This study investigates the different Argonaute proteins and assesses their contribution to the miRNA pathway during mammalian development. The endogenous Argonautes associate with similar pools of miRNAs, independent of slicer or passenger strand cleavage activity. Quantitative expression of the individual Argonautes is found to be a critical determinant of their function; e.g., Ago2's importance is primarily because it is expressed at the highest levels.
Keywords: Argonaute proteins, microRNAs, mammalian development, skin, proteomics
Abstract
Argonaute proteins (Ago1–4) are essential components of the microRNA-induced silencing complex and play important roles in both microRNA biogenesis and function. Although Ago2 is the only one with the slicer activity, it is not clear whether the slicer activity is a universally critical determinant for Ago2's function in mammals. Furthermore, functional specificities associated with different Argonautes remain elusive. Here we report that microRNAs are randomly sorted to individual Argonautes in mammals, independent of the slicer activity. When both Ago1 and Ago2, but not either Ago1 or Ago2 alone, are ablated in the skin, the global expression of microRNAs is significantly compromised and it causes severe defects in skin morphogenesis. Surprisingly, Ago3 is able to load microRNAs efficiently in the absence of Ago1 and Ago2, despite a significant loss of global microRNA expression. Quantitative analyses reveal that Ago2 interacts with a majority of microRNAs (60%) in the skin, compared with Ago1 (30%) and Ago3 (<10%). This distribution is highly correlated with the abundance of each Argonaute, as quantified by shotgun proteomics. The quantitative correlation between Argonautes and their associated microRNAs is conserved in human cells. Finally, we measure the absolute expression of Argonaute proteins and determine that their copy number is ∼1.4 × 105 to 1.7 × 105 molecules per cell. Together, our results reveal a quantitative picture for microRNA activity in mammals.
MicroRNA (miRNA) is a class of tiny (∼19- to 24-nucleotide [nt]), noncoding RNA species that play important roles in the regulation of mammalian gene expression at post-transcriptional levels (Bartel 2009). To execute their diverse physiological functions, miRNAs are loaded into the miRNA-induced silencing complex (miRISC) and, in turn, guide the miRISC to their cognate mRNA targets by base-pairing between the miRNAs and their mRNA targets, usually located within the 3′ untranslated region (3′ UTR) (Bartel 2009). Argonaute proteins are the essential components of the miRISC that directly recruit miRNAs and function as the interface between miRNAs and their mRNA targets (Hock and Meister 2008; Czech and Hannon 2011). Mammals have four Argonautes (Ago1–4) that are involved in the miRNA pathway (Hock and Meister 2008; Czech and Hannon 2011). Among them, Ago2 is unique, with the slicer activity that mediates the cleavage of perfectly matched targets for miRNAs and siRNAs (Liu et al. 2004; Meister et al. 2004; Yekta et al. 2004; Hock and Meister 2008; Czech and Hannon 2011). When individual Argonautes are ablated constitutively in mice, only the loss of Ago2 causes embryonic lethality, whereas single losses of Ago1, Ago3, or Ago4 are dispensable for animal development (Liu et al. 2004; Morita et al. 2007). In addition, the biogenesis of endogenous siRNA (endo-siRNA) in the oocytes and miR-451, a blood-specific miRNA, has been shown to require the slicer activity of Ago2 (Tam et al. 2008; Watanabe et al. 2008; Cheloufi et al. 2010; Cifuentes et al. 2010). Collectively, these observations lead to a widely recognized view that Ago2 is a specialized Argonaute, probably due to its slicer activity. However, exhaustive bioinformatic and experimental analyses have failed to identify a large number of perfectly matched miRNA:mRNA regulatory sequences. It remains unclear why Ago2 is universally important for miRNA functions in diverse organs and tissues of mammals. In addition, the functional significance of Ago1, Ago3, and Ago4 for miRNA activity is poorly understood, probably due to the lack of developmental phenotypes in knockout (KO) mouse models. Interestingly, novel regulatory mechanisms begin to emerge in which Ago1 and their associated miRISCs are asymmetrically segregated to dictate cell fate decision during stem cell division (Neumuller et al. 2008; Schwamborn et al. 2009). Finally, recent studies suggest that miRNA-mediated regulation is highly quantitative and dependent on the relative concentration of miRNA and their target mRNAs (Mukherji et al. 2011; Tay et al. 2011). Therefore, it is critical to determine the function of individual Argonautes and understand the quantitative nature of their contribution to the miRNA pathway during mammalian development.
In this study, we investigate the functions of individual Argonautes by quantitatively cloning and sequencing miRNAs that associate with endogenous Argonautes and examining the developmental defects in Ago1 and Ago2 single- or double-KO mouse models in the skin. Surprisingly, the slicer activity of Ago2 is dispensable for both miRNA biogenesis and function in the skin. Instead, miRNAs are randomly sorted to miRISCs that contain individual Argonautes in proportion to the abundance of each protein. Genome-wide shotgun proteomics and absolute quantification of Argonautes demonstrates that Ago2 is the most abundant Argonaute and so associates with the largest pool of miRNAs in both mouse epidermal cells and human melanoma cells. Finally, genome-wide protein quantification reveals a quantitative landscape for the key components of the miRNA pathway. The demonstration of the quantitative distribution of miRNAs to individual Argonautes in mammals has important implications for our understanding of the function of Argonautes in diverse biological events, and these findings raise a possibility that miRNA activity can be quantitatively manipulated by controlling individual Argonautes.
Results
Individual Argonautes associate with a similar pool of miRNAs in the skin
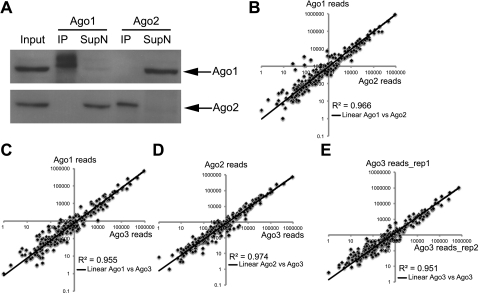
To gain quantitative insights for endogenous Argonautes and their associated miRNAs, we set out to isolate the Ago-specific miRISC. We first determined the efficiency and specificity of primary Ago antibodies for immunoprecipitation (IP). As shown in Figure 1A and Supplemental Figure 1, we specifically isolated Ago1, Ago2, and Ago3 by IP from the total lysates and depleted each protein from the supernatant, respectively, by using two newly developed antibodies to Ago1 and Ago2 and a previously characterized Ago3 antibody (see the Materials and Methods; Azuma-Mukai et al. 2008). Furthermore, we validated the specificity of the antibodies by using Ago1 and Ago2 KO samples (see Fig. 2A). Using these antibodies, we specifically enriched miRNAs and depleted nonspecific small RNA species (e.g., sno25) with our Ago-IP (Supplemental Fig. 2).
Figure 1.
Argonautes associate with a similar pool of miRNAs. (A) Specification and efficiency characterization of Ago1 and Ago2 antibodies. Ago1 or Ago2 antibody specifically pulls down Ago1 or Ago2 in IP, depleting the proteins in the supernatant. (B) Comparison of miRNA cloning frequency of Ago1 and Ago2 IP. (C) Comparison of miRNA cloning frequency of Ago1 and Ago3 IP. (D) Comparison of miRNA cloning frequency of Ago2 and Ago3 IP. (E) Two biological duplicates of Ago3 IP show high correlations, confirming the reproducibility of small RNA cloning.
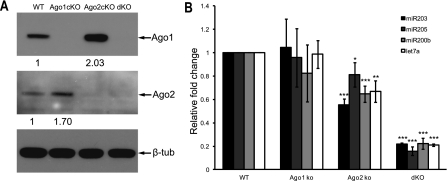
Figure 2.
Genetic deletion of individual Argonautes results in a quantitative loss of global miRNA expression in the skin. (A) Validation of individual or combinatorial Ago conditional knockout (cKO) by Western blotting. (dKO) Ago1/2 double cKO. Ago1 expression increases in Ago2 cKO, whereas Ago2 expression increases in Ago1 cKO. (B) The reduction of miRNA expression in Ago1 cKO, Ago2 cKO, and Ago1/2 dKO. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; n = 3.
Having established the Ago-specific IP, we asked whether individual Argonautes associate with a similar or different pool of miRNAs in vivo. We examined all miRNA species associated with individual Argonautes by cloning and sequencing small RNAs isolated from the Ago-IP. We harvested total cell lysates from postnatal day 4 (P4) wild-type epidermis and divided them into three identical aliquots for the IP and miRNA profiling. Remarkably, the miRNA profile associated with each Argonaute was nearly identical. When plotting with the total reads for each miRNA, the R2 coefficiency had a narrow range between 0.955 and 0.974 in the two-way comparison among the miRNA pools from Ago1-, Ago2-, and Ago3-specific IP samples (Fig. 1B–D; Supplemental Table 1). To determine the reproducibility of our experiments, we generated another small RNA library with an Ago3 IP from another P4 wild-type epidermal sample as the biological duplicate. The R2 coefficiency between the two independent Ago3 miRNA libraries was 0.951, confirming a high consistency of our assay (Fig. 1E). Together, these findings reveal that a similar pool of miRNAs associates with endogenous Argonautes to form the miRISC in the skin.
Genetic ablation of individual Argonautes leads to a quantitative loss of global miRNA expression in the skin
Previous studies suggest an active role of Argonautes in miRNA biogenesis; in particular, the passenger strand cleavage activity of Ago1 and Ago2 is important for miRNA production (Diederichs and Haber 2007; Wang et al. 2009). To examine the requirement of Ago1 and Ago2 for miRNA biogenesis in vivo, we knocked out Ago1 and Ago2 individually (Ago1fl/fl;K14-Cre and Ago2fl/fl;K14-Cre) or combinatorially (Ago1fl/fl;Ago2fl/fl;K14-Cre) in the skin with a well-established K14-Cre line (Vasioukhin et al. 1999; Yi et al. 2006, 2009). By Western blotting, we determined the specific depletion of Ago1 and Ago2 by K14-Cre-mediated conditional KO (cKO) in the skin (Fig. 2A). We observed a mild compensatory effect with enhanced Ago1 or Ago2 expression when the other Argonaute was knocked out (Fig. 2A). In addition, the expression level of Ago3 was also slightly up-regulated in all three mutants (Supplemental Fig. 3). We then examined the impact of the loss of Ago1 and/or Ago2 on the global expression of miRNAs in the skin. When we quantified expression levels of several abundantly expressed miRNAs in the cKO epidermis, we observed an average of 10% reduction in the Ago1 cKO and 30% reduction in the Ago2 cKO (Fig. 2B). Strikingly, when we examined the Ago1/2 double KO (dKO), we observed a severe reduction of miRNA expression with an average loss of 80% (Fig. 2B). The mild reduction of miRNA expression in the Ago1 and Ago2 single cKO suggests that the expression of the remaining Argonautes could largely compensate for the loss of a single Ago in the skin. In contrast, the loss of Ago1 and Ago2 significantly impaired miRNA accumulation despite an increase in the Ago3 level. By using genetic KO mouse models and performing accurate IP-quantitative real-time PCR (IP-qPCR) analysis for miRNAs, our studies provide compelling evidence for the quantitative loss of global miRNA expression when individual Argonautes are genetically ablated in the skin.
Combinatorial deletion of Ago1 and Ago2 leads to severe skin defects
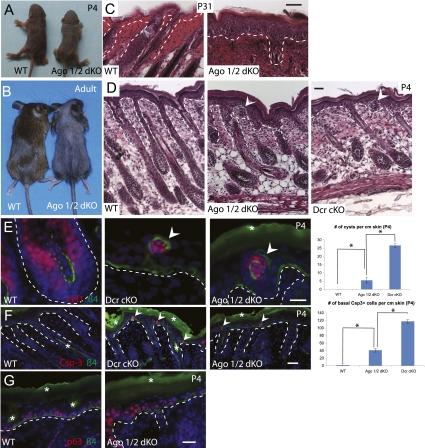
Among four Argonautes in mammals, Ago2 is the only member with slicer activity, and the constitutive inactivation of Ago2 but not Ago1, Ago3, or Ago4 leads to embryonic lethality (Liu et al. 2004; Morita et al. 2007). Because the loss of Ago2 resulted in an ∼30% loss of the total miRNAs in the skin and it uniquely possesses slicer activity, it seemed likely that deletion of Ago2 would reveal a unique requirement for Ago2 in mammalian skin development. Surprisingly, we were not able to identify any developmental defects in either Ago1 or Ago2 single cKO in the skin (data not shown). To probe more deeply into the functions of Argonautes in the skin, we turned to the Ago1/2 dKO animals. We previously generated skin-specific KO models for Dicer and Dgcr8, two essential components in miRNA biogenesis, and observed a complete loss of miRNA production and severe developmental defects in the skin (Yi et al. 2006, 2009). Unlike either Dicer or Dgcr8 skin cKO animals that all died within a week after birth (Andl et al. 2006; Yi et al. 2006, 2009), Ago1/2 dKO animals usually lived several weeks and most of them survived to adulthood (Fig. 3A,B). However, despite their ability to survive postnatally, Ago1/2 dKO mice had sparse hair coat (Fig. 3B). Upon close examination, we found that the hair follicle lineage in the Ago1/2 dKO mice was largely degenerated and the epidermis became hyperthickened (Fig. 3B,C). Because these phenotypes were reminiscent of Dicer cKO skin graft (Yi et al. 2006, 2008) and global miRNA levels were significantly down-regulated (Fig. 2B), we examined neonatal Ago1/2 dKO animals and compared them with the Dicer cKO for skin developmental defects. Remarkably, the compound loss of Ago1 and Ago2 and the subsequent reduction of global miRNA expression resulted in striking skin developmental defects that were very similar to the Dicer cKO (Fig. 3D–G). At P4, we observed evaginating hair follicle cysts in the epidermis, a hallmark of Dicer cKO skin, as well as shortened and misangled hair follicle growth (Fig. 3D; Andl et al. 2006; Yi et al. 2006, 2009). We confirmed the hair follicle origin of the cyst by detecting the expression of Lef1, a marker for hair follicle Matrix cells (Fig. 3E, left panels). When we compared the number of hair follicle cysts in the epidermis of Ago1/2 dKO and Dicer cKO skin, we found that the frequency was less in the Ago1/2 dKO than in the Dicer cKO skin (Fig. 3E, right panel). We also observed apoptotic cells in the proliferative, basal epidermis, and their frequency was also lower in the Ago1/2 dKO skin than the Dicer cKO skin (Fig. 3F). Finally, we observed a slight expansion of p63-positive cells in the Ago1/2 dKO skin, consistent with the hyperproliferative defects observed in the Dicer cKO skin (Fig. 3G; Yi et al. 2008). Thus, our comparative studies between Ago1/2 dKO and Dicer cKO with qualitative and quantitative analyses strongly argue that the skin defects in the Ago1/2 dKO skin are caused by a quantitative reduction (∼80%) of global miRNA expression in the mutant animals. Taken together, our genetic analysis provides unambiguous evidence that neither Ago1 nor Ago2 (with its unique slicer activity) individually was essential for the functions of miRNAs during skin development.
Figure 3.
Combinatorial deletion of Ago1 and Ago2 significantly impairs the miRNA pathway and results in severe skin defects. (A) At birth, Ago1/2 dKO is slightly smaller than wild-type (WT) littermate. (B) Ago1/2 dKO lives to adulthood. Adult dKO shows sparse hair coat with a body size similar to wild-type littermate. (C) Degenerated hair follicles and thickened epidermis in adult dKO. (D) Abnormal hair follicles and cysts (arrowhead) are detected in dKO, as indicated by H&E staining. (E) Validation of the hair follicle original of the cyst by Lef1 immunofluorescence staining. Ago1/2 dKO have less cyst than Dicer cKO (Dcr cKO). (F) Apoptosis (marked by cleaved caspase-3 staining) is detected in dKO epidermis, comparable with Dcr cKO with a lower frequency. (G) Expansion of p63-positive cells in dKO epidermis. The dotted lines mark the boundary between the epidermis and dermis. White asterisks denote autofluorescence. Bar, 20 μm. (*) P < 0.05; n = 3.
Quantitative analysis reveals a random sorting mechanism of miRNAs to individual Argonautes
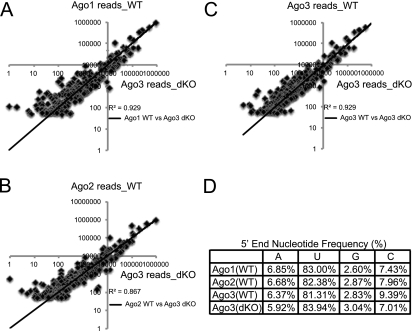
These biochemical and genetic studies provide strong evidence for the requirement for a high level of Argonautes for maintaining miRNAs during mammalian skin development. However, it was not clear whether each Argonaute plays similar roles during miRISC formation and whether the passenger strand cleavage activity of Ago1 and Ago2 is the basis for the significant loss of total miRNAs in the dKO skin. In previous biochemical studies, individual Argonautes—in particular, Ago2 with its unique slicer activity—were shown to play active roles in miRNA loading and miRISC formation in fly, mouse, and human cells (Matranga et al. 2005; Miyoshi et al. 2005; Diederichs and Haber 2007; Wang et al. 2009). In addition, the slicer activity of Ago2 is uniquely required for the biogenesis of miR-451, a blood-specific miRNA, in both zebrafish and mammals (Cheloufi et al. 2010; Cifuentes et al. 2010). This raises a possibility that Ago2 and, to a lesser extent, Ago1 might be uniquely required for processing and loading of some miRNA species; e.g., those that require passenger strand cleavage activity. To examine this possibility, we constructed and sequenced Ago3 IP small RNA libraries from two P4 Ago1/2 dKO epidermal samples. Between these two biological duplicates, the R2 coefficiency was highly consistent (R2 = 0.984) (Supplemental Fig. 4). Thus, we used one data set for comparative analysis. We reasoned that if Ago1 and Ago2 are uniquely involved in miRNA loading and miRISC formation through their passenger strand cleavage activity, we should observe depletion of a subset of miRNA species whose biogenesis depends on Ago1 and Ago2 but not Ago3 and Ago4. Strikingly, when we compared the profile of Ago3-associated miRNAs in the absence of both Ago1 and Ago2 with that of individual Argonautes from the wild-type samples, we observed a remarkable consistency, and the R2 coefficiency was between 0.867 and 0.929 among all samples (Fig. 4A–C; Supplemental Table 2). These findings suggest that, despite the global loss of ∼80% of miRNAs in the Ago1/2 dKO skin, all miRNA species were still similarly processed and loaded onto Ago3. We conclude that Ago3 is equally competent in the processing and loading of miRNAs as Ago1 and Ago2 and that the passenger strand cleavage activity of Ago1 and Ago2 is dispensable for the biogenesis of hundreds of miRNAs in the skin.
Figure 4.
miRNAs are randomly distributed among individual Argonautes. (A) Comparison of miRNA cloning frequency of Ago1 IP in wild-type (WT) cells with Ago3 IP in dKO cells shows high correlations. (B) Comparison of miRNA cloning frequency of Ago2 IP in wild-type cells with Ago3 IP in dKO cells shows high correlations. (C) Comparison of miRNA cloning frequency of Ago3 IP in wild-type cells with Ago3 IP in dKO cells shows high correlations. (D) Cloning frequencies show no distinguishable preferences for 5′-nucleotide identity in individual Ago-IPs in wild-type cells compared with Ago3 IP in dKO cells.
Our findings that each Argonaute associates with a similar pool of miRNAs (Fig. 1) and that Ago3 can also load the same pool of miRNAs in the absence of Ago1 and Ago2 (Fig. 4A–C) provide strong evidence to support a random sorting of miRNAs to individual Argonautes in mammals. In worms and flies, as well as plants, different small RNA species that are sorted to specific Argonautes have distinct 5′-end nucleotide signatures (Czech and Hannon 2011). To further confirm the random sorting mechanism, we analyzed the nucleotide frequency for the 5′ end of miRNAs that associate with individual Argonautes in the wild-type and Ago1/2 dKO libraries. Consistent with the random-sorting hypothesis, the 5′ end of all miRNAs shows a dominant preference for U (∼82%), followed by C (∼7.5%), A (∼6.5%), and G (3%). Importantly, the preference is indistinguishable not only among all Argonautes in the wild type, but also in Ago3 when Ago1 and Ago2 are deleted (Fig. 4D). Taken together, we conclude that mammalian Argonautes show no preference for miRNA species and that miRNAs are randomly sorted into endogenous Argonautes in vivo.
The ability of individual Argonautes to associate with miRNAs is determined by their expression level
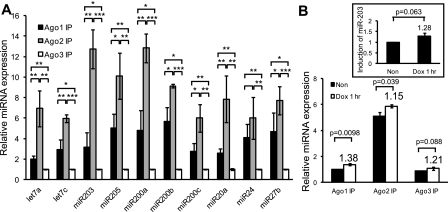
To address why the compound loss of Ago1 and Ago2 caused the significant loss of total miRNAs, we compared the abundance of miRNAs that were associated with individual Argonautes (Ago1–3) by qPCR following Ago-IP from four equally divided pools of P4 epidermal total cell lysates. Three pools were used for individual Ago-IP, and one pool was used to extract total RNA as control. We observed that Ago2 always associated with most miRNAs, followed by Ago1 and Ago3 for pools of 10 abundantly expressed miRNAs (Fig. 5A; Yi et al. 2006, 2009). Overall, the relative amounts of miRNAs associated with Ago1, Ago2, and Ago3 were 3.8, 8.5, and 1.0, respectively. These results demonstrated that most copies of any miRNA were associated with Ago2, followed by Ago1, while a few were associated with Ago3.
Figure 5.
Quantitative distributions of miRNAs among Argonautes are determined by their expression level in the skin. (A) By IP-qPCR, Ago2 associates with the most miRNAs, followed by Ago1, then Ago3. Shown are 10 abundantly expressed miRNAs in mouse epidermis. When normalized to Ago3-associated miRNAs (set as 1.0 U), Ago1 associates with an average of 3.8 U of miRNAs, and Ago2 associates with an average of 8.5 U of miRNAs. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; n = 3. (B) Newly synthesized miR-203 distributes proportionally to three Argonautes in cultured keratinocytes. (Inset) The overall miR-203 level is increased to 1.28-fold after 1 h of induction. P-value is shown (n = 3).
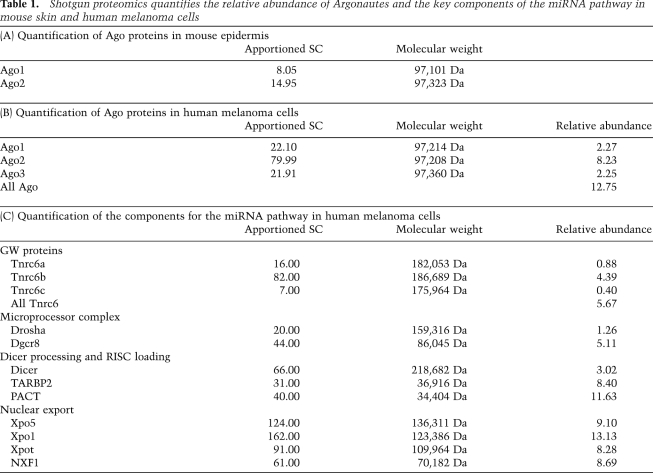
Our analysis for the quantitative distribution of miRNAs among individual Argonautes (Fig. 5A) and the significant loss of miRNAs in the Ago1/2 dKO skin suggested that Ago1 and Ago2 bind a majority of miRNAs. These observations raised two hypotheses: (1) Ago1 and Ago2 are more efficient than Ago3 and Ago4 in loading miRNAs, or (2) Ago1 and Ago2 are more abundant than Ago3 and Ago4 in skin. To distinguish these possibilities, we performed a shotgun proteomics experiment to quantify the relative abundances of individual Argonautes in skin. Of 38,987 peptide counts (total spectral counts, SCs) representing 4993 proteins, we observed 13 peptides that distinguish Ago2 and four peptides that distinguish Ago1 (Table 1A; Supplemental Table 3). We also detected six “bridge” peptides that could not distinguish Ago1–4 (Supplemental Table 3) due to sequence identity among these closely related proteins (Supplemental Fig. 5). Ago3 and Ago4 failed to generate any distinguishing peptides despite the similar probability for each Argonaute; therefore, it was unlikely that all six indistinguishable counts were derived from Ago3 and Ago4. We concluded that Ago3 and Ago4 are significantly less abundant than either Ago1 or Ago2 and estimated that the number of Ago3/4-derived peptides was one or less. Our estimate of apportioned SCs was 8:15 (1.0:1.9) between Ago1 and Ago2, comparable with our analysis of Ago-IP miRNAs (Ago1:Ago2 = 1.0:2.2). This correlation suggests that the abundance of individual Argonautes is a primary determinant for the quantitative distribution of miRNAs, implying random and independent sorting to individual Argonautes.
Table 1.
Shotgun proteomics quantifies the relative abundance of Argonautes and the key components of the miRNA pathway in mouse skin and human melanoma cells
Although our results so far demonstrated that the homeostasis of miRISC is quantitatively determined by the expression of individual Argonautes, it remained possible that newly synthesized miRNAs are preferentially processed by Ago2 and then distributed to other Argonautes. To test this hypothesis, we employed a miR-203-inducible cell line (K14-rtTA;TRE-miR-203) and examined the distribution of this miRNA after a short pulse of doxycyclin in the cultured keratinocytes. After 1 h of induction, we performed Ago-IPs and quantified the amount of miR-203 associated among Ago1–3. Consistent with a random sorting mechanism, we observed that Ago1, Ago2, and Ago3 all loaded miR-203 in a proportion similar to uninduced cells (Fig. 5B). This finding reveals that, within this time scale, newly synthesized miRNAs are not preferentially loaded onto Ago2, but distribute among Argonautes comparably. All together, our data reveal that the formation of the miRISC is quantitatively correlated with the expression levels of Argonautes.
Quantitative analysis of the miRNA pathway in human melanoma cells
Our studies thus far focused on mouse skin. To extend our findings to human cells with different developmental origins, we examined a human melanoma cell line (WM239A). Although usually characterized as skin cancer, melanoma is derived from melanocytes with neuronal origin that is distinct from skin epithelium. We first measured the quantity of miRNAs associated with individual Argonautes using Ago-IP and qPCR. Similar to mouse skin, Ago2 emerged as the isoform that bound the highest levels of all miRNAs tested (Fig. 6A). However, unlike mouse skin, the miRNAs associated equally with Ago1 and Ago3 (Fig. 6A). Overall, miRNAs associated with Ago1, Ago2, and Ago3 was 0.95, 2.8, and 1.0, respectively. We also performed shotgun proteomics to quantify the abundance of each Argonaute in the WM239A cell line (Table 1B). We detected 389,398 peptides (total SCs) that represented 10,021 proteins. We obtained 16 unique counts for Ago1, 67 unique counts for Ago2, and 16 unique counts for Ago3, as well as 18 counts indistinguishable among Ago1–4, six counts indistinguishable among Ago1/3/4, and one count indistinguishable between Ago1 and Ago2 (Supplemental Table 4). Because Ago4 failed to generate any unique counts, we concluded that it is expressed at a very low level and we apportioned all “bridge” peptides to Ago1/2/3. We determined that the ratios among individual Argonautes, based on the normalized SCs, were Ago1:Ago2:Ago3 = 1.0:3.6:1.0 (22:80:22), comparable with our quantitative analysis for miRNAs (Fig. 6A).
Figure 6.
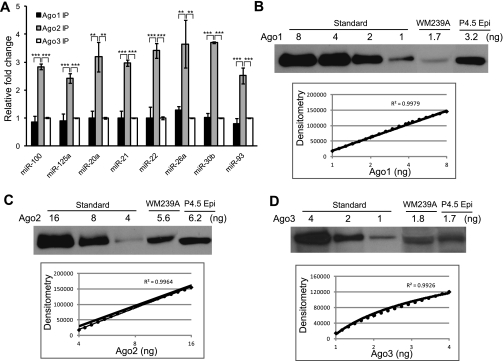
Quantitative distributions of miRNAs among Argonautes are determined by their expression level in human melanoma cells. (A) Quantitative analysis of miRNAs binding to three Argonautes in human melanoma cell line WM239A. When normalized to Ago3-associated miRNAs (set as 1.0 U), Ago1 associates with an average of 0.95 U of miRNAs, and Ago2 associates with an average of 2.8 U of miRNAs. No significant difference was observed between Ago1 IP and Ago3 IP for any miRNA. (**) P < 0.01; (***) P < 0.001; n = 3. (B) Absolute quantification of Ago1 expression in WM239A and P4.5 epidermis using synthetic Ago1 protein. (C) Absolute quantification of Ago2 expression in WM239A and P4.5 epidermis using synthetic Ago2 protein. (D) Absolute quantification of Ago3 expression in WM239A and P4.5 epidermis using synthetic Ago3 protein. (P4.5 Epi) P4.5 epidermis.
To validate our proteomics analysis and quantify the absolute expression level of Argonautes, we obtained synthetic Ago proteins with a minimum purity of >88% (see the Materials and Methods). By using the synthetic proteins, we generated a standard curve for each Argonaute and determined their expression level in both mouse epidermis and human melanoma cells (Fig. 6B–D). We determined, for 0.5 × 106 cells, that 3.2 ng of Ago1 and 6.2 ng of Ago2 are expressed in P4 epidermis and 1.7 ng of Ago1, 5.6 ng of Ago2, and 1.8 ng of Ago3 are expressed in WM239A cells. The ratio among Argonautes, determined by the absolute quantification—e.g., Ago1:Ago2 = 1.0:1.9 in the epidermis, and Ago1:Ago2:Ago3 = 0.94:3.3:1.0 in WM239A cells—showed a remarkable consistency with miRNA quantification determined by IP-qPCR (Figs. 5A, 6A), as well as Argonaute quantification determined by proteomics (Table 1A,B). Together, we conclude that in both mouse epidermis and human melanoma cells, the distribution of miRNA species among individual Argonautes is determined by the abundance of Argonautes and that Ago2 is the most abundant Argonaute. Importantly, with the absolute quantification, we estimate (with ∼80% efficiency in protein extraction) that the total copy number of Argonaute proteins is ∼1.4 × 105 to 1.7 × 105 molecules per cell in mice and humans.
The large-scale and quantitative proteomics data also afforded us an unprecedented opportunity to examine quantitative features of the miRNA pathway (Table 1B,C). It has been previously demonstrated that the relative protein abundance, estimated by correcting SCs by protein molecular weight, correlates well with protein concentrations (Ishihama et al. 2005). Furthermore, our absolute measurement for Argonautes directly validated the quantitative feature of our proteomics data set (Fig. 6). We then calculated the relative abundance of each protein in the miRNA pathway in the melanoma cells (see the Materials and Methods; Weiss et al. 2010; Meyer-Arendt et al. 2011). The total for Argonautes summed to 12.75, approximately twofold higher than the total of 5.67 for GW proteins (Tnrc6a–c). This suggests an interesting possibility that ∼50% of Argonautes are not associated with GW proteins. The ratio between Drosha and Dgcr8, two essential components of the nuclear miRNA microprocessor complex, was ∼1:4 (1.26:5.11). This suggests that multiple Dgcr8 molecules work with a single Drosha molecule to form the microprocessor complex for the nuclear cleavage of primary miRNA (pri-miRNA). The relative abundance of Dicer was 3.02, ∼25% that of the Argonautes, whereas the total for TRBP (TARBP2) and PACT, two critical cofactors of Dicer for dsRNA loading from Dicer to Ago (Chendrimada et al. 2005; Lee et al. 2006; Noland et al. 2011), were 8.40 and 11.63, respectively. Together, both RNase III enzymes, Drosha and Dicer, were significantly less abundant than the Ago and GW proteins, consistent with their role as the processing enzyme, whereas the Ago and GW proteins are core components of the mature miRISC. The single most abundant component that is involved in the miRNA pathway, however, was Xpo5, with a relative abundance of 9.10. The abundance of Xpo5 is thus comparable with Xpot (8.28), an abundantly expressed factor for the nuclear export of tRNA, and Xpo1 (13.13), a highly expressed export factor for ribosomal RNA.
Overexpression of Ago2 or Ago3 rescues the growth defects of Ago1/2 dKO cells
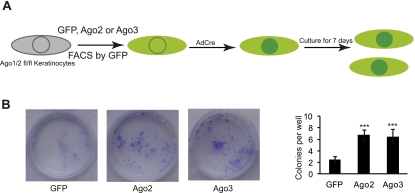
Our biochemical and genetic analyses suggest that Ago proteins function similarly to load miRNAs and the amount of miRNAs associated with each Ago is correlated with their expression level. Thus, we surmised that the overexpression of individual Ago proteins might rescue the Ago1/2 dKO defects. Because the miRNA pathway is required for the survival of epidermal keratinocytes in culture (e.g., keratinocytes derived from both Dicer cKO and Ago1/2 dKO fail to survive) (data not shown), we reasoned that we could initially test the hypothesis by examining whether the overexpression of individual Ago proteins could rescue the growth defects. We first established keratinocytes from Ago1/2fl/fl epidermis and infected them with MSCV-GFP, MSCV-Ago2-GFP, or MSCV-Ago3-GFP, respectively. We then infected these cells (GFP+) with AdCre that mediates the complete deletion of Ago1/2fl/fl alleles within 48 h and measured the colony formation capacity (Fig. 7A). As expected, MSCV-GFP-infected cells only had a few abortive colonies 7 d after the removal of Ago1/2. In contrast, both Ago2- and Ago3-infected cells gave rise to many productive colonies (Fig. 7B). Furthermore, Ago2- and Ago3-infected cells showed a similar rescue to the growth defects, as judged by the number and size of the colonies (Fig. 7B). These results indicate that when overexpressed, both Ago2 and Ago3 are able to rescue the growth defects of Ago1/2 dKO keratinocytes.
Figure 7.
Overexpression of individual Ago proteins rescues the growth defects in Ago1/2 dKO cells. (A) Schematic illustration of the experimental design. (B) Most of GFP-infected Ago1/2fl/fl keratinocytes die after the removal of Ago1/2 by AdCre. In contrast, overexpression of either Ago2 or Ago3 rescues the growth defects. (***) P < 0.001, compare with the GFP control; n = 4.
Discussion
At the heart of the miRISC, Argonautes have attracted considerable interest for their functions in miRNA biogenesis and function (Hock and Meister 2008; Czech and Hannon 2011). Our study, using comprehensive approaches including Ago-specific miRISC isolation, miRNA cloning and sequencing, mouse genetics, and proteomics, now provides the first quantitative view for the function of Argonautes in mammalian development. We demonstrated that Ago2 is the most abundant Argonaute and associates with a majority of miRNAs in both mouse skin and human melanoma cells. Surprisingly, despite its unique slicer activity, Ago2 is dispensable for skin development. In mammals, the requirement for slicer activity has been demonstrated in (1) endo-siRNA biogenesis in oocytes and embryonic stem cells (Babiarz et al. 2008; Tam et al. 2008; Watanabe et al. 2008), (2) miR-451 biogenesis in blood (Christodoulou et al. 2010; Cifuentes et al. 2010; Yang et al. 2010), and (3) rare mRNA cleavage by miRNA (Yekta et al. 2004; Karginov et al. 2010). Because >100 miRNAs are highly expressed in the skin, many of which are universally expressed in all organs and tissues (e.g., the let-7 family, miR17∼92 cluster, and miR21) (Yi et al. 2006), our results suggest a limited requirement for slicer activity of Ago2 in somatic tissues; e.g., skin.
Our data also show that the passenger strand cleavage activity of Ago1 and Ago2 has no major impact for the pool of miRNAs in the skin. When we delete both Ago1 and Ago2, the content of the remaining miRNAs still closely resembles the wild type despite a significant reduction of miRNAs globally. Thus, our results lend support to the notion that the passenger strand cleavage activity is dispensable for miRISC formation (Matranga et al. 2005) and extend the finding to hundreds of miRNAs in vivo. However, the dramatic loss of global miRNAs indeed leads to severe developmental defects in the skin, reminiscent of both Dicer- and Dgcr8-null skin. Notably, conditional ablation of Ago2 has been linked to significant down-regulation of global miRNAs in blood and the brain (O'Carroll et al. 2007; Schaefer et al. 2010), suggesting an even higher percentage of Ago2-miRISC in these tissues. Interestingly, Ago2 is specifically stabilized by hydroxylation (Qi et al. 2008), and this may contribute to the observed high abundance. We propose that a high level of Argonautes is important to maintain the miRNA pathway. In this regard, Ago2 still emerges as the most important Argonaute because of its abundant expression. We also note that the differential expression of individual Argonautes provides an opportunity to control miRNA activities by manipulating each Argonaute and their associated miRISC.
The quantitative proteomics data provide a wealth of knowledge for the miRNA pathway in mammals. For example, our results demonstrate an ∼2:1 ratio between Argonautes (12.75) and GW proteins (5.67). Because Argonautes function in both miRNA loading and miRISC targeting, whereas GW proteins only function in mature miRISC, this observation suggests that significant amounts of Argonautes are not in the final miRISC. More importantly, although miRNAs are randomly sorted to individual Argonautes, each Argonaute could associate differently with GW proteins to execute dynamic functions. We also identified a 1:4 ratio between Drosha (1.26) and Dgcr8 (5.11). Recently, it was shown that two Dgcr8 molecules dimerize to work with one Drosha molecule for pri-miRNA cleavage (Sohn et al. 2007; Senturia et al. 2010). Interestingly, a two-step model has been proposed in which Dgcr8 preferentially and directly interacts with pri-miRNA, followed by transient Drosha cleavage (Han et al. 2006). Our data thus provide a stoichiometric picture for this process. The data also shed light on Dicer processing of dsRNA. TRBP and PACT have been identified as key cofactors for dsRNA loading from Dicer to Ago (Chendrimada et al. 2005; Lee et al. 2006). It was recently proposed, based on biochemical studies, that TRBP or PACT forms a heterodimer with Dicer to sense the thermodynamic asymmetry of dsRNA; e.g., siRNA or miRNA (Noland et al. 2011). Both TRBP and PACT exist in excess to Dicer, probably reflecting their diverse roles in other pathways. It will be of great interest to determine whether the Dicer:TRBP complex functions similarly or differently compared with the Dicer:PACT complex in vivo. Finally, Xpo5 (9.10) emerges as a highly expressed protein in the miRNA biogenesis pathway. Its abundance is on a par with Xpot (8.28), which exports tRNA, and Xpo1 (13.13), which exports rRNA, as well as NXF1 (also known as TAP [8.69]), which exports cellular mRNA. Because Xpo5 is easily saturated, and this dampens miRNA biogenesis (Yi et al. 2005; Grimm et al. 2006), it indicates a stringent requirement for the high level of Xpo5 expression by the miRNA pathway.
Finally, our quantification with synthetic Ago proteins enables us to estimate the absolute expression level of Argonautes in both mice and humans. With an estimated 80% efficiency for protein extraction, we determined that the total copy number of Argonautes is ∼1.4 × 105 to 1.7 × 105 molecules per cell. Because all mature miRNAs associate with Argonautes in a 1:1 manner, our estimate likely represents an upper limit for the copy number of total mature miRNAs. With recent appreciation for the quantitative nature of miRNAs’ functions (Mukherji et al. 2011; Tay et al. 2011), our results thus provide a framework to quantitatively decipher complex regulation of gene expression mediated by the miRNA pathway.
Materials and methods
Ribonucleicprotein IP (RIP) assay
RIP assay was performed following the protocol published previously (Keene et al. 2006), with minor modifications. Briefly, antibodies were first incubated with 10% protein G slurry for 2 h at 4°C, and then the cell lysates were added to the antibody complex and incubated for 4 h. After incubation, the supernatant was saved for the analysis of the IP efficiency. The agarose beads were treated with 30 μg of proteinase K to release the ribonucleicprotein components. For extraction of RNA, Trizol reagent (Invitrogen) was used by following the manufacturer's protocol. For each IP, 2 × 106 to 5 × 106 epidermal keratinocytes, 5 μL of Ago1 Ab (1 μg/μL; MBL), 5 μL of Ago2 Ab (1 μg/μL; Wako), and 2 μL of Ago3 Ab ( 3.3 μg/μL; a gift from M. Siomi) were used. For miR-203-inducible experiments, doxycycline was added to K14-rtTA:pTRE-miR-203 MK cell for 1 h, and then 2 × 106 cells were collected and lysed for IP. With the human melanoma cell line WM239A, 5 × 106 cells were used for each IP; Ago1 and Ago3 Ab were the same as above, except Ago2 Ab was human-specific and was characterized previously (Azuma-Mukai et al. 2008) (2.5 μg/μL; a gift from M. Siomi).
Normalization for AgoIP-qPCR
For all experiments, a 10% amount of lysate of that used for IP was used as the “total RNA” control and subjected to RNA extraction and qRT–PCR together with the IP samples. After qRT–PCR, for each miRNA, individual Ago-IP was first compared with the “total RNA” directly, then divided by 10 to obtain the value for “normalized enrichment in each IP.”
Precipitation of protein from the supernatant after IP
After IP, ∼1 mL of supernatant was collected. Two percent sodium-deoxycholate (1/100 vol) was added, followed by 15 min of incubation on ice; then 1/10 vol of 100% trichloroacetic acid was added, followed by 1 h of incubation on ice. Proteins were pelleted by centrifugation and washed with acetone. After drying, the pellet was resolubilized with Laemmli sample buffer and used directly for Western blotting.
Small RNA cloning, sequencing, and annotation
Construction of small RNA libraries was performed as described previously (Yi et al. 2006, 2009). The small RNA libraries were sequenced using an Illumina HiSeq2000 sequencer and annotated as previously described (Yi et al. 2006, 2009).
Reverse transcription and real-time PCR miRNA expression analysis
Reverse transcriptions were performed using miScript reverse transcription kit (Qiagen). Real-time PCR reagents and miScript primers for miRNAs and control Sno25 were from Qiagen. Reactions were performed according to the manufacturer's manual and on a CFX384 real-time system (Bio-Rad). Differences between samples and controls were calculated based on the 2−ΔΔC(t) method.
Mice and animal maintenance
Ago1 and Ago2 floxed models were described previously (O'Carroll et al. 2007) and bred to K14-Cre for the skin conditional deletion. Dicer1 and Dgcr8 skin cKO models were described previously (Yi et al. 2006, 2009). Mice were bred and housed according to the guidelines of the Institutional Animal Care and Use Committee at a pathogen-free facility at the University of Colorado at Boulder.
Immunofluorescence, immunoblot. and antibodies
Immunofluorescence and immunoblot were performed as described (Yi et al. 2006), with the following primary antibodies: Lef1 (1:100; a gift from E. Fuchs), p63 (1:500; Cell Signaling), β4-integrin (β4, 1:200; BD Biosciences), cleaved Caspase-3 (1:500; Cell Signaling); Ago1 (1:2000; MBL), Ago2 (1:1000; Wako), and β-tubulin (1:2000; Developmental Studies Hybridoma Bank).
Proteomics
Tryptic digests of 250 μg of human melanoma cell (WM239A) or mouse keratinocyte proteins were generated using the filter-aided sample preparation (FASP) protocol described by Wiśiniewski et al. (2009). LC-MS/MS was carried out using a Thermo LTQ-Orbitrap Velos mass spectrometer interfaced with a Waters 2D nanoAcquity UPLC outfitted with Waters BEH C18 reversed-phase columns for the first (pH 10, 5 cm × 300 mm i.d, 5 mm) and second (pH 2.5, 25 cm × 75 mm i.d, 1.7 mm, 100 Å) dimensions. For each cell type, one-third of each sample was separated into 18 steps of increasing acetonitrile (4%–65%) in the first dimension, each followed by gradient elution (8%–32% acetonitrile) in the second dimension. MS/MS were collected enabling settings for monoisotopic precursor and charge selection. Ions with charge state unassigned or equal to 1 were excluded. For each MS scan, the 10 most intense ions were targeted with dynamic exclusion for 180 sec at 20 ppm exclusion width and repeat count = 1. The maximum injection time for Orbitrap parent scans was 500 msec, allowing one microscan and AGC = 1 × 106. The maximum injection time for the LTQ MS/MS was 250 msec, with one microscan and AGC 1 × 104. The normalized collision energy was 35%, with activation Q = 0.25 for 30 msec.
Data sets of keratinocytes or melanoma cells, respectively, consisted of one or four complete 2D-LC-MS/MS runs. For each data set, MS/MS were searched against a human protein database (IPI version 3.65) using MASCOT, with MOWSE thresholds set to false discovery rate (FDR) = 1% using decoy database searching. MOWSE thresholds for MH2+2, MH3+3, and MH4+4 and above were 26.2, 16.8, and 15.7, respectively. Parent ion tolerances were set to 50 ppm on the monoisotopic peak (A0) and the first isotopic peak (A1), and the fragment ion tolerance was set to 0.5 Da, allowing one missed cleavage. Protein inference was carried out using the peptide-centric algorithm IsoformResolver, which generates protein profiles displaying all identified and inferred proteins, listing SCs for “bridge” peptides, which are found in two or more primary proteins, versus “nonbridge” peptides, whose presence distinguishes primary proteins (Meyer-Arendt et al. 2011). Relative protein abundances were estimated by first apportioning SCs (apportioned SC) for bridge peptides according to the distribution SCs for nonbridge peptides and then normalizing by protein mass. Specifically, it was calculated as relative abundance = (apportioned SC/MW) × 10,000.
The keratinocyte data set comprised 4993 proteins, 1505 with only one unique peptide; 21,770 unique peptides; 38,987 total peptides (total SC); 157,648 total MS/MS attempts.
The melanoma data set comprised 10,021 proteins, 1590 with only one unique peptide (16%); 87,527 unique peptides; 389,398 total peptides (total SC); 1,097,985 total MS/MS attempts.
Absolute quantification of Ago proteins
His-tagged recombinant Ago1 protein (NP_036331.1; >97% purity), Ago2 protein (Q8CJG0; >88% purity), and Ago3 protein (Q9H9G7-1; >88% purity) were purchased from Sino Biological, Inc. The concentration of the proteins was provided by the supplier and confirmed by Nanodrop. Standard curves were determined experimentally. A half-million WM239A cells or P4.5 epidermal keratinocytes were lysed and loaded for each Western blotting. Both standard curve generation and protein expression quantification were determined by ImageJ. The molecular weight of Ago1 is 97.2 kDa, Ago2 is 97.3 kDa, and Ago3 is 97.4 kDa. For a half-million cells, 8 ng of Ago protein represents 1 × 105 molecules per cell.
In vitro rescue assay
Ago1/2 double-floxed (Ago1/2fl/fl) keratinocytes were isolated from P4.5 mouse epidermis. After infection with MSCV-GFP, MSCV-Ago2-IRES-GFP, or MSCV-Ago3-IRES-GFP, GFP-positive cells were isolated using Moflo XDP cell sorter (Beckman Coulter). GFP-positive cells were further infected with Adeno-CreGFP (AdCre) to delete endogenous Ago1 and Ago2. The multiplicity of infection (MOI) = 50 was experimentally determined to convert floxed alleles to null alleles with >99% efficiency. AdCre-infected cells were cultured for 7 d to assay the growth phenotype.
Statistical analysis
Unpaired Student's t-test was performed and the P-value was calculated for each experiment. For all experiments with error bars, standard deviation was calculated to indicate the variation within each experiment.
Acknowledgments
We thank Drs. T. Blumenthal and M. Han for critical reading of the manuscript; the members of the Yi laboratory for discussion; L. Greiner for maintaining the mice; Y. Han for cell sorting; J. Huntley, J. Castoe, and J. Dover for Illumina sequencing; E. Fuchs for providing K14-Cre mice and antibodies; and M. Siomi for sharing Ago2 and Ago3 antibodies. This publication was made possible by grant numbers R00AR054704 and R01AR059697 and a startup fund provided by the University of Colorado (to R.Y.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.182758.111.
References
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE 2006. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 16: 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D 2010. Ancient animal microRNAs and the evolution of tissue identity. Nature 463: 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. 2010. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328: 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ 2011. Small RNA sorting: Matchmaking for Argonautes. Nat Rev Genet 12: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441: 537–541 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Hock J, Meister G 2008. The Argonaute protein family. Genome Biol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ 2010. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell 38: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB 2006. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307 [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN 2006. The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Meyer-Arendt K, Old WM, Houel S, Renganathan K, Eichelberger B, Resing KA, Ahn NG 2011. IsoformResolver: A peptide-centric algorithm for protein inference. J Proteome Res 10: 3060–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I 2007. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89: 687–696 [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A 2011. MicroRNAs can generate thresholds in target gene expression. Nat Genet 43: 854–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Ma E, Doudna JA 2011. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell 43: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A 2007. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21: 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J 2008. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455: 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Veno MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O'Carroll D, Greengard P 2010. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med 207: 1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F 2010. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci 19: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y 2007. Crystal structure of human DGCR8 core. Nat Struct Mol Biol 14: 847–853 [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. 2011. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E 1999. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci 96: 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD 2009. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol 16: 1259–1266 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Weiss M, Schrimpf S, Hengartner MO, Lercher MJ, von Mering C 2010. Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics 10: 1297–1306 [DOI] [PubMed] [Google Scholar]
- Wiśiniewski JR, Zougman A, Nagaraj N, Mann M 2009. Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC 2010. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci 107: 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596 [DOI] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR 2005. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11: 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E 2006. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38: 356–362 [DOI] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E 2008. A skin microRNA promotes differentiation by repressing ‘stemness.’ Nature 452: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, et al. 2009. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci 106: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]