The Drosophila midblastula transition (MBT) is accompanied by a dramatic change in replication timing, when the period of replication lengthens nearly fourfold. O'Farrell and colleagues find that down-regulation of the mitotic regulator Cdc25 is required for this S-phase lengthening. Cdc25 activity, via its target Cdk1, can speed S-phase progression and promote premature replication of late-replicating sequences, even after their specification. The study thus uncovers the mechanism underlying the MBT developmental switch that affects replication timing.
Keywords: Drosophila, midblastula transition, Cdc25, Cdk1, DNA replication, late replication
Abstract
The Drosophila midblastula transition (MBT), a major event in embryogenesis, remodels and slows the cell cycle. In the pre-MBT cycles, all genomic regions replicate simultaneously in rapid S phases that alternate with mitosis, skipping gap phases. At the MBT, down-regulation of Cdc25 phosphatase and the resulting inhibitory phosphorylation of the mitotic kinase Cdk1 create a G2 pause in interphase 14. However, an earlier change in interphase 14 is the prolongation of S phase. While the signals modifying S phase are unknown, the onset of late replication—where replication of constitutively heterochromatic satellite sequences is delayed—extends S-phase 14. We injected Cdc25 mRNA to bypass the developmentally programmed down-regulation of Cdc25 at the MBT. Introduction of either Cdc25 isoform (String or Twine) or enhanced Cdk1 activity triggered premature replication of late-replicating sequences, even after their specification, and thereby shortened S phase. Reciprocally, reduction of Cdk1 activity by knockdown of mitotic cyclins extended pre-MBT S phase. These findings suggest that high Cdc25 and Cdk1 contribute to the speed of the rapid, pre-MBT S phases and that down-regulation of these activities plays a broader role in MBT-associated changes than was previously suspected.
Following fertilization, the first order of business for large, externally developing eggs is to increase cell number, often by specialized post-fertilization mitotic cycles dedicated to this increase. In Drosophila, the early mitotic cycles are extremely fast (8.6–21 min) and synchronous (Foe 1989) and lack most transcription (Edgar and Schubiger 1986). In these cycles, S phase alternates with mitosis without intervening gap phases or cytokinesis, so nuclei exponentially amplify in a syncytial cytoplasm (Edgar and Schubiger 1986). At cycle 14, the cell cycle slows and morphogenesis replaces nuclear proliferation as the key feature of embryonic development (Foe and Alberts 1983). Numerous events accompany the cell cycle slowing, including the destruction of many maternal mRNAs, cellularization of the cortical nuclei, and the onset of gastrulation. Most importantly, transcription is activated and the zygotic genome begins to control the cell cycle and embryonic development (Edgar and Schubiger 1986; Edgar and O'Farrell 1989). This major embryonic transformation is the midblastula transition (MBT), named after analogous events in early frog development. We explored the regulation that slows the cell cycle at the Drosophila MBT.
Earlier studies of cell cycle slowing at the MBT focused on the cycle 14 introduction of a G2 phase, which results from changing regulation of Cdk1, the major coordinator of mitosis. Until the MBT, Cdk1 is highly active; thus, S phase occupies the entirety of interphase, and embryos are driven into mitosis immediately following replication. After the MBT, however, Cdk1 is inhibited by phosphorylation, causing cells to pause in G2 until the phosphorylation is removed (Edgar et al. 1994), meaning that in these cycles, S phase does not occupy all of interphase. This change in Cdk1 activity results from the down-regulation of Cdc25 (Edgar et al. 1994; Edgar and Datar 1996), the phosphatase that activates preformed cyclin–Cdk complexes by removing inhibitory phosphates from the roof of the ATP-binding pocket (Edgar and O'Farrell 1990; Nurse 1990; Jeffrey et al. 1995). Cdc25 phosphatase activity is encoded by two Drosophila homologs, string (stg) (Edgar and O'Farrell 1989) and twine (twe) (Alphey et al. 1992; Courtot et al. 1992), whose transcripts are both present at a high level until the MBT, when they are down-regulated (Edgar et al. 1994; Edgar and Datar 1996). Cdc25 down-regulation results in the accumulation of Cdk1 inhibitory phosphorylation (Edgar et al. 1994), and thus cells pause in G2 until spatially and temporally regulated pulses of zygotic string expression allow them to enter mitosis (Edgar and O'Farrell 1989).
Recent work has emphasized a second contribution to the slowing of the cell cycle at the MBT. Cycle 14 is distinguished from pre-MBT cycles in having a 50-min S phase versus the S phase of the preceding cycle, which lasts only 15 min (Edgar and O'Farrell 1990; Shermoen et al. 2010). This prolongation of S phase is the result of a shift from a mode where all regions of the DNA are replicated nearly simultaneously (Blumenthal et al. 1974) to a mode where the replication of repetitive satellite sequences is delayed until later in S phase (Shermoen et al. 2010). Shifts in replication timing occur in many metazoans as cells become more specified, including Drosophila, Xenopus (Walter and Newport 1997), and mammals (Gilbert et al. 2010). In the new mode of replication, the bulk of the DNA replicates in the first 15 min of S phase, but satellite sequences exhibit an ordered sequence of replication that occurs later, from 5 to 50 min in S phase (Shermoen et al. 2010). These satellite sequences are long stretches of short, repetitive sequence that are primarily heterochromatic. While the importance of the longer S phase is shown by evidence that it is required for the completion of other critical MBT events, such as cellularization (Edgar et al. 1986; McCleland et al. 2009), many questions remain about the regulation of S-phase length.
What developmental changes underlie the shift in replication timing of the satellite sequences? One possibility is a local change to the late-replicating sequences, such as a modification in chromatin structure. Indeed, progressive introduction of heterochromatic features during early development led us to think of late replication as part of the progression (Shermoen et al. 2010). However, some aspects, such as compaction, begin in the pre-MBT cycles, while other aspects, such as HP1 binding, occur after the introduction of late replication (Shermoen et al. 2010). Since these structural changes were not coincident with the onset of late replication, we entertained another possibility, which was that a global regulator of DNA replication changed and exposed differences in the late-replicating sequences that had been set up well before the MBT. This model, however, presented several questions still. What was the regulator? How is the change coupled to the MBT?
We wondered whether the developmental down-regulation of Cdc25 had an important function at the MBT aside from regulating the G2 phase. Cdc25 reintroduction in cycle 14 had previously been shown to eliminate the G2 (Edgar and O'Farrell 1990). However, it was reintroduced late in cycle 14, after many other MBT changes—notably, the extension of S phase—had occurred. To truly study the effect of Cdc25 down-regulation at the MBT, especially on earlier events in cycle 14, we used a more acute approach to reintroduce Cdc25 at the time of the MBT. This uncovered a role for Cdc25 in the regulation of S-phase length.
Results
Cdc25 causes an early mitosis 14 during the time normally occupied by S-phase 14
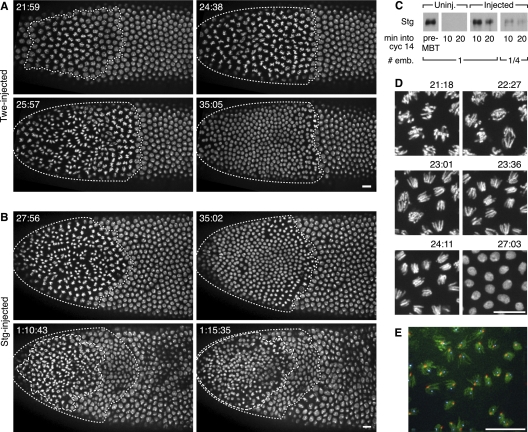
To study the role of Cdc25 down-regulation on post-MBT events prior to the G2 phase, we overexpressed Cdc25 at the end of cycle 13 so that Cdc25 would persist into cycle 14 longer than normal (Fig. 1C) and allow us to examine its effect during a previously inaccessible developmental time. We did this by injecting embryos with in vitro transcribed mRNA encoding either Twe or Stg. We found that addition of Cdc25—either Twe or Stg—induced an early mitosis in the treated portion of the embryo (Fig. 1A,B; Supplemental Movies S1, S2), though injection of mRNA encoding phosphatase-dead mutants of Twe and Stg had no phenotype (data not shown). The Cdc25-induced mitoses originated at the point of injection and proceeded in quick waves like pre-MBT blastoderm divisions. Extra divisions generally left the embryo well organized, unless they occurred near the time of gastrulation (Fig. 1B, bottom right panel). After the extra cycle (or in the case of Stg-injected embryos, often two extra cycles), embryos executed the patterned mitosis that would normally occur in cycle 14 (data not shown). The average length of the shortened interphase 14 in treated embryos was 28 min, though S-phase 14 is normally 50 min (Shermoen et al. 2010) in a wild-type embryo. This indicated that Cdc25 could shorten interphase 14, though by more than the previously reported removal of G2 phase, and was causing embryos to enter mitosis during what would ordinarily be S-phase 14.
Figure 1.
Cdc25 mRNA injection induces a successful, early mitosis in cycle 14 during what would ordinarily be S phase. (A,B) Stills from movies following His2AvD-GFP embryos injected during cycle 13 with Twe (A) or Stg (B) mRNA. Times displayed are time after mitosis 13. The region undergoing division (dotted line) is localized near the point of injection (left pole). (A) A single mitosis after a 21-min interphase (see Supplemental Movie S1). (B) Two successive induced mitoses—the first (dotted line) after a 23-min interphase, and the second (dashed line in bottom panels) ∼35 min later. The normal onset of gastrulation movements has shifted nuclear positions in the last panel (see Supplemental Movie S2). (C) Western blot showing Stg expression in injected embryos is similar to that in pre-MBT embryos. Lanes were loaded with aliquots equivalent to either one or one-quarter embryo (# emb.) from pooled extracts of three embryos of the indicated type. The pre-MBT sample is from cycle 11 embryos. Time into cycle 14 was estimated by nuclear length. (D) Series of stills from Supplemental Movie S3. These high-magnification views of the induced mitosis in an embryo injected with Stg mRNA show no DNA bridging at anaphase (see especially panels 23:01 and 23:36). Times are those displayed in Supplemental Movie S3. (E) Induced anaphases in a Stg mRNA-injected embryo that was fixed 22 min after mitosis 13. Hoechst staining of the DNA (green) shows no evident bridging of the DNA, and the in situ hybridization signals for the X-chromosomal 359 satellite (red) and the Y-chromosomal AATAC satellite (cyan) show distinct foci of these late-replicating sequences in each segregating complement. Bars, 10 μm.
Cdc25 shortens S-phase 14
Since Cdc25 expression caused embryos to enter mitosis during what would normally be S phase, they had either finished S phase early or entered mitosis without completing replication. Entry into mitosis before chromosomes finish replicating leads to catastrophe upon attempting to separate sister chromatids (Heald et al. 1993) that are still held together by unreplicated regions. However, mitosis proceeded normally in embryos injected with Cdc25 mRNA, without bridged DNA during anaphase (Fig. 1D; Supplemental Movie S3), indicating that DNA replication was complete. As an alternative assay of the completeness of S phase, we examined normally late-replicating sequences by in situ hybridization. An embryo undergoing an early, Stg-induced mitosis showed cleanly separated anaphase sister chromatids (Fig. 1E), each with its own nonbridged focus for the X-chromosomal 359 satellite and the late-replicating, Y-chromosomal AATAC satellite (Shermoen et al. 2010). We conclude that, in embryos injected with Cdc25 mRNA, satellites that usually replicate later in S phase are successfully duplicated early, and genome duplication is complete before the induced mitosis. Thus, introduction of Cdc25 is sufficient to accelerate the progress of S-phase 14 so that it is completed in less than the normal time.
Cdc25 causes early replication of normally late-replicating sequences
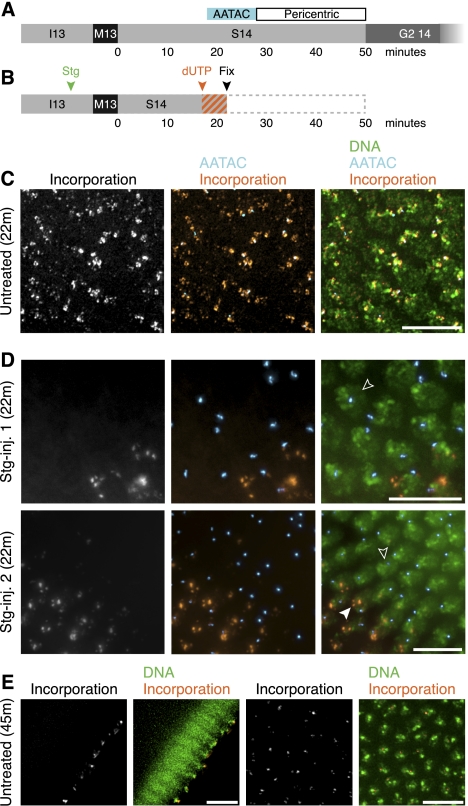
To more directly assay replication in Cdc25-treated embryos, and do so in a manner that would allow us to assay replication of particular sequences, we injected Stg-treated embryos with fluorescent deoxynucleotides, fixed the embryos, and then assessed fluorescent incorporation into replicated DNA. Using in situ hybridization to localize particular blocks of satellite sequences, we could define the timing of incorporation into distinct sequences. Alterations in incorporation after injection of Stg mRNA showed that the accelerated S phase it induced was associated with earlier replication of ordinarily late-replicating satellite sequences. At 22 min into interphase 14, normally all nuclei would be midway through replication; they would be in the process of replicating the AATAC satellite sequence and would not have begun to replicate the latest-replicating sequences, such as the pericentric heterochromatin (Fig. 2A). Thus, they showed nucleotide incorporation as a faint, punctate pattern with a large focus that colocalizes with the AATAC satellite (Fig. 2C). In Stg-treated embryos that were also 22 min into interphase 14 (Fig. 2B), replication was unusually asynchronous (Fig. 2D). Many of their nuclei exhibited no incorporation (Fig. 2D, open arrowheads), indicating that they completed replication when normal embryos would still be replicating heavily. Those nuclei with incorporation showed a pattern representative of a more advanced stage in S phase (Fig. 2E). In particular, (1) the labeling was confined to the pericentric sequences (Fig. 2D, closed arrowhead), evident because of their apical localization (Wilkie et al. 1999) and which usually would not begin replicating for at least six more minutes, and (2) the AATAC satellite sequence lacked incorporation, although it would ordinarily still be midreplication at that time (Shermoen et al. 2010). These embryos show that introduction of Cdc25 shortened S phase, causing them to complete replication and stop incorporating nucleotides earlier than normal. Moreover, most nuclei had finished replicating the pericentric sequences at 22 min—before these sequences would normally begin to replicate—showing that Cdc25 advanced the time of replication of late-replicating sequences.
Figure 2.
Late-replicating sequences replicate prematurely during a Cdc25-accelerated S phase. (A) A timeline illustrating when the AATAC satellite sequence and the pericentric sequences replicate in S-phase 14 of an untreated embryo (Shermoen et al. 2010). (B) A schematic of the experiment shown in C and D. Stg mRNA was injected (as shown in D) in cycle 13 (green arrowhead), fluorescent nucleotide (dUTP) was injected in mid-S-phase 14 (orange arrowhead), and embryos were fixed shortly after (black arrowhead) to give a brief labeling period (orange/gray stripes). (C) A region of an untreated embryo fixed 22 min after mitosis 13. All nuclei exhibit multiple puncta of incorporation (orange), and in most nuclei, incorporation colocalizes with the late-replicating AATAC satellite sequence (cyan/white in overlap). (D) Regions of two Stg mRNA-injected embryos fixed 22 min after mitosis 13. Many nuclei (open arrowheads) lack incorporation (orange). Labeled nuclei show restricted apical foci (closed arrowhead) that do not colocalize with the AATAC satellite (cyan). (E) Cross-sectional (left) and en face (right) views of fluorescent nucleotides incorporated late (∼40–45 min post mitosis 13) during S-phase 14 in an untreated embryo. Note the focal, apically located incorporation pattern (orange), which is typical of late S-phase 14. Bars, 10 μm.
Cdc25 acts on sequences that are already specified as late-replicating
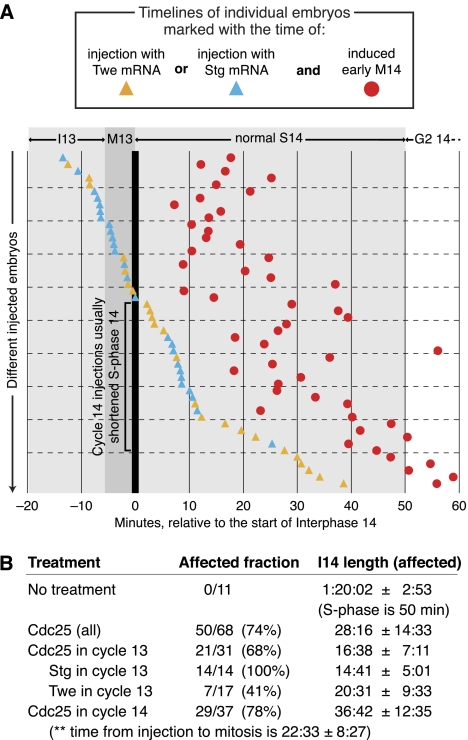
Cdc25 could advance replication of late-replicating sequences by either preventing them from being specified as late-replicating sequences or overriding that specification and causing a sequence that has been specified as late-replicating to replicate early anyway. To differentiate between these possibilities, we tested the relative effectiveness of Cdc25 injection in cycles 13 and 14. Replication is widespread by 1 min into cycle 14 (Shermoen et al. 2010), but the late-replicating sequences do not initiate their replication until later. Thus, late sequences must have been distinctly specified by the beginning of cycle 14, when S-phase 14 begins, otherwise their replication would have initiated with the early-replicating sequences. So, if Cdc25 introduced after the onset of S-phase 14 was able to accelerate that S phase, it would show that Cdc25 could advance the replication of sequences that had already been specified as late. We watched embryos on the microscope to precisely time injections with Stg or Twe mRNA in either cycle 13 or cycle 14. Injections in both cycles proved similarly effective at inducing a shortened S-phase 14 (Fig. 3A), and most embryos that were injected with Cdc25 mRNA, even several minutes after the beginning of S-phase 14 (Fig. 3A, embryos highlighted with black bracket), exhibited a shortened S phase. Not surprisingly, embryos injected later in cycle 14 were accelerated less than those injected earlier, with the extra mitosis occurring, on average, 23 min after injection (Fig. 3A). We conclude that Cdc25 can shorten S phase even after establishment of the post-MBT replication program. Since Cdc25 shortens replication by advancing the replication of late-replicating sequences (Fig. 2), these experiments suggest that Cdc25 can promote the replication of satellite sequences even after they have been specified as late-replicating.
Figure 3.
Injection of Cdc25 mRNA after establishment of late replication can shorten S-phase 14. (A) Time courses for 50 individual His2AvD-GFP embryos (each represented as a separate position on the Y-axis) that were induced into an extra mitosis (red circles) by injection of Stg mRNA (blue triangles) or Twe mRNA (yellow triangles). Other experimental embryos (18) that did not undergo an induced mitosis are omitted from the graph, but are tabulated in B. A live record of each embryo spanned the time course except for a brief interruption around the time of injection. Time (X-axis) is recorded in relation to the end of mitosis 13 (thick vertical line). A black bracket highlights embryos injected during S-phase 14 with shortened S phases. (B) Tabulation of interphase times. Treatments are grouped in several ways to illustrate their relative efficacy. For Cdc25 in cycle 14, the time between injection and entry into mitosis is also given (**). “Affected fraction” is affected/total injected, and “I14 length” is in hours:minutes:seconds ± the standard deviation and is calculated from only the affected embryos.
Cdk1 activity also drives a successful mitosis during normal S-phase 14
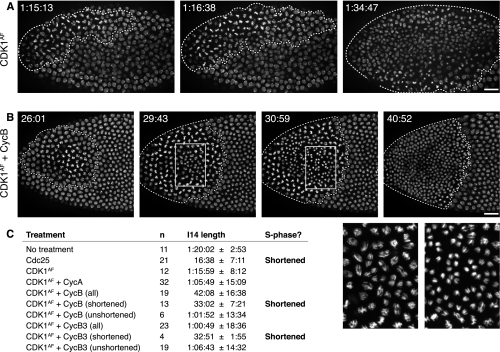
The only previously described roles for String and Twine are in mitosis or meiosis. Our discovery that these traditionally mitotic proteins had a role in replication led us to test whether other mitotic activators could cause early entry into mitosis during what would normally be S-phase 14. Cdk1 is both a mitotic activator and a traditional target of Cdc25, so we began with it. We made a construct of Cdk1 with mutations that eliminate the phosphorylation sites that usually result in its inhibition and are dephosphorylated by Cdc25 (referred to as “Cdk1AF”). Injection of Cdk1AF mRNA advanced mitosis, but not to an extent that would suggest a shortened S phase; Cdk1AF triggered mitosis, on average, 76 min into cycle 14, after S phase would normally finish (Fig. 4A,C). In Cdk1AF-treated embryos, neighboring mitoses did not always occur concurrently, and the zone of effect spread further (Supplemental Movie S4) than the local, concerted waves of mitosis in Cdc25-treated embryos.
Figure 4.
Coinjection of Cdk1AF mRNA with CycB or CycB3 mRNA induces an early, successful mitosis during what would normally be S phase. (A,B) His2AvD-GFP embryos were injected on the left with Cdk1AF mRNA, either alone or in combination with CycB mRNA. Dotted lines surround regions of the embryo during or after an early mitosis. Times displayed are time within interphase 14. Bars, 20 μm. (A) Stills from Supplemental Movie S4. An embryo injected with only Cdk1AF mRNA entered an induced mitosis, but only after an interphase that is longer (72 min) than a normal S phase. (B) Stills from Supplemental Movie S5. An embryo injected with Cdk1AF and CycB mRNA that entered a successful induced mitosis 24 min after mitosis 13, indicating a shortened S phase. White boxes highlight regions that are shown in closeup view in the panels at the bottom right. (C) Average length of interphase 14 (±standard deviation) following expression of different mitotic inducers. Cdk1AF + CycB and Cdk1AF + CycB3 are shown as both the entire treated population (all) and subpopulations with and without shortened S phases (shortened/unshortened). Data for Cdc25 and no treatment are reproduced from Figure 3 for purposes of comparison.
Since Cdk1, regardless of phosphorylation, requires cyclin to be active, it was possible that newly produced Cdk1AF in our experiments did not have sufficient cyclin to bind until later in the cell cycle. So, we tested whether coexpression of a mitotic cyclin with Cdk1AF would further advance mitosis. Embryos coinjected with CycA and Cdk1AF mRNA did not further shorten interphase (Supplemental Fig. S1A). However, coinjection of CycB3 mRNA or CycB mRNA with Cdk1AF mRNA further shortened interphase in a fraction of the injected embryos—17% and 68%, respectively (Fig. 4B,C; Supplemental Fig. S1B,C; Supplemental Movie S5). When injected alone, none of the cyclin mRNAs advanced mitosis (data not shown), indicating that it was the Cdk1AF–cyclin complex that shortened S phase. Those embryos with shortened interphases following Cdk1AF and cyclin mRNA coinjections had an average interphase time of 33 min and a coordinated wave of mitosis resembling the embryos injected with Cdc25 mRNA (Fig. 4B; Supplemental Movie S5). Thus, injection with Cdk1AF and CycB or CycB3 can phenocopy injection with Cdc25 and is sufficient to shorten S phase. Given that Cdk1 is the target of Cdc25 (Edgar et al. 1994), we suggest that Cdk1–cyclin activation mediates the shortening of S phase induced by Cdc25 in our experiments.
Visualizing Cdc25 and Cdk1 acceleration of replication
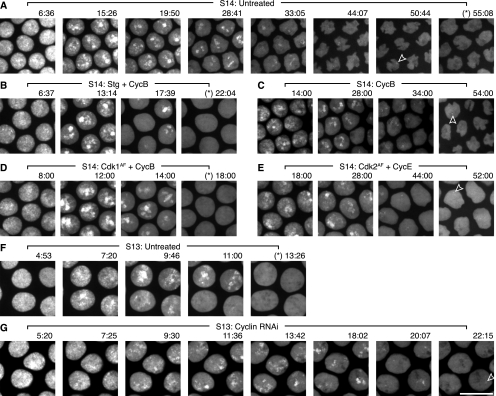
We also tested the effect of Cdc25 and Cdk1 on replication by injecting embryos with purified GFP-PCNA protein to use as a live marker. PCNA associates with DNA polymerase at replication forks, and GFP-tagged PCNA accumulates into visible foci during replication. The formation of these foci requires replication, as none are observed in embryos that are prevented from licensing their origins and initiating replication (McCleland et al. 2009). Thus, we used GFP-PCNA foci as an approximation of the progress of replication. In embryos injected solely with GFP-PCNA, foci were detectable for an average of 51 min (Fig. 5A), similar to the previously reported 50-min S phase determined by nucleotide incorporation (Shermoen et al. 2010). PCNA exhibited a widespread pattern during the beginning of S phase (Fig. 5A, 6:36), corresponding to the bulk replication of euchromatin that occurs during early cycle 14 (Shermoen et al. 2010). Then, as S-phase 14 progressed, the GFP-PCNA pattern became focal, with the foci becoming fewer, smaller, and fainter (Fig. 5A, 28:41–50:44) at the same time as replication becomes progressively restricted to the late-replicating satellite sequences (Shermoen et al. 2010).
Figure 5.
Stg or Cdk1AF mRNA, coinjected with CycB mRNA, accelerate replication, while dsRNA against mitotic cyclins slows replication. Stills from Supplemental Movie S6. Embryos were filmed after injection with purified GFP-PCNA protein and nothing (A,F), Stg and CycB mRNAs (B), CycB mRNA (C), Cdk1AF and CycB mRNAs (D), Cdk2AF and CycE mRNAs (E), or dsRNA against CycA, CycB, and CycB3 (G). Time into interphase 14 is marked above the micrographs. An asterisk (*) before the time indicates a micrograph in which no PCNA foci are visible. In some panels, an open arrowhead highlights the last visible late PCNA focus. Also, in some later time-point micrographs (e.g., A, 44:07), the nuclei can be observed changing shape, which is a normal consequence of cellularization that is happening during cycle 14. Bar, 10 μm.
Injection of Stg mRNA did not change the dynamics of the GFP-PCNA foci in embryos injected with GFP-PCNA protein and only triggered mitosis after sufficient time to complete an unaccelerated S-phase 14 (data not shown). This reveals a peculiarity of PCNA overexpression through either injection of purified protein or transgenic expression. We injected Stg mRNA into five strains, all descended from different, unconnected backgrounds; a strain expressing YFP-PCNA was the only tested strain in which Stg mRNA did not trigger an early mitosis, consistent with a shortened S phase (data not shown). Moreover, injection of GFP-PCNA protein prevented Stg mRNA from triggering a shortened S phase in other strains where it would in the absence of PCNA overexpression (data not shown). Thus, we suspect there may be an interaction between PCNA overexpression and Cdc25, which is a subject of interest for future study. However, since mitotic cyclin had increased the efficacy of Cdk1AF in previous experiments, we tried coinjecting CycB mRNA and Stg mRNA into embryos also injected with GFP-PCNA.
Embryos injected with a mixture of Stg and CycB mRNAs displayed a clear acceleration in the dynamics of GFP-PCNA foci, which disappeared after an average of 22 min (Fig. 5B). Moreover, the foci did not become as few or as restricted in the last frames of the shortened S phase (Fig. 5B, 17:39), suggesting that a greater number of sequences were still replicating concurrently at the end of S phase. CycB mRNA injected alone did not change the dynamics of GFP-PCNA foci, which remained detectable as late as 54 min (Fig. 5C, open arrowhead). We conclude that increased expression of Stg and CycB can accelerate the progress of S phase, as marked by GFP-PCNA.
Coinjection of Cdk1AF and CycB mRNAs produced a similar result, where the dynamics of GFP-PCNA foci were accelerated so that they disappeared after an average of 22 min (Fig. 5D), and the foci did not become as small or faint in the last minutes of replication as in control embryos (Fig. 5D, 14:00). Based on this, we conclude that combined Cdk1 and CycB can also accelerate replication. In our hands, however, a similar coinjection (Supplemental Fig. S2A) of Cdk2AF and CycE mRNAs did not have an effect on replication timing, as GFP-PCNA foci were still visible as late as 52 min (Fig. 5E, open arrowhead). While we cannot conclude that Cdk2 cannot accelerate S phase, it does not do so in our experimental setting, while Cdk1 is effective. Since Stg with CycB and Cdk1AF with CycB accelerated the dynamics of GFP-PCNA foci in S-phase 14 and caused those foci to disappear before the embryos entered early mitoses, we conclude that increasing Cdc25 or Cdk1 activity can accelerate progression through S phase.
Reduction of Cdk1 activity prolongs pre-MBT S phase
Since the replication of the late-firing sequences could be advanced by increasing Cdk1 activity after the MBT, this suggested that the short S phases before the MBT may be due to high Cdk1 activity during S phase. So, we tested whether reducing Cdk1 activity before the MBT could prolong S-phase 13, the last of the rapid S phases. To dampen Cdk1, we injected a cocktail of dsRNAs against its three cyclin partners (CycA, CycB, and CycB3), and to assay replication, we injected purified GFP-PCNA protein. We analyzed embryos that were prevented from entering mitosis 13 by the dsRNAs, which indicated that Cdk1 activity was reduced in these embryos, although it was almost surely not eliminated.
In embryos injected with only GFP-PCNA, PCNA foci persisted for an average of 12 min during S-phase 13 (Fig. 5F). PCNA showed a pattern of initially widespread staining (e.g., Fig. 5F, 6:06) that became slightly restricted and focal as S phase progressed (e.g., Fig. 5F, 9:46–13:26), although it never became as restricted as in late S-phase 14 (e.g., Fig. 5A, 33:05–50:44). In embryos also injected with cyclin dsRNA, however, PCNA foci persisted for an average of 19 min (Fig. 5G). The PCNA pattern progressed in these embryos similarly to that in unperturbed S13: PCNA was initially widespread (e.g., Fig. 5G, 5:20) and then became progressively more focal and restricted (Fig. 5G, 11:36–22:15). However, the widespread pattern seemed to persist for approximately the same amount of time (Fig. 5, F [through 9:46] vs. G [through 9:30]), while the more focal-restricted pattern seemed to lengthen considerably (Fig. 5, F [11:00–13:26] vs. G [11:36–22:15]). While this experiment does not indicate the timing of replication of individual sequences, the extension of the focal pattern, but not the widespread pattern, suggests that these results represent the introduction of a small amount of late replication in these embryos. The observed extension of S phase does not approach the full length of S-phase 14, but it does show that Cdk1 activity plays a role in the rapidity of the pre-MBT S phases. Moreover, it suggests that the late replication program is already in place before the MBT, since reduction in Cdk1 seems to partially uncover the program.
An unexpected checkpoint
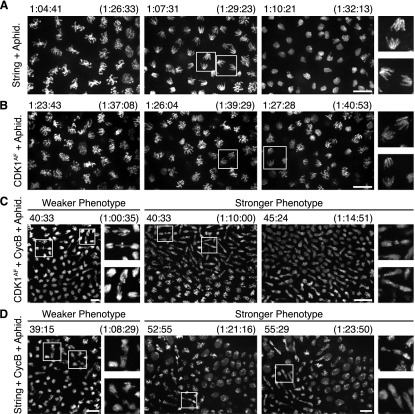
Our findings presented us with one further puzzle. Why did expression of the mitotic activators Cdc25 and Cdk1AF not induce an immediate mitosis with unreplicated DNA? The well-recognized checkpoint that guards against entry into mitosis with incompletely replicated DNA is thought to act by Chk1 promotion of inhibitory phosphorylation of Cdk1 (Sibon et al. 1997). However, Cdc25 expression should override inhibitory phosphorylation by removing it, and Cdk1AF expression should bypass inhibitory phosphorylation entirely. Thus, it seemed unlikely that the canonical S-phase checkpoint could protect against premature mitosis in our experiments. To test whether a different checkpoint was involved, we prolonged S phase by injecting embryos with aphidicolin, a competitive inhibitor of DNA replication (Spadari and Sala 1982), at a dose that slowed but did not halt replication. Aphidicolin delayed the mitosis induced by Stg mRNA (Supplemental Fig. S3C), and this mitosis showed no anaphase bridging (Fig. 6A; Supplemental Movie S7). This indicated that replication could still delay entry into mitosis, even after Cdc25 injection. Similarly, treatment with aphidicolin delayed the mitosis induced by injection of Cdk1AF mRNA (Supplemental Fig. S3C; Supplemental Movie S8). Although the Cdk1AF-induced mitoses in aphidicolin exhibited a prolonged prophase and some disorganization of the nuclei, anaphase separation of chromosomes was successful, which indicated complete replication prior to mitosis (Fig. 6B; Supplemental Movie S8). We conclude that ongoing replication delays mitosis, even after induction of Cdc25 or Cdk1AF. Thus, we suggest that a secondary replication checkpoint, independent of inhibitory phosphorylation of Cdk1, prevents catastrophic entry into mitosis.
Figure 6.
Limited cyclin acts as a replication checkpoint in the absence of inhibitory Cdk1 phosphorylation. His2AvD-GFP embryos injected with mRNA in cycle 13 and aphidicolin in cycle 14. Times in parentheses are time within interphase 14, and those without parentheses are the times displayed in the corresponding Supplemental Movies. Bars, 10 μm. (A) Stills from Supplemental Movie S7 of an embryo injected with Stg mRNA undergoing an induced but aphidicolin-delayed mitosis. Representative unbridged anaphase chromosomes are boxed and shown at high magnification. (B) Stills from Supplemental Movie S8 of an embryo injected with Cdk1AF mRNA and then aphidicolin with a successful anaphase. Entry into mitosis is highly asynchronous in these embryos, but representative unbridged anaphase chromosomes are boxed and shown at high magnification. (C) Stills from Supplemental Movie S9 of embryos injected with Cdk1AF and CycB mRNAs, then aphidicolin. A range of phenotypes resulted, and embryos with weaker and stronger anaphase-bridging phenotypes are displayed. Representative chromosomes with bridged anaphases are boxed and shown at high magnification. (D) Stills from Supplemental Movie S10 of embryos injected with Stg and CycB mRNAs, then aphidicolin. A range of phenotypes resulted, and embryos with weaker and stronger anaphase-bridging phenotypes are displayed. Representative chromosomes with bridged anaphases are boxed and shown at high magnification.
In these experimental contexts where phosphorylation control was disturbed, we considered the possibility that cyclin limitation might underlie the alternative checkpoint. Indeed, embryos injected with aphidicolin, Cdk1AF mRNA, and any mitotic cyclin mRNA entered mitosis with catastrophic anaphase bridging (Fig. 6C; Supplemental Fig. S3A,B; Supplemental Movie S9). Moreover, in aphidicolin-injected embryos, coinjection of Stg mRNA and CycB mRNA induced some anaphase bridging (Fig. 6D; Supplemental Movie S10). We conclude, therefore, that low cyclin function limits the ability of Cdk1AF and Cdc25 to induce mitosis during an ongoing S phase. Thus, by definition, low cyclin function would constitute a secondary checkpoint in these situations, but we have not defined how ongoing replication would act to limit cyclin function.
Discussion
Cdc25 decline is a developmental switch involved in changing replication timing
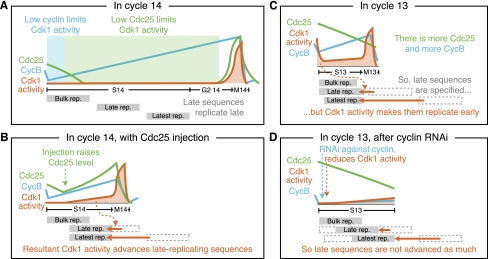
Our experiments overrode the normal down-regulation of Cdc25 and Cdk1 activity at the MBT and showed that this developmental down-regulation is required for the introduction of late replication at S-phase 14 (Fig. 7A,B). We showed that increased Cdc25 or Cdk1 activity during cycle 14 abbreviates the late replication program normally active at the time. Conversely, we found that decreasing Cdk1 activity during cycle 13 lengthens the rapid replication program that is active before the MBT (Fig. 7C,D). This suggests that Cdc25 and Cdk1 activity are regulating the length of S phase. Moreover, the pre-MBT S phases are unusual in having a rapid replication program and also in having high Cdc25 and Cdk1 activity during S phase. Then, cycle 14—when late replication begins in earnest—is the first cycle in which Cdc25 is effectively down-regulated and Cdk1 is inhibited by phosphorylation (Edgar et al. 1994; Edgar and Datar 1996). Given our findings, we propose that this high Cdc25 and Cdk1 activity is actually the reason the pre-MBT S phases are rapid and the removal of these activities by down-regulation at the MBT is the developmental switch that lengthens S phase.
Figure 7.
A model of how declining Cdc25 and Cdk1 activity results in S-phase 14 prolongation. (A) Cdk1 activity is low throughout S-phase 14, first due to low cyclin, then due to low Cdc25 (as a result of its developmental down-regulation), which allows late-replicating sequences to replicate late and S-phase 14 to be long. (B) Our injections of Cdc25 mRNA raise the level of Cdc25 during S-phase 14, causing higher Cdk1 activity, which causes the late-replicating sequences to replicate early, leading to a shortened S phase. (C) In the pre-MBT S phases, such as cycle 13 depicted here, all sequences basically replicate early, with only a couple of minutes difference in the time that individual sequences replicate. We propose that, while the late-replicating sequences are specified, Cdk1 activity during the pre-MBT S phases results in the early replication of late-replicating sequences during those cycles as well, leading to their short S phases. (D) Our injections of RNAi against the mitotic cyclins extend the length of S-phase 13. We propose that this is due to a reduction in Cdk1 activity, which is necessary to advance the late-firing sequences in the pre-MBT S phases.
While these results do not address mechanistic aspects of the ordered firing of late-replicating sequences, they do indicate mechanistic aspects of the activation of the late-replication program. Our findings suggest that the satellite sequences do not become late-replicating at the MBT due to local changes at the origins. Instead, it seems that those origins may already be specified as late-replicating. For instance, satellite sequences are selectively compacted before the MBT (Shermoen et al. 2010), suggesting that the onset of late replication is not due to de novo acquisition of heterochromatic structure at the MBT. Moreover, reduction of Cdk1 activity even before the normal onset of late replication can lengthen replication, which suggests that the embryo is already programmed with its post-MBT replication program. However, our experiments show that Cdc25 can advance replication of late-replicating sequences when introduced after the beginning of S-phase 14 and thus demonstrate the capacity of increased Cdc25 to override local signals that specify genomic regions as late-replicating. Thus, we propose that late-replicating sequences are specified as late long before the MBT but that high Cdc25 and Cdk1 activity drives them to fire early anyway during the pre-MBT S phases. Once Cdk1 is inactivated at the MBT by Cdc25 destruction and Cdk inhibitor (CKI) induction (Grosshans and Wieschaus 2000), satellite sequences would no longer be driven to replicate at the beginning of S phase and so could follow the replication program that will drive them to replicate late thereafter. Thus, the onset of late replication and lengthening of S phase would result not from local changes specific to the late-replicating sequences, but rather from a global change with an impact on the replication machinery: the down-regulation of Cdc25 and Cdk1.
Cdc25 down-regulation feeds into an integrated Cdk1 switch
Our experiments allowed us to understand features of previous studies that were otherwise unexplained. Although none of these studies examined or commented on S-phase length, they presented other manipulations that advanced mitosis 14. In light of our observations, we can infer from the timing and success of these mitoses that S phase may have been shortened. For instance, mutants in frühstart, an inhibitor of Cdk1 (Gawliński et al. 2007) that is expressed in cycle 14 (Grosshans et al. 2003), have a low-penetrance shortening of interphase 14 that indicates, we infer, a short S-phase 14. Another study revealed that mutations that increased CycB levels in early interphase 14 caused a successful, early mitosis (Papoulas et al. 2010) that we interpret as indicative of a shortened S phase. Moreover, there is a 5-min window at the beginning of interphase 14, when injections of stable CycB protein can drive embryos into an early mitosis (Royou et al. 2008). The success of the mitosis was not assayed due to interference from the stable cyclin, but S phase was probably shortened, given the previously cited result.
Based on our observations and those from the literature, we propose that it is ultimately the down-regulation of Cdk1 that causes the lenthening of S-phase 14, and Cdc25 down-regulation is an important component of this. In fact, we show that Cdk1 activity during S-phase 14 will keep S phase short. The brief window of time when CycB can shorten S phase suggests that Cdk1 is probably inactive in the very beginning of cycle 14 because of insufficient cyclin. This results from the standard mitotic destruction of cyclins by the APC and provides time for the induction of the Cdk1 inhibitor Frühstart and the down-regulation of Cdc25—the developmental switches that further inactivate Cdk1 (Fig. 7A). These changes prevent Cdk1 activity during S-phase 14 even after the accumulation (or injection) of cyclin, thereby allowing S phase to be longer than those that preceded it. Furthermore, the features of a Cdk1-controlled switch may also explain earlier changes in S phase. There is a progressive, slight extension of S phase during the pre-MBT cycles (Shermoen et al. 2010). There is also a slow decline of Cdc25 during this time (though not much increase in inhibitory Cdk1 phosphorylation) as well as progressively strong destruction of cyclin during these cycles (Edgar et al. 1994). One or both of these may result in slight reductions in Cdk1 function during these cycles that explains the slight extension of S phase in the pre-MBT cycles.
A role for mitotic inducers in S phase
Our results provide an intriguing new view of proteins traditionally considered to be mitotic activators. Although vertebrate Cdc25A has previously been shown to have a role in the G1/S transition by activating Cdk2 (Blomberg and Hoffmann 1999), our results are substantially different. First, Stg and Twe are homologs of vertebrate Cdc25C, a mitotic inducer, rather than Cdc25A, an inducer of S phase. Second, Cdc25A acts in vertebrates to stimulate the G1-to-S transition, but here we show action of Cdc25 during an ongoing S phase to stimulate the late-replicating sequences. We believe that this is the first indication of a role for Cdc25 during the progression of S phase. Experiments transplanting mammalian nuclei into Xenopus extracts have shown that replication timing can be affected by general Cdk levels (Thomson et al. 2010). However, in our hands, we found that it is Cdk1 activity, traditionally associated with entry into mitosis, rather than Cdk2 activity, traditionally associated with stimulation of S phase, that advances late replication. This raises the intriguing possibility that late-replicating sequences are sensitive to Cdk1 when they are not sensitive to Cdk2.
We do not know how Cdk1 advances late replication; it may involve a novel mechanism, or it may parallel established roles for Cdk in firing origins. Work in yeast has established that Cdk has an essential role in origin firing by enabling the recruitment of required components to the fork after phosphorylating the replication proteins Sld2 and Sld3 (Zegerman and Diffley 2007; Labib 2010; Lopez-Mosqueda et al. 2010). The phosphorylation control of Sld3 by Cdk is conserved to mammals, where presumably Cdk2—the major S-phase kinase—acts on it (Boos et al. 2011). Perhaps the major difference in the late-firing origins is that, when the early-firing origins can be acted on by Cdk2, the late-firing origins are resistant at either the as-yet-undiscovered Drosophila homolog of Sld3 or another component. However, we propose that these origins are sensitive to stimulation by Cdk1, as we think happens in the pre-MBT cycles. In this case, Cdk1 may act in place of Cdk2 on the components it would usually phosphorylate; after all, in mice, the mitotic Cdks can compensate for deletions of the S-phase Cdks (Santamaría et al. 2007). Another possibility is that Cdk1 removes the impediment to Cdk2 at the late-firing origins and allows Cdk2 to act on them. Once Cdk1 is down-regulated at the MBT, perhaps an alternate mechanism removes the impediment to Cdk2 activity and allows the late origins to fire.
A second DNA replication checkpoint?
A surprising feature of our experiments is that ectopic induction of mitotic activators does not provoke mitosis before completion of S phase, which would lead to mitotic catastrophe. This is further highlighted by our results that even in aphidicolin-slowed S phases, treatment with Cdc25 or Cdk1AF does not generate mitotic catastrophe, which means that it is probably not simply that the kinetics of finishing replication are faster than those of entering mitosis. The previously described replication checkpoint that protects embryos treated with aphidicolin acts through Chk1 kinase (Sibon et al. 1997), but our treatments should override or bypass Chk1-mediated inhibitory phosphorylation of Cdk1. Thus, we believe that we exposed a second layer of regulation that protects embryos from premature entry into mitosis during DNA replication. Because this regulation can be bypassed by expression of mitotic cyclins, we believe that this checkpoint may act by limiting cyclin activity or levels.
We are still left with the challenge of explaining the finding that the S-phase checkpoint that we uncovered prevents entry into mitosis with incompletely replicated DNA but does not prevent Cdk1 from accelerating S phase. We note, however, that it has been shown that the Cdk activity required to trigger S phase is less than that required to activate mitosis (Coudreuse and Nurse 2010). Thus, this checkpoint might be effective enough to block mitosis, but permit a rise in Cdk1 activity sufficient to stimulate the replication of late-replicating sequences and thus promote the completion of S phase. This organization could help safeguard against disruptions in DNA replication. The dramatic developmental changes in S phase that we described expose unappreciated aspects of its regulation that should provide a context for their further investigation.
Materials and methods
Creation of constructs
All inserts were amplified from a general cDNA library made with SuperScript II reverse transcriptase and oligo(dT) on RNA isolated from 0- to 3-h embryos, unless otherwise noted. A description of the construction of the plasmids pJFStg, pJFTwe, pJFCdk1AF, pJFCdk2, pCycA-GFP, pCycB-GFP, pCycB3-GFP, and pCycE-GFP is given in Supplemental Table S1. The primers used are described in Supplemental Table S2.
Phosphatase-dead Cdc25 mutants (StgPD and TwePD) were produced by PCR site-directed mutagenesis. pJFStgPD was created by introducing into pJFStg the Cys379Ala mutation (C1137T and G1138C) with the primers Stg C379A Fwd and Rev. pJFTwePD was created by introducing into pJFTwe the Cys318Ala mutation (T952G and G953C) with the primers Twe C318A Fwd and Rev. The mutations contained in CDK1AF are T14A and Y15F (in the DNA sequence, these changes are A40G and A44T). PCR site-directed mutagenesis was then used to introduce into pJFCdk2 the mutations T17A and Y18F (in the DNA sequence, these changes are A49G and A53T) with the primers CDK2AF Fwd and Rev.
In vitro transcription of mRNA
Supplemental Table S3 describes the specifics of each particular mRNA used. Generally, mRNAs were transcribed for injection using CellScript's T7 mScript mRNA production system (C-MSC11610). Constructs were first linearized by incubating 2 μg of plasmid in 8 μL of water with 1 μL of NEBuffer, 1 μL of 10× New England Biolabs BSA, and 0.25 μL of restriction enzyme for 3 h at 37°C. In a modification of CellScript's protocol, to transcribe, 1.7 μL of digested template, 1.8 μL of NTP solution, 0.5 μL of 10× transcription buffer, 0.5 μL of 100 mM DTT, and 0.5 μL of T7 enzyme mix were combined at room temperature and incubated for 90 min at 37°C. An exception was the Stg and Stg-PD RNAs, which were transcribed using Promega T3 RNA Polymerase (#P2083) according to the manufacturer's protocol for “synthesis of nonlabeled RNA.” The transcription reaction was cleaned up using the Qiagen RNEasy MinElute Cleanup kit as described by the manufacturer, but with the drying spin (step 6) extended to 7 min and elution volume (step 7) increased to 18 μL and incubated for 2 min at room temperature before spinning. RNA was quantitated using a Thermo Scientific Nanodrop 1000. In a modified version of CellScript's protocol, RNA was converted into mRNA by denaturing 22.5 μg of RNA in 28 μL of water for 5 min at 65°C, icing for 1 min, and adding 3.75 μL of 10× capping buffer, 1.9 μL of 20 mM GTP, 0.38 μL of 20 mM SAM, 0.95 μL of ScriptGuard, 1.5 μL of mScript capping enzyme, and 1.5 μL of mScript 2′-O-methyltransferase. This was incubated for 1 h at 37°C, then 4.5 μL of 10× Tailing buffer, 2.25 μL of 20 mM ATP, and 1.9 μL of poly(A) polymerase were added and incubated for exactly 30 min at 37°C. The mRNA was cleaned up and quantitated as described above. Integrity of the mRNA was verified by electrophoresing 1 μL of the product on a standard 1% agarose gel with ethidium bromide and looking for a sharp, well-defined band.
Embryo injections and imaging
Drosophila melanogaster expressing histone H2AvD-GFP (Clarkson and Saint 1999) were cultured on standard yeast–cornmeal–agar medium. Embryos were collected on grape–agar plates for 20–30 min and then aged for 60–90 min at 25°C to the appropriate stage. They were dechorionated for 2 min in 50% bleach and washed thoroughly with water, then aligned under a dissecting scope on a grape–agar plate and affixed to a coverslip with glue made from Scotch 3M doublestick tape dissolved in heptane. They were desiccated for 8–9 min and then covered in halocarbon oil (Sigma-Aldrich).
Microinjection needles were made using a P-87 Micropipette Puller (Sutter Instruments). They were backloaded with solution using Eppendorf Microloader pipettes (5242 956.003). The needles were then dragged against broken coverslips to produce a beveled, ∼2-um opening. All injections were performed in 1× PBS. Embryos were imaged on a spinning-disk confocal microscope, as described (McCleland and O'Farrell 2008). For some movies, individual embryos were filmed as two fields of view that were stitched together manually.
Unless otherwise noted, embryos were staged by watching slides with ∼40 embryos until the majority of embryos were in cycle 13; the slide was removed from the microscope, the embryos were injected, and then those embryos that were injected in the latter half of cycle 13 were filmed.
For the timed injections used in Figure 3, embryos were filmed to capture mitosis 12 or mitosis 13. Filming was then stopped, and embryos were injected without removing the slide from the microscope. An image was taken as each embryo was injected to mark the time, and filming was then resumed. The time of injection relative to the beginning of interphase 14 was then back-calculated using each embryo's time of injection, time of mitosis 12 or 13, and our observed lengths of interphase 13 and mitosis 13. We estimate that this gave us accuracy to within 3 min.
For Figure 5, embryos were injected with mRNA in cycle 12 or dsRNA against mitotic cyclins in cycle 11, prepared as previously described (McCleland and O'Farrell 2008). They were then injected with previously described purified GFP-PCNA protein (Shermoen et al. 2010).
For embryos injected with aphidicolin in Figure 6, rows of 40 embryos affixed to a slide were first injected with mRNA while the embryos were in cycle 13, as described above. They were then filmed to capture the time of mitosis 13 in several embryos; all embryos were then injected with 0.295 mM aphidicolin in 1% DMSO. Filming was resumed for those embryos that had progressed into cycle 14 by the time of injection.
Western blotting
For Figure 1, histone H2AvD-GFP embryos were injected at one pole with either Stg mRNA or buffer and observed on the microscope. Embryo age was measured by internuclear distance and nuclear length in cycle 14, which were matched to the standard progression (Shermoen et al. 2010). Pre-MBT embryos were in cycle 11, while the post-MBT embryos were 10 and 20 min into cycle 14, with nuclear lengths of 6 and 9 um, respectively. When selected, individual embryos were released from the slide with a paintbrush and immediately fixed in methanol-heptane. Pools of three embryos were smashed by vortexing with glass beads in 2× SDS buffer (0.1 M Tris at pH 6.8, 4% SDS, 20% glycerol, 2% 2-mercaptoethanol, bromophenol blue) and boiled for 8 min. A fraction of the extract, 1/12 or one-third, was loaded per lane (representing one-quarter or one embryo, respectively) on a 10% acrylamide gel and electrophoresed, transferred to PVDF membrane, and blocked for 2 h with 5% nonfat dry milk in PBST. It was probed overnight with rat anti-Stg antibody raised by Bruce Edgar during his tenure in the laboratory (that was affinity-purified against 6xHis-Stg) at 1:1000 in 5% nonfat dry milk and then for 1 h with 1:10000 HRP-conjugated donkey anti-rat (Jackson Laboratories). The blot was washed four times for 15 min in PBST and once for 5 min in PBS, then treated with Pierce Supersignal West Femto Maximum Sensitivity ECL (#34095), and used to expose Phenix Blue X-Ray Film (F-BX810).
For Supplemental Figures 1 and 2, 30 histone H2AvD-GFP embryos in cycle 13 were injected at one pole with either buffer or Cdk1AF or Cdk2AF mRNA. These embryos were then aged 40 min, released from the slides with heptane, and transferred to 45 μL of 1× NEBuffer with Roche Complete EDTA-free protease inhibitor. They were smashed using a pestle and incubated with 10 U of calf intestinal phosphatase (New England Biolabs) for 30 min at 37°C; then, an equal volume of 2× SDS buffer was added, and the samples were boiled for 10 min. Five embryos worth of pooled extract (15 μL) was loaded on a gel, electrophoresed, and blotted as above, with a few changes: The primary antibody was either 1:4000 Sigma monoclonal anti-PSTAIR (Supplemental Fig. S1) or 1:500 Roche 12CA5 anti-HA (Supplemental Fig. S2), and the secondary antibody was 1:10000 HRP-conjugated donkey anti-mouse (Jackson Laboratories).
Monitoring replication and fluorescent in situ hybridizations
Embryos were injected in cycle 13 with Stg mRNA as described above. They were then injected with Alexa 546-dUTP as described (Shermoen et al. 2010). Hand devitellinization and in situ hybridizations were as described (Shermoen et al. 2010).
Acknowledgments
We thank Mark McCleland for sharing plasmids and purified proteins. We thank members of the O'Farrell laboratory, Barbara Panning, and Stefano Di Talia for helpful comments. This research was supported by a National Science Foundation Graduate Research Fellowship (to J.A.F.) and National Institutes of Health grant GM037193 (to P.H.O.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.186429.111.
References
- Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM 1992. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 69: 977–988 [DOI] [PubMed] [Google Scholar]
- Blomberg I, Hoffmann I 1999. Ectopic expression of Cdc25A accelerates the G(1)/S transition and leads to premature activation of cyclin E- and cyclin A-dependent kinases. Mol Cell Biol 19: 6183–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 205–223 [DOI] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JFX 2011. Regulation of DNA replication through Sld3–Dpb11 interaction is conserved from yeast to humans. Curr Biol 21: 1152–1157 [DOI] [PubMed] [Google Scholar]
- Clarkson M, Saint R 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol 18: 457–462 [DOI] [PubMed] [Google Scholar]
- Coudreuse D, Nurse P 2010. Driving the cell cycle with a minimal CDK control network. Nature 468: 1074–1079 [DOI] [PubMed] [Google Scholar]
- Courtot C, Fankhauser C, Simanis V, Lehner CF 1992. The Drosophila cdc25 homolog twine is required for meiosis. Development 116: 405–416 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Datar SA 1996. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes Dev 10: 1966–1977 [DOI] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH 1989. Genetic control of cell division patterns in the Drosophila embryo. Cell 57: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH 1990. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell 62: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G 1986. Parameters controlling transcriptional activation during early Drosophila development. Cell 44: 871–877 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G 1986. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44: 365–372 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell PH 1994. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev 8: 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE 1989. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development 107: 1–22 [PubMed] [Google Scholar]
- Foe VE, Alberts BM 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci 61: 31–70 [DOI] [PubMed] [Google Scholar]
- Gawliński P, Nikolay R, Goursot C, Lawo S, Chaurasia B, Herz H-M, Kussler-Schneider Y, Ruppert T, Mayer M, Grosshans J 2007. The Drosophila mitotic inhibitor Frühstart specifically binds to the hydrophobic patch of cyclins. EMBO Rep 8: 490–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi S-I, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I 2010. Space and time in the nucleus: Developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol 75: 143–153 [DOI] [PubMed] [Google Scholar]
- Grosshans J, Wieschaus E 2000. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell 101: 523–531 [DOI] [PubMed] [Google Scholar]
- Grosshans J, Müller HAJ, Wieschaus E 2003. Control of cleavage cycles in Drosophila embryos by frühstart. Dev Cell 5: 285–294 [DOI] [PubMed] [Google Scholar]
- Heald R, McLoughlin M, McKeon F 1993. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell 74: 463–474 [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP 1995. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376: 313–320 [DOI] [PubMed] [Google Scholar]
- Labib K 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP 2010. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, O'Farrell PH 2008. RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle. Curr Biol 18: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Shermoen AW, O'Farrell PH 2009. DNA replication times the cell cycle and contributes to the mid-blastula transition in Drosophila embryos. J Cell Biol 187: 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508 [DOI] [PubMed] [Google Scholar]
- Papoulas O, Monzo KF, Cantin GT, Ruse C, Yates JR, Ryu YH, Sisson JC 2010. dFMRP and Caprin, translational regulators of synaptic plasticity, control the cell cycle at the Drosophila mid-blastula transition. Development 137: 4201–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A, McCusker D, Kellogg DR, Sullivan W 2008. Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation. J Cell Biol 183: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448: 811–815 [DOI] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O'Farrell PH 2010. Developmental control of late replication and S phase length. Curr Biol 20: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon OC, Stevenson VA, Theurkauf WE 1997. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388: 93–97 [DOI] [PubMed] [Google Scholar]
- Spadari S, Sala F 1982. Aphidicolin: A specific inhibitor of nuclear DNA replication in eukaryotes. Trends Biochem Sci 7: 29–32 [Google Scholar]
- Thomson AM, Gillespie PJ, Blow JJ 2010. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol 188: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport JW 1997. Regulation of replicon size in Xenopus egg extracts. Science 275: 993–995 [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Shermoen AW, O'Farrell PH, Davis I 1999. Transcribed genes are localized according to chromosomal position within polarized Drosophila embryonic nuclei. Curr Biol 9: 1263–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]