrease in the size of PNA-positive GCs and in the staining for IgG-positive cells (Fig. 6B). However, the staining for IgM was similar in Ebf1fl/fl and Ebf1+/fl spleens. Quantitative RT–PCR analysis of genes characteristic of GC B cells revealed down-regulation of Cr2, IgCγ1, and Irf4 in Ebf1fl/fl GC B cells (Fig. 6C). Cr2, encoding CD21, has been implicated in the maintenance of GCs (Fischer et al. 1998; Srinivasan et al. 2009), and Irf4 is a key transcription factor for GC B-cell formation (Klein and Dalla-Favera 2008). In ChIP-seq and ChIP analyses, we detected binding of Ebf1 to an intronic DNase hypersensitivity site (DHS) of Irf4 in mature B cells but not in pro-B cells (Fig. 1G). However, no significant alteration in somatic hypermutation was detected in immunized Ebf1fl/flRERTCre mice (Fig. 6D). Finally, we examined the secretion of different classes of antibodies by immunizing tamoxifen-treated mice with either T-cell-independent (TI) antigen (NP-LPS) or TD antigen (NP-CGG). Analysis of secreted antibodies by ELISA indicated that the IgM titers were similar in Ebf1+/fl and Ebf1fl/fl mice, whereas the levels of IgG3 and IgG1 were reduced in Ebf1fl/fl mice by a factor of three and a factor of five, respectively (Fig. 6E).
Similar numbers of CD138+CD19− cells were detected in the spleen of Ebf1fl/fl and Ebf1+/fl mice 2 d after immunization, and the expression of genes characteristic of plasma cells was similar in these cells (Supplemental Fig. S7D,E), suggesting that Ebf1 is dispensable for the differentiation of antibody-secreting cells.
Ebf1 regulates Cγ1 and Cγ3 germline transcription and binds to the IgH 3′ LCR
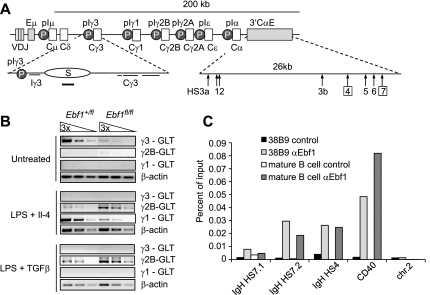
IgH class switch recombination is dependent on cytokine-induced generation of germline transcription that initiates from intronic promoters that are located upstream of the constant region gene segments (Stavnezer et al. 2008). In addition, class switching requires multiple cycles of cell division and the expression of Aid (Rush et al. 2005; Stavnezer et al. 2008). We examined the generation of GLTs as an early marker for class switch recombination in sorted CD21lo splenic B cells from Ebf1fl/fl and Ebf+/fl mice because the proliferation defect of Ebf1-deficient B cells precluded an analysis of class switching in vitro (Rush et al. 2005). In unstimulated and LPS-stimulated Ebf1fl/fl cells, the generation of γ3 GLTs was markedly impaired (Fig. 7A,B; data not shown). In Ebf1fl/fl cells stimulated with LPS and IL-4 to favor switching to IgG1 and IgE, we observed a decrease in γ1 GLTs. In contrast, the production of γ2B GLTs was more efficient in Ebf1fl/fl cells than in Ebf1+/fl cells, suggesting that the response to IL-4 stimulation is not impaired. Stimulation of cells with LPS, in combination with TGFβ to augment switching to IgG2b, resulted in enhanced generation of γ2B GLT in Ebf1fl/fl cells (Fig. 7B).
Figure 7.
Ebf1 regulates γ3 and γ1 germline transcription by binding to the IgH 3′ Eα LCR. (A) Schematic representation of the IgH locus. Light-gray bars depict rearranged variable gene segments, dark gray bars represent enhancers, and gray circles represent the promoters (P) of the constant region (C) gene segments. The bottom part depicts a magnified view of the γ3 gene segment and indicates regulatory sequences, switch (S) region, and 3′ Eα LCR. DHSs are marked with arrows and numbers. Numbers in squares represent HSs that are directly bound by Ebf1. (B) Semiquantitative RT–PCR analysis of IgH constant region GLTs in unstimulated B cells and B cells stimulated with LPS + IL4 or LPS + TGFβ for 16 h. (C) ChIP analysis to examine binding of Ebf1 in 38B9 pro-B cells or MACS-enriched splenic B cells. Binding is represented as percentage of input chromatin. (See also Supplemental Fig. S7.)
Previous studies determined a pivotal role for the 3′ Eα LCR in the regulation of GLT expression and class switching to most isotypes (Dunnick et al. 2005). Genome-wide ChIP-seq analysis indicated that Ebf1 binds to HS4 and HS7 of the 3′ Eα LCR already in pro-B cells (Treiber et al. 2010). We confirmed binding of Ebf1 to these sequences in both pro-B cells and mature B cells by quantitative ChIP analysis (Fig. 7C). Regulation of GLT expression also involves changes in the accessibility of chromatin at the I promoters and the LCR (Garrett et al. 2005). Analysis of the acetylation and methylation status of histones H3 and H4 at the IgH constant region in resting and LPS- plus IL-4-stimulated splenic B cells showed that H3K4me2 and H3Ac modifications are reduced at the γ3 promoter and switch region in Ebf1fl/fl cells (Supplemental Fig. S7F). Thus, Ebf1 regulates class switching to Igγ1 and Igγ3 by binding to the 3′ LCR and inducing changes in the chromatin structure of the γ3 switch region.
Discussion
B lymphopoiesis requires the coordination of general processes, such as cell proliferation and survival, with a complex program of differentiation, including the activation of a B-cell-specific expression program, immunoglobulin gene rearrangement, BCR signaling, class switch recombination, and terminal differentiation into plasma cells. Transcriptional regulation of these events is governed by elaborate networks of transcription factors. Here, we show that Ebf1 acts as a molecular node in multiple regulatory networks that operate at different developmental stages.
Functions of Ebf1 in early stage B cells
In early stage B cells, Ebf1 exerts functions beyond the previously noted roles in specification and commitment of the lineage (Maier et al. 2004; Medina et al. 2004; Pongubala et al. 2008; Treiber et al. 2010). In particular, Ebf1 regulates cell proliferation and survival, whereby our data suggest that Ebf1 regulates both processes via different targets and pathways. Transformation of Ebf1-deficient pro-B cells with A-MuLV rescues their proliferation defect, but does not overcome their survival defect. Primary Ebf1-deficient pro-B cells arrest in G1 of the cell cycle and show impaired expression of direct Ebf1 targets, including the E2F2 and E2F8 transcriptional regulators of the cell cycle, cyclin D3 (Ccnd3), and the replication initiation factor Cdc6. With the exception of Cdc6, these Ebf1 target genes are almost normally expressed in A-MuLV-transformed Ebf1fl/fl pro-B cells. Activated Abl kinase has been found to increase the expression of D-type cyclins and E2F transcription factors (Coutts et al. 2000). Moreover, cyclin D2-deficient bone marrow cells fail to proliferate in response to infection with BCR/ABL-expressing retroviruses (Jena et al. 2002). Therefore, A-MuLV-mediated transformation may compensate for the Ebf1 dependence of D cyclins and E2Fs. In B-cell leukemias carrying the BCR–ABL1 chromosomal translocation and in A-MuLV-transformed pro-B cells, cyclin D3 transcription is augmented through the constitutive activation of Stat5 (Mandal et al. 2009; Hoelbl et al. 2010). Thus, cyclin D3 transcription is under the control of both Ebf1 and Stat5. Cyclin D2 and cyclin D3 are abundantly expressed in early stage B cells, and targeted gene inactivation showed that cyclin D3 is preferentially required for pre-B-cell development (Cooper et al. 2006). Moreover, the marked down-regulation of cyclin D3 is required for the proliferative expansion of GC B cells (Cato et al. 2011). However, the marked down-regulation of cyclin D3 in primary Ebf1-deficient pro-B cells alone does not account for their proliferation defect because this defect is not overcome by the forced expression of cyclin D3. Therefore, we favor the view that Ebf1 regulates at least two steps in cell cycle progression: the activation of D cyclins and the function of an origin activation checkpoint.
The role of Ebf1 in pro-B-cell survival could, in principle, involve the regulation of IL-7Rα by Ebf1 because IL-7Rα signaling and Stat5 expression regulate pro-B-cell survival and prevent Igκ rearrangement (Malin et al. 2010). However, it is unlikely that Ebf1-mediated survival of pro-B cells is linked to the function of IL-7Rα. First, IL-7Rα is indirectly regulated by Ebf1, and second, forced expression of a constitutive form of Stat5 or A-MuLV-mediated transformation failed to overcome the survival defect of Ebf1-deficient pro-B cells. In contrast, forced expression of Bcl2l1, a direct Ebf1 target, rescued, to a large extent, the survival defect of A-MuLV-transformed Ebf1-deficient pro-B cells. Bcl2l1 has been implicated in early stage B-cell survival (Fang et al. 1996; Malin et al. 2010). Mcl1 expression is not altered in Ebf1-deficient pro-B cells (data not shown), and therefore Bcl2l1 may act as an Ebf1-regulated determinant of early B-cell survival. However, the survival defect of A-MuLV-transformed Ebf1-deficient pro-B cells is even more efficiently rescued by the forced expression of c-Myb, which plays a major role in B lymphopoiesis and has been shown to regulate cell survival and IL-7 signaling (Fahl et al. 2009). Myb proteins regulate both G1/S transition and the G2/M phase of the cell cycle and act in a complex with E2F and Rb proteins (Litovchick et al. 2007). Therefore, the overexpression of Myb in transformed Ebf1-deficient pro-B cells may compensate for the impaired expression of E2Fs and phosphorylation of Rb.
Ebf1 has also been implicated in the restriction of alternative cell fates (Pongubala et al. 2008; Lukin et al. 2011). In contrast to gain-of-function experiments or the analysis of a heterozygous germline mutation, we did not observe a major change in the expression of alternative lineage determinants after conditional inactivation of Ebf1. Likewise, the expression of Pax5 and many other prominent Ebf1 targets remained virtually unchanged, suggesting that Ebf1 is required for the establishment but not the maintenance of the B-cell fate. However, we were not able to monitor gene expression during multiple rounds of cell divisions, and therefore it is possible that the expression of Pax5 and alternative lineage determinants would be unstable in the absence of Ebf1 and deregulated with prolonged cell culture.
The question also arises as to mechanisms by which Ebf1 regulates different genes in pro-B cells and mature B cells. Stage-specific expression in pro-B cells may be accomplished by competitive recruitment of Ikaros, which is up-regulated in mature B cells and binds to a sequence that overlaps with the Ebf1-binding site. The pro-B-cell- and pre-B-cell-specific expression of λ5 (Igll1) has been shown to involve a replacement of promoter-bound Ebf1 by Ikaros proteins at later stages of differentiation (Thompson et al. 2007). By ChIP-seq analysis, we could confirm the loss of Ebf1 binding in mature B cells (see Fig. 5C). Conversely, we found that many B-cell-specific genes are bound by Ebf1 specifically in mature B cells, suggesting that the Ebf1-binding site is not accessible due to competitive binding of a repressor or nonpermissive chromatin structure. We showed previously that Ebf1 binding requires a permissive chromatin context (Treiber et al. 2010). Therefore, we favor the view that in genes specifically bound by Ebf1 in mature B cells, a permissive chromatin context is generated by the binding and/or function of collaborating transcription factors specifically in B cells.
Functions of Ebf1 in mature and GC B cells
At the mature B-cell stage, Ebf1 is also required for proliferation and survival of stimulated cells. Stimulation of Ebf1fl/fl B cells with LPS, CpG, or anti-IgM fails to induce proliferation, as determined by dilution of CFSE and incorporation of EDU. As these stimuli act via different signaling pathways, we conclude that the inactivation of Ebf1 results in a proliferation and survival defect, rather than in a defect in cell stimulation. Similar to early stage B cells, the effects of Ebf1 inactivation on the survival of mature B cells is less pronounced than that observed in pro-B cells. Survival of resting B cells is regulated by the BAFF-R and PI3K signaling pathways (Kraus et al. 2004; Mecklenbrauker et al. 2004; Srinivasan et al. 2009). In Ebf1-deficient B cells, the surface expression of BAFF-R is diminished, and we observed a modest rescue of the survival defect by the addition of excess BAFF. We also observed a marked defect of Akt phosphorylation in anti-BCR-stimulated Ebf1-deficient B cells. However, Ebf1 may be dispensable for tonic BCR/Akt signaling, as we observed neither changes in the number of resting Ebf1-deficient B cells nor alterations in the expression of genes associated with tonic Akt signaling (Supplemental Fig. S4E; Srinivasan et al. 2009).
Many components of the (pre-)BCR signaling pathway were previously identified in genome-wide ChIP-seq analyses as Ebf1-bound targets (Lin et al. 2010; Treiber et al. 2010). Consistent with a role of Ebf1 in BCR signaling, we observed defects in Ca2+ mobilization and tyrosine phosphorylation of anti-IgM-stimulated Ebf1-deficient B cells. In these cells, we observed reduced phosphorylation of Igα (Cd79a), despite normal protein expression. Moreover, we detected a reduction of tyrosine phosphorylation of Cd19 that exceeded the modest twofold decrease in Cd19 RNA and protein expression. CD19 plays an important role in B-cell activation by regulating the formation of BCR microclusters (Depoil et al. 2008). Therefore, it is possible that Ebf1-deficient B cells show a more pronounced defect in canonical BCR signaling in the context of membrane-bound ligands. In anti-IgM-stimulated Ebf1-deficient B cells, the expression of the Src family kinase Lyn was normal, but we detected a marked decrease of the expression of the Src family kinase gene Blk. We also examined the expression of Ebf1 targets implicated in the regulation of the PI3K arm of the BCR. Although many components of the Akt signaling pathway, including the catalytic and regulatory subunits of PI3K, Pik3cγ, Pik3, Pik3r1, and Pik3r5, as well as the adaptor BCAP, are bound by Ebf1, we only detect a deregulation of Pik3c2b (Supplemental Tables S2, S5A; Treiber et al. 2010). Consistent with a BCR-proximal defect of Akt signaling, the decrease in CD19 surface expression and phosphorylation could account for an impaired recruitment of PI3K to the BCR. However, most Ebf1 targets in the BCR pathway are only modestly deregulated in Ebf1-deficient B cells, suggesting that Ebf1 plays an important role in the establishment but not the maintenance of the BCR and Akt signaling pathways.
The reduced phosphorylation of Akt and impaired Ca2+ mobilization in anti-IgM-stimulated Ebf1fl/fl B cells is reminiscent of the defects observed in B cells from Foxo1fl/f mice. Although Foxo1 expression is reduced in Ebf1-deficient pro-B cells, no changes in Foxo1 and Foxo3 expression are observed in mature Ebf1-deficient B cells. Ebf1fl/fl and Foxo1fl/fl B cells differ in the proliferative response to stimulation with LPS and the defects in GC B-cell functions (Dengler et al. 2008). In particular, Foxo1 regulates the expression of Aicda, but does not affect locus accessibility and germline transcription of constant region segments (Dengler et al. 2008). In contrast, Ebf1 does not regulate Aicda expression, but is required for the generation of Cγ1 and Cγ3 GLTs. Thus, Ebf1 and Foxo1, which coregulate many genes in early stage B cells (Lin et al. 2010), appear to have only partially overlapping functions in mature B cells.
In summary, our analysis showed that Ebf1 regulates a diverse set of processes in early and late stages of B-cell differentiation. By regulating important components of transcription factor and signaling networks, Ebf1 may act as a molecular node that helps to coordinate proliferation, survival, and differentiation of B-lineage cells.
Materials and methods
Generation of Ebf1+/fl mice
The targeting construct was designed to introduce loxP sites flanking exons 2 and 3, which encode part of the DNA-binding domain. The flanks and “deletable” fragment of the Ebf1 gene were PCR-cloned with the Expand High-Fidelity PCR system (Roche) using genomic DNA from D3V embryonic stem cells as a template. The targeting vector allowed the FLP recombinase-mediated excision of the neomycin selection cassette (gift of R. Fässler). Chimeras were obtained after injection of the targeted embryonic D3V stem cell clones into blastocysts derived from C57BL/6 mice. The chimeras were crossed with C57BL/6ACTFLPe mice (Rodriguez et al. 2000) to generate an frt-Neo-null allele, referred to as Ebf1+/fl mice.
Induced deletion of Ebf1 in vitro and in vivo
Ebf1+/fl and Ebf1fl/flRERTCre fetal liver pro-B cells were cultured as described (Treiber et al. 2010). For inactivation of Ebf1, the B cells were treated with 2 μM 4-hydroxy-tamoxifen for 24 h, washed, and cultured for an additional 4 d. Three independent cultures of each genotype were analyzed for their survival and gene expression pattern. The oral gavage method of tamoxifen-citrate administration was performed as described (Hobeika et al. 2006). Ebf1+/flRERTCre and Ebf1fl/flRERTCre mice received 3 mg of tamoxifen-citrate on four consecutive days and were immunized or sacrificed for analysis on the 10th day after the start of the tamoxifen treatment.
Expression profiling and quantitative RT–PCR
For expression profiling, B220+AA4.1− and B220+AA4.1−CD21lo splenic B cells were sorted from tamoxifen-treated Ebf1+/flRERTCre and Ebf1fl/flRERTCre mice in triplicates, and total RNA was prepared using the RNeasy minikit (Qiagen) according to the manufacturer's protocol. Labeling, hybridization to the Agilent Technologies Microarray Agilent-014868 4x44K, and standard analysis of gene expression were performed by ImaGenes.
For quantitative RT–PCR analysis, RNA was prepared using Trizol reagent (Invitrogen) according to the manufacturer's instructions. SuperScript II Reverse Transcriptase (Invitrogen) was used to reverse-transcribe 500 ng of RNA. Real-time RT–PCR was performed using Power SYBR Green master mix and a 7500 Fast Real-Time PCR system (Applied Biosystems). All quantitative PCR primer sequences are available on request.
Immunization and plasma cell analysis
SRBCs (5 × 108 cells), NP-CGG, or NP-LPS (100 μg in Alum) were intraperitoneally injected into adult Ebf1+/fl and Ebf1fl/fl mice. Serum immunoglobulin titers were determined by enzyme-linked immunosorbant (ELISA) assays. Plasma cells were FACS-sorted and analyzed after 3 d of SRBC immunization.
ChIP
Anti-EBF ChIP was performed as previously described (Pongubala et al. 2008). The histone modification analysis was performed as previously described (Gyory et al. 2004). Antibodies specific for histone modifications were anti-H3K4me3 (Abcam, #8580), anti-H3K4me2 (Millipore, #07-030), anti-acetyl H3 (Millipore, #06-599), and anti-acetyl H4 (Millipore, #06-598).
Generation and analysis of ChIP-seq libraries
ChIP-seq libraries in splenic B cells were prepared as previously described (Treiber et al. 2010) and sequenced on the Illumina Genome Analyzer platform. The Ebf1 ChIP-seq library was processed as described, and Ebf1-binding regions in pro-B cells (data from Treiber et al. 2010) and splenic B cells (this study) were identified using CCAT with default settings (version 3.0) (Xu et al. 2010). A high-confidence subset of binding sites was obtained by retaining the top 25% most-enriched peaks of each library. H3K4me2 and H3K4me3 ChIP-seq data in pro-B cells (Lin et al. 2010; obtained from http://biowhat.ucsd.edu/homer/data/index.html), H3K4me2 and H3K4me3 ChIP-seq data in splenic B cells (this study), and DHS sequencing data in murine CD19+ B cells and naive T cells (downloaded from http://genome.ucsc.edu/ENCODE; Sabo et al. 2006) were mapped using Bowtie (Langmead et al. 2009). All libraries were aligned to the mouse reference genome (mm8, NCBI build 36). BedGraph files of each sequencing library were generated using Homer with default settings for visualization of sequencing data on the University of California at Santa Cruz (UCSC) Genome Browser. All libraries were normalized to 10 million reads.
H3K4me2 modifications at Ebf1-binding sites in splenic B cells were assessed by generating a histogram of normalized H3K4me2 tag counts across 6-kb intervals in 25-base-pair (bp) bins around Ebf1-binding sites using Homer. Binding regions were clustered with Cluster 3.0 and visualized with Java TreeView.
The data mentioned in this study have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GSE36061, which includes ChIP-seq data (GSE35915) and gene expression microarray data (GSE36061). Data may be viewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36061.
Acknowledgments
We thank Drs. Klaus Rajewsky and Stefano Casola for generously providing the Cγ1-Cre and CD21-Cre mouse strains. We are grateful to Drs. Michael Reth, Klaus Rajewsky, and Alan Harris for providing the mb1-Cre, Cd19-Cre, and TgBcl2 mouse strains, respectively. We thank Ingrid Falk and Stefanie Fietze for their expert technical help, and Elias Hobeika and Hassan Jumaa for stimulating discussions.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.187328.112.
References
- Amin RH, Schlissel MS 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol 9: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79: 885–892 [DOI] [PubMed] [Google Scholar]
- Beck K, Peak MM, Ota T, Nemazee D, Murre C 2009. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med 206: 2271–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K 2006. Tracking germinal center B cells expressing germline immunoglobulin γ1 transcripts by conditional gene targeting. Proc Natl Acad Sci 103: 7396–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato MH, Chintalapati SK, Yau IW, Omori SA, Rickert RC 2011. Cyclin D3 is selectively required for proliferative expansion of germinal center B cells. Mol Cell Biol 31: 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Hsia CY, Leone G, Liou HC 2003. Cyclin E and Bcl-xL cooperatively induce cell cycle progression in c-Rel−/− B cells. Oncogene 22: 8472–8486 [DOI] [PubMed] [Google Scholar]
- Cooper AB, Sawai CM, Sicinska E, Powers SE, Sicinski P, Clark MR, Aifantis I 2006. A unique function for cyclin D3 in early B cell development. Nat Immunol 7: 489–497 [DOI] [PubMed] [Google Scholar]
- Coutts M, Zou X, Calame K 2000. v-Abl utilizes multiple mechanisms to drive G1/S progression in fibroblasts. Oncogene 19: 801–809 [DOI] [PubMed] [Google Scholar]
- Danial NN, Pernis A, Rothman PB 1995. Jak–STAT signaling induced by the v-abl oncogene. Science 269: 1875–1877 [DOI] [PubMed] [Google Scholar]
- Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC 2008. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol 9: 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depoil D, Fleire S, Treanor BL, Weber M, Harwood NE, Marchbank KL, Tybulewicz VL, Batista FD 2008. CD19 is essential for B cell activation by promoting B cell receptor-antigen microcluster formation in response to membrane-bound ligand. Nat Immunol 9: 63–72 [DOI] [PubMed] [Google Scholar]
- Dias S, Silva H Jr, Cumano A, Vieira P 2005. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J Exp Med 201: 971–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick WA, Shi J, Graves KA, Collins JT 2005. The 3′ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four γ genes. J Exp Med 201: 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahl SP, Crittenden RB, Allman D, Bender TP 2009. c-Myb is required for pro-B cell differentiation. J Immunol 183: 5582–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Mueller DL, Pennell CA, Rivard JJ, Li YS, Hardy RR, Schlissel MS, Behrens TW 1996. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity 4: 291–299 [DOI] [PubMed] [Google Scholar]
- Fischer MB, Goerg S, Shen L, Prodeus AP, Goodnow CC, Kelsoe G, Carroll MC 1998. Dependence of germinal center B cells on expression of CD21/CD35 for survival. Science 280: 582–585 [DOI] [PubMed] [Google Scholar]
- Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK 2005. Chromatin architecture near a potential 3′ end of the igh locus involves modular regulation of histone modifications during B-cell development and in vivo occupancy at CTCF sites. Mol Cell Biol 25: 1511–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig KT, de Graaf CA, Murphy JM, Carpinelli MR, Pang SH, Frampton J, Kile BT, Hilton DJ, Nutt SL 2010. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood 115: 2796–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillot DA, Merino R, Pena JC, Fanslow WC, Finkelman FD, Thompson CB, Nunez G 1996. bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. J Exp Med 183: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M 2003. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4: 111–120 [DOI] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5: 299–308 [DOI] [PubMed] [Google Scholar]
- Herzog S, Hug E, Meixlsperger S, Paik JH, DePinho RA, Reth M, Jumaa H 2008. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K–PKB–Foxo pathway. Nat Immunol 9: 623–631 [DOI] [PubMed] [Google Scholar]
- Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci 103: 13789–13794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA, Fajmann S, Grebien F, Warsch W, Stengl G, et al. 2010. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med 2: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcher M, Souabni A, Busslinger M 2001. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity 14: 779–790 [DOI] [PubMed] [Google Scholar]
- Jena N, Deng M, Sicinska E, Sicinski P, Daley GQ 2002. Critical role for cyclin D2 in BCR/ABL-induced proliferation of hematopoietic cells. Cancer Res 62: 535–541 [PubMed] [Google Scholar]
- Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity 28: 335–345 [DOI] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol 10: 176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R 2008. Germinal centres: Role in B-cell physiology and malignancy. Nat Rev Immunol 8: 22–33 [DOI] [PubMed] [Google Scholar]
- Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K 2004. Survival of resting mature B lymphocytes depends on BCR signaling via the Igα/β heterodimer. Cell 117: 787–800 [DOI] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28: 751–762 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorchak AS, Schlissel MS, Zhuang Y 2006. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin κ locus in pre-B cells. Mol Cell Biol 26: 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Grosschedl R 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376: 263–267 [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Lu R, Medina KL, Lancki DW, Singh H 2003. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev 17: 1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin K, Fields S, Guerrettaz L, Straign D, Rodriguez V, Zandi S, Mansson R, Cambier JC, Sigvardsson M, Hagman J 2011. A dose-dependent role for EBF1 in repressing non-B-cell-specific genes. Eur J Immunol 41: 1787–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, et al. 2004. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol 5: 1069–1077 [DOI] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M 2010. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol 11: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H, Clark MR 2009. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol 10: 1110–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel EM, Grosschedl R 2010. Transcription control of early B cell differentiation. Curr Opin Immunol 22: 161–167 [DOI] [PubMed] [Google Scholar]
- Mecklenbrauker I, Kalled SL, Leitges M, Mackay F, Tarakhovsky A 2004. Regulation of B-cell survival by BAFF-dependent PKCδ-mediated nuclear signalling. Nature 431: 456–461 [DOI] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H 2004. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell 7: 607–617 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Kee BL 2007. The transcriptional regulation of B cell lineage commitment. Immunity 26: 715–725 [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, Fruman DA 2010. PI3Ks in lymphocyte signaling and development. Curr Top Microbiol Immunol 346: 57–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada Y, Banerji L, Glassford J, Lea NC, Collado M, Rivas C, Lewis JL, Gordon MY, Thomas NS, Lam EW 2001. BCR–ABL and interleukin 3 promote haematopoietic cell proliferation and survival through modulation of cyclin D2 and p27Kip1 expression. J Biol Chem 276: 23572–23580 [DOI] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol 9: 203–215 [DOI] [PubMed] [Google Scholar]
- Quong MW, Martensson A, Langerak AW, Rivera RR, Nemazee D, Murre C 2004. Receptor editing and marginal zone B cell development are regulated by the helix–loop–helix protein, E2A. J Exp Med 199: 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol 9: 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25: 1317–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25: 139–140 [DOI] [PubMed] [Google Scholar]
- Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG 2005. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc Natl Acad Sci 102: 13242–13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. 2006. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods 3: 511–518 [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K 2009. PI3 kinase signals BCR-dependent mature B cell survival. Cell 139: 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE 2008. Mechanism and regulation of class switch recombination. Annu Rev Immunol 26: 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal MA, Carvalho TL, He T, Kim HG, Gao H, Hagman J, Klug CA 2009. Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proc Natl Acad Sci 106: 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP 2005. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity 23: 275–286 [DOI] [PubMed] [Google Scholar]
- Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26: 335–344 [DOI] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, Grosschedl R 2010. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity 32: 714–725 [DOI] [PubMed] [Google Scholar]
- Tudzarova S, Trotter MW, Wollenschlaeger A, Mulvey C, Godovac-Zimmermann J, Williams GH, Stoeber K 2010. Molecular architecture of the DNA replication origin activation checkpoint. EMBO J 29: 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki Y, Neurath MF, Max EE, Strober W 1994. The B cell-specific transcription factor BSAP regulates B cell proliferation. J Exp Med 179: 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Beaty N, Chen S, Qi CF, Masiuk M, Shin DM, Morse HC III 2012. The CXCR7 chemokine receptor promotes B-cell retention in the splenic marginal zone and serves as a sink for CXCL12. Blood 119: 465–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Hobeika E, Jumaa H 2010. Role of PI3K in the generation and survival of B cells. Immunol Rev 237: 55–71 [DOI] [PubMed] [Google Scholar]
- Wolniak KL, Shinall SM, Waldschmidt TJ 2004. The germinal center response. Crit Rev Immunol 24: 39–65 [DOI] [PubMed] [Google Scholar]
- Xu H, Handoko L, Wei X, Ye C, Sheng J, Wei CL, Lin F, Sung WK 2010. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics 26: 1199–1204 [DOI] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M 2008. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol 181: 3364–3372 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Cotta CV, Stephan RP, deGuzman CG, Klug CA 2003. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. EMBO J 22: 4759–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H 1994. The helix–loop–helix gene E2A is required for B cell formation. Cell 79: 875–884 [DOI] [PubMed] [Google Scholar]