Abstract
Enhanced nuclear localization of nuclear factor κB (NF-κB) in prostate cancer (PCa) samples and constitutive NF-κB signaling in a class of PCa cell lines with low androgen receptor (AR) expression (PC3 and DU-145) imply an important role of the IκB kinase (IKK)/NF-κB system in PCa. However, most PCa and PCa cell lines depend on the activity of the AR, and the role of NF-κB in these AR-expressing PCa remains unclear. Here, we demonstrate that inhibition of NF-κB signaling by the IKK inhibitor BMS345541 reduced proliferation and increased apoptosis in AR-expressing PCa cell lines. Furthermore, AR activity and target gene expression were distinctively reduced, whereas AR protein levels remained unaltered on BMS345541 treatment. Similar effects were observed particularly after small interfering RNA (siRNA)-mediated knockdown of IKK1, but not by siRNA-mediated suppression of IKK2. Moreover, IKK1 overexpression augmented 5α-dihydrotestosterone-induced nuclear AR translocation, whereas nuclear AR was reduced by IKK1 knockdown or BMS345541. However, because IKK1 also enhances the activity of a chronically nuclear AR mutant, modulation of the subcellular distribution seems not to be the only mechanism by which IKK1 enhances AR activity. Finally, reduced in vivo AR phosphorylation after BMS345541 treatment and in vitro AR phosphorylation by IKK1 or IKK2 imply that AR constitutes a novel IKK target. Taken together, our data identify IKK1 as a potentially target structure for future therapeutic intervention in PCa.
Introduction
Prostate cancer (PCa) is one of the most common malignancies and one of the leading causes of cancer deaths in men in the United States and in Europe. Therapy includes prostatectomy or radiation therapy in case of organ-confined PCa and chemical or surgical castration causing testosterone ablation in case of metastatic PCa. Most PCa cells respond well to hormone ablation; however, after 18 to 36 months, castration-resistant recurrence evolves eventually, which is unresponsive to hormone ablation or chemotherapeutic intervention. Because of this unresponsiveness, there is a growing need for novel therapeutic options for the treatment of castration-resistant PCa (CRPCa).
The molecular target of castration therapy is the androgen receptor (AR), a member of the superfamily of nuclear receptors, which is responsible for the development and function of the prostate epithelium. On binding of its ligand—5α-dihydrotestosterone (DHT)—the AR is released from a complex with heat shock proteins and translocates into the nucleus where it induces a set of target genes necessary for cell proliferation and survival. Therefore, castration-sensitive PCa cells die on androgen deprivation. However, most of the CRPCa cells remain dependent on AR activity, which is maintained by alternative mechanisms including AR gene amplifications, expression of specific AR splice variants, and AR mutations either causing a higher sensitivity toward DHT or creating a broader ligand repertoire [1,2]. AR activation can also be achieved by site-specific phosphorylations, a mechanism generally referred to as “outlaw pathway” of AR activation [3].
Besides this alternative AR activation, AR-independent pathways have also been identified, which confer apoptosis resistance to PCa cells. One of those pathways is nuclear factor κB (NF-κB) signaling, known to regulate immune and inflammatory responses or developmental processes. NF-κB is a dimer composed of nearly all combinations of the NF-κB family members p65/RelA, RelB, c-Rel, NF-κB1/p50, or NF-κB2/p52. Uninduced NF-κB dimers are restrained in the cytoplasm by complex formation with a member of the IκB family [4]. On stimulation, IκB proteins are phosphorylated by the multisubunit IκB kinase (IKK) complex, subsequently ubiquitinated and degraded through the proteasomal pathway [5]. Liberated NF-κB translocates to the nucleus and induces the expression of a wide variety of NF-κB target genes including BCL2, BCLXL, and BIRC5 as well as growth factors like interleukin 6 [6,7]. The IKK complex consists of two closely related kinases (IKK1/IKKα and IKK2/IKKβ) and an adaptor protein (NEMO/IKKγ). More recently, the role of NF-κB in tumorigenesis including PCa has been highlighted. For instance, elevated NF-κB levels have been identified in primary PCa [8–13], and NF-κB seems to be involved in the regulation of AR expression [14]. In addition, several AR target genes important for pathogenesis of PCa—like PSA—are also regulated by NF-κB. By contrast, inactivation of IKK2 in prostate epithelial cells in a mouse model of chemically induced CRPCa had an impact neither on the initiation and progression of PCa nor on the formation of metastases [15]. CRPCa cell lines can be subdivided in two groups, with PC3 and DU145 cells characterized by constitutive IKK- and NF-κB activity and low or absent AR expression (designated here as AR-negative CRPCa) and AR-positive VCaP, 22Rv1, and LNCaP-SSR cells displaying low basal NF-κB activity. The growth arrest of AR-negative CRPCa cells after IKK blockage by pharmacological inhibitors suggests that NF-κB is a main survival pathway that might completely bypass the need for AR in these PCa cell lines [16]. By contrast, the role of NF-κB in AR-positive CRPCa is far less clear.
To define the role of IKK1 and IKK2 in AR-expressing PCa cells more precisely, we analyzed proliferation and survival of a panel of PCa cells by using either specific pharmacological IKK inhibitors or by using small interfering RNA (siRNA)-mediated suppression of IKK1 and/or IKK2. Pharmacological IKK inhibition or siRNA-mediated IKK suppression diminished proliferation and survival of PCa cell lines. In addition, knockdown of IKK1 attenuated expression of AR target genes. However, AR protein levels remained largely unchanged under any of these conditions. In conclusion, we present here a novel regulatory mechanism by which IKK1 controls the activity of AR.
Materials and Methods
Antibodies, Reagents, and Plasmids
The antibodies used were anti-phospho-serine (clone 1C8; Biomol GmbH, Hamburg, Germany), anti-IKK1 (sc7218), anti-IKK1/2 (sc7607), anti-NKX3.1 (15022), HDAC1 (sc7872), and ERK2 (sc15022) from Santa Cruz Biotechnology (Santa Cruz, CA); anti-AR (441; Dako, Glostrup, Denmark); anti-tubulin (B-5-1-2; Sigma-Aldrich, St Louis, MO); and anti-IκBα (44D4, no. 4812; Cell Signaling Technology, Inc, Danvers, MA). Cleaved caspase 3, cleaved caspase 9, and cleaved poly (ADP-ribose) polymerase (PARP) antibodies were obtained from the Cleaved Caspase Sampler Kit (no. 9929; Cell Signaling). Anti-NF-κB2 antibody recognizing p100 and p52 are from Cell Signaling (no. 4882). Wedelolactone, BMS345541, and DHT were purchased from Sigma, and tumor necrosis factor α (TNFα) was purchased from ImmunoTools (Friesoyte, Germany). Xpress-IKK1WT and Xpress-IKK2WT have been described elsewhere [17]. The 3xκB-luc firefly, the MMTVluc luciferase reporter, and the Renilla luciferase reporter have been described previously [18]. Vectors for FLAG-His-tagged IKK1CA or FLAG-tagged IKK1DN were generated by inserting the complementary DNA (cDNA) sequence in the pcDNA3 or pFLAG-CMV2 vectors. Expression vectors encoding FLAG-tagged AR5, AR1, AR6, and AR7 were generated by insertion of the appropriate polymerase chain reaction (PCR)-amplified cDNA sequence into the NotI and XbaI sites of the pFLAG-CMV2 vector (Sigma).
Cell Culture and Transfection
Human PCa cell lines (LNCaP, VCaP, and 22Rv1) were obtained from ATCC (Manassas, VA), and LNCaP-SSR cells were kindly provided by Dr Ralph Buttyan (Columbia University, New York, NY [3]). The monkey fibroblastic COS7 cell line was kindly provided by Dr Barabara Moepps (Institute of Pharmacology, University of Ulm, Ulm, Germany). Unless otherwise indicated, cells were maintained at 37°C in 5% CO2 in RPMI medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% glutamine. In all experiments designated to establish the effect of hormone on AR, cells were cultivated in medium with charcoal-stripped serum (CSS). Stock solution of BMS345541 (Sigma) was prepared in dimethylsulfoxide, and DHT (Sigma) was prepared in ethanol. For siRNA transfection of LNCaP, Hi-Perfect Transfection Reagent (Qiagen, Hilden, Germany) was used, and COS7 cells were transfected with Lipofectamine2000 according to the manufacturer's protocol (Invitrogen, Life Technologies, Ltd, Paisley, United Kingdom). The following siRNA for IKK1, IKK2, or RelA was obtained by eurogentec (Seraing, Belgium): IKK1si1-GGAAAGAAAUGGAAGAAAA-A55, IKK1si2-GGAUGUUGGUGGAAAGAU-A55, IKK2si1-GGUU AGGGCUUCUGAACUA55, IKK2si2-GGGAACAAGACCAGAGUU-U55, Si1RelA-CAACCAUGGCUUAAGGAA-A55, Si2RelA-GCAUUAACUUCUCUGGAA-A55, and control siRNA duplex negative control (5 nmol; no. SR-CL000-005). The siRNA for human NF-κB2 p100/p52 was from Qiagen (Hs_NFkB2_1).
Luciferase Reporter Assay
To determine the activity of NF-κB or AR, cells were transiently transfected with either an AR reporter construct (MMTV-Luc) or an NF-κB-dependent reporter (3xκB-luc) along with a plasmid encoding a Renilla luciferase under the control of the human ubiquitin promoter as the internal control. Attractene transfection reagent (Qiagen) was used for transfection of LNCaP cells, Lipofectamine2000 (Invitrogen) was used for transfection of COS7 cells, and CaPO4 method was used for transfection of HEK 293 cells. In brief, 1 x 106 cells/well were seeded. The next day, transfection was performed using 200 ng of a Firefly luciferase plasmid along with 30 ng of the Renilla vector per sample. Twenty-four hours after transfection, samples were treated with 20 nM DHT for at least 24 hours or 10 ng of TNFα for 4 hours (stimulated) or the appropriate vehicle (unstimulated) as indicated. Pretreatment with BMS345541 or DMSO was started 2 hours before stimulation. Luciferase activity was measured according to the Dual Luciferase Reporter Assay System from Promega (Madison, WI). Firefly luciferase activity was normalized to the appropriate Renilla luciferase values. The experiments were done in duplicates and were repeated at least three times.
Cell Growth and Viability Assay
A total of 5 x 105 cells/well were seeded in 24-well plates and were pretreated with inhibitor as indicated. After 18 to 24 hours, the medium was exchanged, and cell proliferation was measured by adding AlamarBlue dye (AbD Serotec, Kidlington, United Kingdom) for 1 to 4 hours. Fluorescence intensity (excitation = 560 nm, emission = 590 nm) was measured with a SPECTRAMaxGeminiEM fluorometer (Molecular Devices, Sunnyvale, CA) using the SOFTMAXPro software. All measurements were done in triplicates and were repeated at least three times. Statistical analysis was performed as described here. To determine survival of LNCaP and LNCaP-SSR cells, cells were cultivated in RPMI + CSS for 2 hours before BMS345541 treatment for another 18 hours under standard conditions either without or with an additional treatment with DHT. Cell numbers were counted using a Neubauer chamber (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany). Experiment was performed with triplicates, and statistical analysis was done as described later.
Immunoprecipitation and Immunoblot Analysis
Whole-cell extracts were prepared by using TNT buffer (20 mM Tris pH 8.0, 200 mM NaCl, 1% Triton X-100, 1 mM DTT, 50 mM NaF, 50 mM β-glycerophosphate, 50 µM leupeptin, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and isolation of cytoplasmic and nuclear proteins was achieved by subsequent incubation of the cells in buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM PMSF) and buffer C (20 mM HEPES pH 7.9, 0.4 M KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, including protease inhibitors [COMPLETE Protease Inhibitor; Roche Applied Sciences, Mannheim, Germany]). Immunoprecipitation and immunoblot analysis procedures were performed as described previously [17]. In brief, 250 to 500 µg of protein extracts was mixed with 1 µg of antibody/sample, and samples were incubated overnight at 4°C with agitation. Afterwards, 10 µl of 50% protein G slurry was added, and samples were further incubated for 1 hour. Subsequently, precipitates were washed in TNT buffer and subjected to immunoblot analysis. For the immunoblot analysis, either the immunopurified protein complexes or 50 to 100 µg of a protein extract was separated on SDS-PAGE and then transferred to nitrocellulose membranes (Schleicher & Schuell, Whatman, Dassel, Germany) using standard protocols. The membrane was blocked with 5% milk powder in TBS-Tween-20 before incubating with the primary antibody (1:1000 in TBS-Tween-20) and subsequently washed and incubated in a TBS-Tween-20 solution containing horse-radish peroxidase-conjugated secondary antibody (1:5000). The detection was performed using ECL substrates (Pierce, Thermo Scientific, Rockford, IL). For the densitometric quantification, the scanned images were analyzed using the ImageJ software (National Institutes of Health, Bethesda, MD).
Immunoprecipitation and Kinase Assay
HEK 293 cells were transfected with appropriate plasmid using CaPO4 method. Forty-eight hours after transfection, cells were lysed with TNT buffer, and lysates were subjected to anti-FLAG IP for 2 hours at 4°C. Immunoprecipitates were washed thrice with TNT and once with KAB (20 mM HEPES pH7.6, 2 mM EGTA, 10 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, 20 mM β-glycerophosphate, 5 mM NaF, 1 mM PMSF). For kinase reaction, samples were mixed as indicated along with 10 µCi of 32P-labeled γATP per sample in 30 µl of KAB and incubated for 30 minutes at 30°C with agitation. Subsequently, immunoprecipitates were washed once with PBS and were resolved by SDS-PAGE, transferred to nitrocellulose membrane, and visualized by autoradiography. Successful immunoprecipitation was determined with immunoblot analysis.
RNA Extraction and Quantitative Real-time PCR Analysis
Quantitative real-time PCR was performed by using the iCycler PCR instrument (Bio-Rad Laboratories, Hercules, CA). Total RNA was prepared using RNeasy protect kit from Qiagen according to the manufacturer's protocol. One microgram of total RNA was used to generate cDNA using First-Strand Synthesis kit (Invitrogen). Quantitative PCR was performed using 0.1 µl of cDNA reaction mix in the IQSYBR Green SuperMix (Bio-Rad). PCR was carried out as follows: after an initial 3-minute preincubation step at 95°C, 40 amplification cycles were run (95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 15 seconds). Quantification of gene regulation was performed by the ΔΔCp method. Results are presented relative to the expression of the housekeeping gene β-ACTIN. Sequences of primers used for quantitative PCR are available on request. The primers for BIM were obtained from RealTimePrimers (LLC, Elkins Park, PA).
Statistical Analysis
All the results are expressed as the mean ± SD of at least three independent data sets. Student's t test was used to compare the mean of two groups and to calculate P value. P < .05 was considered significant.
Gel Shift Analysis
For the gel shift analysis (electrophoretic mobility shift asay), 5 µg of nuclear proteins or whole-cell extracts (DignamC extracts) from untreated or stimulated cells was incubated on ice for 20 minutes in a reaction containing 0.3 ng of 32P-labeled κB-specific or Oct-specific oligonucleotide, 1 µg pdI:dC and 3 µl of a binding buffer. The samples were separated on a native 5% polyacryl amide gel, and the gel was dried and subjected to autoradiography.
Results
Pharmacological Inhibition of IκB Kinases Impairs Cell Growth and Attenuates AR Signaling in PCa Cell Lines
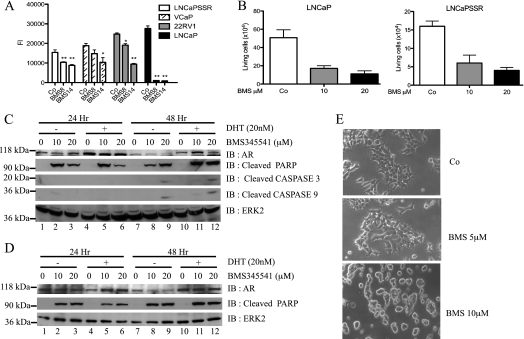
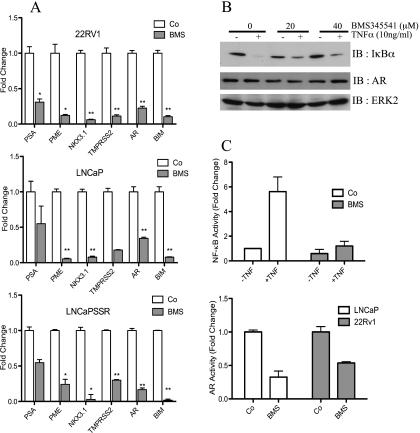
The controversial picture regarding the role of NF-κB in PCa prompted us to explore the function of NF-κB in AR-expressing PCa cell lines. Proliferation of PCa cell lines (LNCaP-SSR, VCaP, 22Rv1, and LNCaP) treated with the IKK inhibitor BMS345541 was significantly attenuated in a dose-dependent manner when compared with untreated PCa cells (Figure 1A). Similar effects were seen with IKK inhibitors wedelolactone and Bay 117082 (data not shown). Further analysis of the BMS345541 effect in androgen-dependent LNCaP and androgen-independent LNCaP-SSR cells revealed a significant reduction in survival of both cell lines (Figure 1B), which was most likely due to an increased apoptosis because levels of cleaved PARP and cleaved caspases 3 and 9 were elevated (Figure 1, C and D). As expected, levels of cleaved PARP were highest in samples cultivated in absence of androgens (Figure 1C, lanes 2 and 3). Morphologic changes induced by BMS345541 treatment of LNCaP cells (Figure 1E) further supported the notion that IKK inhibition caused increased apoptosis. A reduced NF-κB-dependent expression of AR might be responsible for the negative BMS345541 effect on PCa proliferation and survival. However, BMS345541 treatment had no gross effect on AR expression in LNCaP and LNCaP-SSR cells (Figure 1, C and D). Furthermore, AR protein levels remained stable on BMS345541 treatment also in CRPCa cell lines 22Rv1 and VCaP (Figure W1), whereas the expression of the AR target NKX3.1 showed a significant reduction in LNCaP cells. Collectively, these data suggest that pharmacological inhibition of IKKs is deleterious for AR-positive PCa cell lines. The reduced expression of the AR target genes PSA, PME, NKX3.1, and TMPRSS2 and NF-κB target gene BIM (which was included as internal NF-κB control) in BMS345541-treated 22Rv1, LNCaP, and LNCaP-SSR cells (Figure 2A) further underscored these findings.
Figure 1.
Growth inhibition of androgen-dependent and androgen-independent PCa cells by BMS345541. (A) Cells were either left untreated or treated with BMS345541 (8 µM; 14 µM) for 24 hours, and cell proliferation was measured by reduction of AlamarBlue dye (bars depict mean ± SD, **P < .0001, *P < .001, compared with vehicle control). (B) Twenty-four hours after exposure to BMS345541 (0, 10, and 20 µM) of LNCaP and LNCaP-SSR cells, viable cell numbers were determined. Graph indicates mean ± SD. LNCaP (C) and LNCaP-SSR (D) cells were treated with BMS345541 for 24 and 48 hours. Cells were left unstimulated (-) or stimulated with 20 nM DHT (+), and whole-cell extracts were subjected to immunoblot analysis with the indicated antibodies. (E) LNCaP cells were either left untreated (Co) or were treated with indicated concentrations of BMS345541 for 24 hours. Cell morphology is depicted as bright field images (original magnification, x20).
Figure 2.
BMS345541 inhibits transcriptional activity of AR and NF-κB. (A) LNCaP, LNCaP-SSR, and 22Rv1 cells were either left untreated (Co) or treated with 8 µM BMS345541 for 24 hours (BMS). Quantitative PCR analysis of AR, AR target genes (PSA, PME, NKX3.1, and TMPRSS2), and NF-κB target gene (BIM) was performed. Normalized readings were plotted relative to Co of each gene. Bar depicts mean ± SD. *P < .001, **P < .0001. (B) LNCaP cells were pretreated for 4 hours with BMS345541 (0, 20, and 40 µM) followed by TNFα stimulation for 20 minutes. Resulting whole-cell extracts were subjected to immunoblot analysis for the indicated proteins. (C) LNCaP cells were transiently transfected with a 3xκB-luc (upper part) or with the MMTVluc (lower part) and treated with BMS345541 (8 µM) for 24 hours. Cells transfected with 3xκB-luc were additionally stimulated with TNFα (+TNF) or were not further stimulated (-TNF) for 3 more hours. Subsequently, dual luciferase analysis was performed with all samples. Bars depict mean ± SD of normalized luciferase reading relative to untreated samples.
BMS345541 impaired NF-κB signaling in LNCaP cells in a dose-dependent manner as monitored by the TNFα-induced IκBα degradation (Figure 2B). Again, AR protein levels remained unaffected by BMS345541 treatment (Figure 2B, middle panel). BMS345541 pretreatment also caused a significant reduction of the TNFα-induced NF-κB activity by luciferase reporter assays (Figure 2C, upper part). Importantly, also a distinct attenuation of the AR activity on BMS345541 treatment was observed with either LNCaP or 22Rv1 cells (Figure 2C, lower part). Together, these results demonstrate a negative effect of IKK inhibition on NF-κB and AR activity, whereas AR protein levels remain stable.
Knockdown of IKK1 or IKK2 Affects PCa Cell Growth
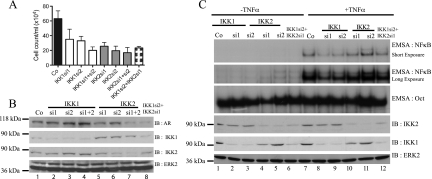
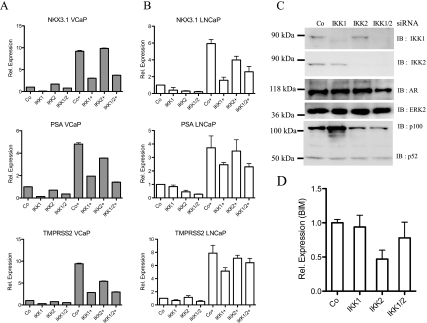
To exclude potential off-target effects of BMS345541, we compared cell proliferation in IKK1 siRNA- as well as IKK2 siRNA-transfected LNCaP cells (Figure 3A). siRNA-mediated suppression of IKK1 or IKK2 caused a distinct reduction of viable cell numbers (Figure 3A). Again, knockdown of IKK1 had no obvious effect on AR protein levels (Figure 3B, lanes 1–4), whereas a reduced AR expression after IKK2 suppression was occasionally detectable (Figure 3B, lanes 5–8). Analysis of TNFα-induced NF-κB DNA binding activity by EMSA showed a more pronounced decrease after IKK1 suppression then by knockdown of IKK2 (Figure 3C, upper panel). Similarly, knockdown of IKK1 or IKK1 + IKK2 caused a considerable reduction in the expression of the AR target genes NKX3.1, PSA, and TMPRSS2 in VCaP or LNCaP cells, although the degree of inhibition showed some variations (Figure 4, A and B). By contrast, suppression of IKK2 alone had only a moderate effect on the expression of the AR target genes, whereas it is essential for the basal expression of the NF-κB target BIM (Figure 4D). To further analyze the potential functional divergence between IKK1 and IKK2 regarding their impact on AR activity, either IKK1CA or IKK2WT was overexpressed in LNCaP cells (in control experiments, IKK2WT was highly active, whereas IKK1CA was required for full IKK1 activity [data not shown]) in conjunction with either a NF-κB-dependent reporter (Figure 5A) or an AR-dependent reporter (Figure 5B). As expected, NF-κB activity was moderately increased by IKK1CA and more pronounced by IKK2WT overexpression (Figure 5A). However, only overexpression of IKK1CA, but not of IKK2WT, led to a distinct increase in AR-dependent activity (Figure 5B).
Figure 3.
Effect of siRNA-mediated suppression of IKK1 and/or IKK2. (A) LNCaP cells were transfected with the indicated IKK siRNA. Four days after transfection, viable cell numbers were determined and plotted as mean ± SD. (B) Efficiency of knockdown corresponding to A was checked by immunoblot analysis. The same membrane was reprobed with antibodies recognizing AR and ERK2. (C) LNCaP cells were transfected with IKK1 and/or IKK2 siRNA, as indicated. After 72 hours, cells either were left untreated (-TNFα) or were stimulated with 10 ng/ml TNFα for 20 minutes. EMSA for NF-κB or Oct probes was performed with nuclear extract of these cells (upper panel). Corresponding cytoplasmic extracts were used for immunoblot analysis with indicated antibodies (lower panel).
Figure 4.
IKK1 siRNA attenuates the expression of AR target genes. VCaP (A) and LNCaP cells (B) were transfected with IKK1 and/or IKK2 siRNA. Forty-eight hours after transfection, cells were either left untreated or treated with 20 nM DHT (+) for an additional 24 hours. Transcript levels of AR, PSA, NKX3.1, and TMPRSS2 were analyzed with real-time PCR. Histogram depicts fold change in transcript levels after siRNA transfection relative to Co-siRNA-transfected samples. (C) IKK1, IKK2, and AR protein levels in corresponding LNCaP samples were determined with immunoblot analysis. (D) Similar quantitative PCR analysis of BIM transcript levels after knockdown of IKK1 and/or IKK2 siRNA in LNCaP cells.
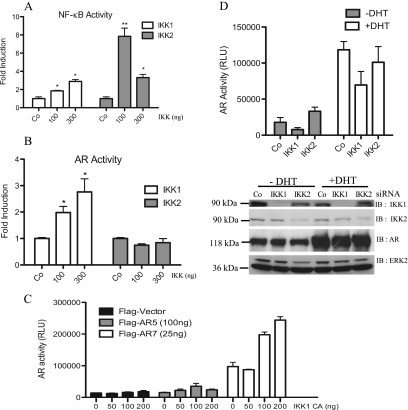
Figure 5.
Effects of IKK1 and IKK2 on AR activity. (A, B) LNCaP cells were transfected with IKK1CA or IKK2WT (0, 100, and 300 ng), along with either 3xκB-luc (A) or MMTVluc (B). After 24 hours, dual luciferase analysis was performed. Bars in histogram represent mean ± SD of normalized relative light unit (RLU) relative to IKK-untransfected samples. *P ≤ .03, **P ≤ .001, compared with control always. (C) COS7 cells were transfected with MMTVluc and Ubi-Renilla and with increasing amounts of IKK1CA along with either full-length AR (AR5) or constitutive active AR (AR7). Luciferase values were determined 24 hours after transfection. The mean values ± SD of the normalized RLU are depicted. (D) 293 cells were cotransfected with IKK siRNA along with MMTVluc and Ubi-Renilla. Twenty-four hours after transfection, cells were stimulated with 20 nM DHT (+DHT) or left untreated (-DHT) for another 24 hours. Mean ± SD of normalized RLU value of respective transfection is depicted (upper part). Knockdown efficiency was controlled by immunoblot analysis with the corresponding samples (lower part).
To further dissect the positive IKK1 effect on AR, reporter gene analysis was performed in COS7 cells ectopically expressing either full-length AR (AR5) or a constitutive active AR mutant harboring a truncated carboxy-terminus (AR7). Again, addition of IKK1CA augmented the activity of AR5 as well as of AR7 (Figure 5C). Furthermore, AR activity in transiently transfected HEK 293 cells was reduced in the absence of IKK1, whereas IKK2 knockdown either had no effect or even led to a slight increase in AR activity in unstimulated cells (Figure 5D, upper part). Accordingly, AR5 protein levels remained stable, whereas IKK1 and IKK2 were efficiently reduced by the corresponding siRNAs as measured by immunoblot analysis (Figure 5D, lower part). A similar effect of siRNA-mediated IKK1 or IKK2 suppression was observed when the constitutive active AR7 deletion mutant was used. The AR7 mutant lacks the carboxy-terminal ligand binding domain and is therefore constitutively located in the nucleus. Again, AR7 activity was reduced after IKK1 knockdown was and moderately increased after IKK2 suppression (Figure W2, right panel), whereas AR7 protein levels remain stable in IKK siRNA-transfected cells (Figure W2, left panel). Collectively, these results imply that especially IKK1 is capable of increasing the activity of full-length AR or a constitutively active AR version.
IKK1 Augments the Hormone-Induced Nuclear Translocation of AR
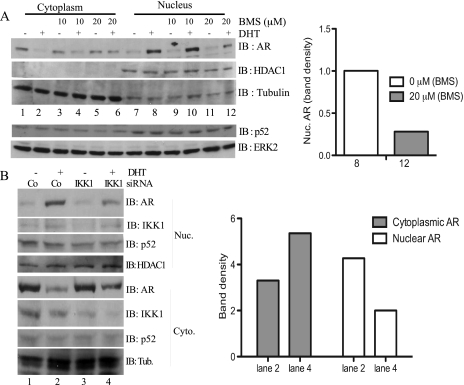
To further characterize the positive IKK1 effect on AR activity, subcellular localization of AR was analyzed in LNCaP cells. Without BMS345541, cytosolic AR levels decreased in hormone-stimulated cells, whereas nuclear AR levels increased (Figure 6A, lanes 1 and 2 and lanes 7 and 8). Pretreatment with 20 µM BMS345541 reduced nuclear AR, whereas the nuclear and cytoplasmic p52 levels remained unaffected (Figure 6A, lanes 3–6 and lanes 9–12; a quantification of the nuclear AR seen in lanes 8 and 12 is given at the right panel) and stabilized cytosolic AR levels. Consistently, siRNA-mediated IKK1 suppression in LNCaP cells led to a moderately reduced DHT-induced nuclear translocation of the AR (Figure 6B, compare lanes 2 and 4).
Figure 6.
Effect of IKK inhibition and IKK1 knockdown on nuclear translocation of the AR. (A) LNCaP cells were cultivated in Dulbecco modified Eagle medium (DMEM) + charcoal-stripped fetal calf serum (CS-FCS) for 2 hours before pretreatment with BMS345541 (0, 10, and 20 µM) for an additional 1 hour. Subsequently, cells were either left untreated (-) or treated (+) with DHT for 1 hour, and nuclear and cytoplasmic proteins were subjected to immunoblot analysis using anti-AR, anti-HDAC1, and anti-tubulin antibodies. The subcellular distribution of p52 was determined with immunoblot analysis in a similar experiment (lower part). (B) LNCaP cells were transiently transfected with either control siRNA (Co) or with IKK1si2 siRNA (IKK1). After 48 hours, the cells were transferred to DMEM + CS-FCS for 3 hours followed by treatment with either vehicle (DMSO, -) or with DHT (5 nM, +) for an additional hour. Subsequently, nuclear and cytoplasmic proteins were extracted and subjected to immunoblot analyses with antibodies for the indicated proteins. A quantification of the cytoplasmic and the nuclear AR protein levels in DHT-stimulated cells (lanes 2 and 4) is given in the right part.
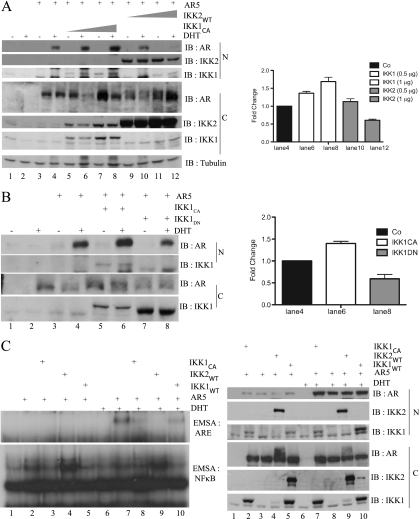
To further delineate the effect on hormone-induced nuclear localization of the AR, COS7 cells were transfected with AR5 either alone or in combination with IKK1CA or IKK2WT as indicated (Figure 7A). Subsequently, the cells were left untreated or were stimulated with DHT before the analysis of the subcellular distribution of the AR. Again, IKK1CA coexpression alone was not sufficient to increase nuclear AR levels in unstimulated cells (Figure 7A, lanes 3, 5, and 7). However, in stimulated cells, nuclear AR was distinctively increased by IKK1CA, whereas coexpression of IKK2WT reduced hormone-induced nuclear AR (Figure 7A, left part, lanes 9–12). Accordingly, coexpression of a dominant-negative IKK1 mutant (IKK1DN) attenuated DHT-induced nuclear translocation of the AR in COS7 cells (Figure 7B, lanes 4 and 8), suggesting that enzymatic activity of IKK1 is required for increased nuclear AR. Studies by Nadiminty et al. [19] revealed a cytokine-induced and IKK1-dependent mobilization of NF-κB2/p52:AR heterodimers as a novel hormone-independent mechanism for AR activation. However, neither BMS345541 treatment (Figure 6A) nor IKK1 suppression (Figure 4C) had a gross effect on either the nuclear p52 levels or the processing of the NF-κB2/p100 precursor, suggesting that the observed IKK1 effect could not only be dependent on NF-κB/p52.
Figure 7.
Differential effects of IKK1 or IKK2 coexpression on nuclear expression and DNA binding of the AR. (A) COS7 cells expressing ectopic AR5 alone or in combination with FLAG-IKK1CA or FLAG-IKK2WT as indicated were either left untreated (-) or stimulated with 20 nM DHT (+) for 1 hour. The resulting nuclear and cytoplasmic extracts were subjected to immunoblot analysis. This experiment was repeated twice, and a quantification of selected samples of all three experiments is given in the right part. (B) COS7 cells, transiently transfected with FLAG-AR5 alone or with FLAG-IKK1CA or HA-IKK1DN, were incubated in DMEM + CSS before stimulation with DHT for 1 hour. Nuclear and cytoplasmic extracts were subjected to immunoblot analysis with the indicated antibodies. This experiment was repeated twice, and a quantification of selected samples of all three experiments is given in the right part. (C) COS7 were transiently transfected with FLAG-AR5, FLAG-IKK1CA, FLAG-IKK2WT, and FLAG-IKK1WT as indicated. Subsequently, cells were treated either with solvent or with DHT (+) for 1 hour. Nuclear extracts were subjected to EMSA experiments using NF-κB (left part, lower panel) or AR (left part, upper panel) specific oligonucleotides. In addition, immunoblot analysis with the indicated antibodies was performed with the cytoplasmic (C) and nuclear (N) proteins (right part).
To analyze a potential effect of IKK1 on the DNA binding of the AR, full-length AR5 was expressed in COS7 cells alone or in combination with IKK1WT, IKK2WT, or IKK1CA. Nuclear proteins from untreated (Figure 7C, lanes 1–5) or from DHT-stimulated cells (Figure 7C, lanes 6–10) were used for EMSA experiments with either an androgen response element (Figure 7C, left part, upper panel) or a κB-specific oligonucleotide (Figure 7C, left part, lower panel). As expected, IKK2WT augmented basal NF-κB activity in COS7 cells, whereas IKK1CA or IKK1WT increased NF-κB activity only moderately (Figure 7C, lanes 1–5). By contrast, neither IKK1CA/IKK1WT nor IKK2WT caused an increased basal DNA binding of the AR (Figure 7C, lanes 1–6). However, IKK1CA led to a distinct increase in the hormone-induced DNA binding of the AR, whereas IKK2WT reduced AR binding (Figure 7C, lanes 7–10). Immunoblot analysis revealed a moderate increase in nuclear levels of the hormone induced AR by IKK1CA (Figure 7C, lower part, lanes 7 and 8), whereas IKK2WT caused slightly reduced nuclear AR levels (lanes 8 and 9). Moreover, corresponding cytoplasmic extracts also showed a moderate decrease in AR levels and additional slower-migrating AR isoforms in cells with IKK2WT coexpression. Although the basis for the additional slower-migrating AR isoforms remains unclear, it is tempting to speculate that these might resemble AR molecules with specific posttranslational modifications.
The AR Is an IKK Target In Vitro
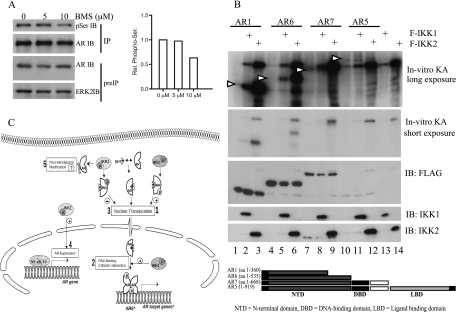
To analyze whether the AR constitutes an IKK substrate, we first determined whether IKK inhibition by BMS345541 treatment affects the phosphorylation status of the AR. Hence, LNCaP cells were pretreated with 5 or 10 µM BMS345541 before cell lysis. Phosphorylation of the AR was determined with anti-AR immunoprecipitation followed by immunoblot analysis with anti-phospho-Ser antibodies. Indeed, IKK inhibition decreased AR phosphorylation, suggesting that AR might resemble an IKK target protein (Figure 8A, compare lanes 1 and 3). To determine whether AR is phosphorylated by IKK1 or IKK2 in vitro, we expressed FLAG-tagged full-length AR (AR5) or a panel of FLAG-tagged deletion mutants of the AR (AR1, AR6, AR7; see Figure 8B, lower part, for a schematic representation) in HEK 293 cells. The FLAG-tagged AR proteins were immunopurified and subjected to an in vitro kinase assay with likewise immunopurified FLAG-tagged IKK1 or IKK2 proteins. Interestingly, full-length AR protein as well as all AR deletion mutants were efficiently phosphorylated by IKK2 and less pronounced by IKK1 (Figure 8B, upper part). Furthermore, the highest phosphorylation was observed with the amino-terminal deletion mutants AR1 (aa 1–360) and AR6 (aa 1–535; Figure 8B, upper part, lanes 2 and 3 and lanes 5 and 6). Furthermore, an interaction of AR either with IKK1 (Figure W3A) or with IKK2 (Figure W3B) was observed in interaction studies with ectopically expressed proteins. These data show that the AR is a target of IKK1 and IKK2 and imply a phosphorylation-dependent regulation of AR by the IKKs.
Figure 8.
The AR is an IKK target in vitro. (A) LNCaP cells were exposed to BMS345541 (0, 5, and 10 µM). After 48 hours, AR was immunoprecipitated (IP), and anti-phospho-serine, AR, and ERK2 immunoblot analysis was performed. Histogram depicts densitometry of anti-phospho-serine signals relative to control. (B) 293 cells expressing AR5, AR deletion mutants, IKK1, or IKK2 were lysed, and anti-FLAG immunoprecipitation was performed. Kinase reaction was performed with AR proteins alone or with either IKK1 or IKK2 and analyzed with autoradiography. Arrows indicate the position of AR variants. A schematic representation of AR variants is given in the lower part. (C) Model depicting the mechanism by which the IKK proteins control the activity of the AR: DHT binding induced nuclear localization, ARE binding, and target gene induction by AR. IKK1 phosphorylates AR in the nucleus and/or cytoplasm causing an increased nuclear localization of AR (1). IKK1 additionally affects transcriptional activity of AR independent of its nuclear localization possibly by altered DNA binding ability or Cofactor interaction (2). IKK2 attenuates ligand-dependent nuclear localization of AR most likely by phosphorylating AR at alternative sites (3). Conversely, IKK2 regulates AR expression (4). In addition, IKK2 seems to affect cytoplasmic AR by modifying AR after translation (5).
Discussion
A prominent role of NF-κB in the initiation or progression of PCa has been proposed previously. This notion is supported by the chronic IKK and NF-κB activity observed in a group of androgen-independent PCa cell lines (PC3 or DU145) [9,16] and the elevated NF-κB activation reported for primary PCa cases [8,11,20]. For instance, Gasparian et al. [9] reported nuclear RelA in approximately 25% of the cells. Therefore, a constitutive NF-κB activity was proposed to generally replace the AR in PCa, and the androgen-independent PCa cell lines PC3 or DU145 were assumed to be model systems for CRPCa. However, most CRPCa cells depend on a functional AR, whereas PC3 and DU145 cells are devoid of measurable AR expression levels. Thus, PC3 and DU145 cells might represent only a subgroup of CRPCa. To analyze the importance of the NF-κB/IKK system in AR-positive PCa, we determined the effect of specific IKK inhibitors on growth and survival of the androgen-dependent PCa cell line LNCaP, and the androgen-independent PCa cell lines 22Rv1, VCaP, and LNCaP-SSR. Irrespective of their AR status or androgen sensitiveness, the growth of all PCa cell lines tested was impaired by the IKK inhibitors BMS345541 and wedelolactone (Figure 1; data not shown), suggesting a prominent role of IKKs in AR-expressing PCa cells. One molecular mechanism may be the regulation of AR expression byNF-κB [14]. Indeed, a reduced AR activity (Figure 3B) and expression of different AR target genes (Figure 2) by BMS345541 treatment was observed. However, although a reduced AR expression might also contribute to the overall picture, it seems not to be the major mechanism because protein levels of the AR were found to be unaffected in BMS345541-treatedLNCaP, VCaP, and 22Rv1 cells (Figures 1, C and D, and W1).
BMS345541 is known to impair both IKK1 and IKK2 [21]. Because NF-κB-dependent and NF-κB-independent functions were reported for both IKKs [4], the altered AR functions on BMS345541 treatment might be due to different mechanisms. Although both IKK isoforms are inhibited by BMS345541, our biochemical analyses suggest that IKK1 is the predominant positive regulator of AR activity, whereas IKK2 seems to have mainly a negative impact on AR activity. For instance, the suppression of IKK1 attenuates the expression of AR target genes (TMPRSS2, PSA, or NKX3.1) in LNCaP and VCaP cells (Figure 4, A and B), and only IKK1CA coexpression, but not the addition of IKK2, augmented the AR activity in reporter gene assays (Figure 5B). Different mechanisms might account for the positive IKK1 effect on AR. One of these mechanisms is the nuclear translocation of the AR. This notion is supported by 1) the reduced hormone-induced nuclear translocation of AR by BMS345541 treatment (Figure 6A), 2) the attenuated nuclear translocation after siRNA-mediated IKK1 knockdown (Figure 6B), 3) the increased nuclear translocation of full-length AR in hormone-induced COS7 cells on IKK1CA coexpression (Figure 7A), and 4) the reduced nuclear expression of AR on IKK1DN coexpression (Figure 7B). A previously reported IKK1-dependent mobilization of NF-κB2/p52:AR heterodimers [19,22] might contribute to the increased nuclear AR expression. However, this seems not to be the exclusive mechanism because neither BMS345541 treatment nor IKK1 knockdown affected NF-κB2 expression or p100 processing (Figures 4C and 6A). Furthermore, the increased activity of the already constitutive nuclear AR mutant AR7 by coexpression of IKK1CA (Figure 5C) and its reduced activity in cells with IKK1 knockdown (Figure W2) suggest that nuclear translocation is not the only level modulated by IKK1. Another level might be an increased DNA binding activity of the AR (Figure 7C).
In contrast to IKK1, IKK2 seems to have divergent roles. IKK2 might have a positive effect on AR activity indirectly by controlling basal AR expression (Figure 3B), also evident by BIM expression (Figure 4D). The basis for this positive IKK2 effect on AR is not completely clear. Although a moderate increase in AR expression by RelA was observed previously [14], we were unable to detect changes in AR protein and mRNA levels after knockdown of either RelA or p100/p52 (Figure W4), suggesting that different IKK2-dependent NF-κB dimers might regulate AR expression in a redundant fashion. Conversely, IKK2 coexpression attenuated the nuclear translocation of AR (Figure 7A, lanes 9–12), and finally, IKK2 reduced the DNA-binding activity of AR (Figure 7C, compare lanes 7 + 9), suggesting a negative effect of IKK2 on AR protein functions. Consistently, knockdown of IKK2 augmented the basal activity of both the full-length AR (Figure 5D, upper panel, left part) and the constitutive active AR7 mutant (Figure W2), further supporting this notion. In addition, IKK2 coexpression caused the formation of slower-migrating AR isoforms in the cytoplasm (Figure 7C, left part). Because the AR is a target for several posttranslational modifications like phosphorylation, ubiquitination, or SUMOylation [23–26], it is very likely that these additional AR isoforms are caused by such posttranslational modifications. Experiments are underway to characterize these posttranslational AR modifications. The various levels at which IKK1 and IKK2 affect the AR is summarized in a model in Figure 8C.
Divergent roles of IKK1 and IKK2 for the initiation and progression of PCa have been reported previously. Suppression of IKK1 by siRNA, for instance, delayed the appearance of castration resistant PCa in the murine myc-CaP allograft model of PCa [15], whereas deletion of IKK2 had no effect. However, nuclear localization of IKK1 was evident for this process as well as for the negative regulation of MASPIN expression [15,27]. A potential mechanism underlying the positive IKK1 effect on AR activity might include the IKK1-mediated phosphorylation of this transcription factor [28,29] illustrated by the IKK-mediated phosphorylation of the amino-terminal DNA binding domain of the AR (Figure 8B). The activity or the stability of other non-IκB targets like RelA or p53 is influenced by IKK-mediated phosphorylation [5]. Furthermore, phosphorylation of the estrogen receptor at Ser118 was found to be impaired by the IKK inhibitor Bay 117082 [30]. Therefore, an IKK-mediated phosphorylation of the AR might be another example of non-IκB IKK targets. In this line, it is important to mention that the AR is targeted by several kinases like the MAPK, protein kinase C, protein kinase A, or glycogen synthase kinase 3. These phosphorylations affect the protein-protein interactions of the AR (including cofactor interaction), the stability of the AR, the nuclear localization, or the DNA binding activity [29,31,32]. One prominent example is the glycogen synthase kinase 3-mediated phosphorylation of the AR, which increases the nuclear localization and, in turn, the activity and stability of the AR. Interestingly, most of the known phospho-acceptor sites in AR are located in its amino-terminus [23,28] similar to the IKK-mediated AR phosphorylation (Figure 8B). Therefore, an IKK1-mediated phosphorylation of the amino-terminus of the AR, which positively affects the activity of this nuclear receptor, is consistent with the previously reported functions of AR phosphorylations. However, several important questions need to be answered in future studies. For instance, the identity of the serine residues serving as target sites for IKK1 and/or IKK2 need to be defined. In this line, it is necessary to define the stimuli causing the activation of IKK1. Interleukin 6- or lymphotoxin β-induced and phosphorylated signal transducer and activator of transcription 3-mediated activation of IKK1 has been reported to play a crucial role in the progression toward castration-independent growth of PCa by enhancing p100 processing and mobilization of p52:AR heterodimers into the nucleus in a hormone-independent fashion [19,33,34]. Although no gross change in p100 or p52 protein levels was observed on BMS345541 treatment or IKK1 knockdown (Figures 4C and 6A), IKK1-mediated AR phosphorylation might act in conjunction with a likewise IKK1-dependent p100 processing to establish the overall positive IKK1 effect on AR activity. Moreover, the nature of the IKK2-dependent additional posttranslational AR modification and its potential role in the regulation of the AR needs to be determined. However, taken together, our data suggest that IKK1-specific pharmacological inhibitors might represent promising tools for therapeutic intervention in PCa.
Supplementary Material
Abbreviations
- AR
androgen receptor
- CRPCa
castration-resistant prostate cancer
- IKK
IκB kinase
- PCa
prostate cancer
Footnotes
This work was supported by a grant from the Dr.-Robert-Pfleger-Stiftung (to R.B.M. and M.V.C.) and by a fellowship from the Deutsche Forschungsgemeinschaft (GRK1041 to G.J.). The authors declare no conflicts of interest.
This article refers to supplementary materials, which are designated by Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–3555. [PubMed] [Google Scholar]
- 2.Zhao XY, Malloy PJ, Krishnan AV, Swami S, Navone NM, Peehl DM, Feldman D. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med. 2000;6:703–706. doi: 10.1038/76287. [DOI] [PubMed] [Google Scholar]
- 3.Schutz SV, Schrader AJ, Zengerling F, Genze F, Cronauer MV, Schrader M. Inhibition of glycogen synthase kinase-3β counteracts ligand-independent activity of the androgen receptor in castration resistant prostate cancer. PLoS One. 2011;6:e25341. doi: 10.1371/journal.pone.0025341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oeckinghaus A, Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheidereit C. IκB kinase complexes: gateways to NF-κB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 6.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 7.Karin M. The IκB kinase—a bridge between inflammation and cancer. Cell Res. 2008;18:334–342. doi: 10.1038/cr.2008.30. [DOI] [PubMed] [Google Scholar]
- 8.Fradet V, Lessard L, Begin LR, Karakiewicz P, Masson AM, Saad F. Nuclear factor-κB nuclear localization is predictive of biochemical recurrence in patients with positive margin prostate cancer. Clin Cancer Res. 2004;10:8460–8464. doi: 10.1158/1078-0432.CCR-04-0764. [DOI] [PubMed] [Google Scholar]
- 9.Gasparian AV, Yao YJ, Kowalczyk D, Lyakh LA, Karseladze A, Slaga TJ, Budunova IV. The role of IKK in constitutive activation of NF-κB transcription factor in prostate carcinoma cells. J Cell Sci. 2002;115:141–151. doi: 10.1242/jcs.115.1.141. [DOI] [PubMed] [Google Scholar]
- 10.Jin RJ, Lho Y, Connelly L, Wang Y, Yu X, Saint Jean L, Case TC, Ellwood-Yen K, Sawyers CL, Bhowmick NA, et al. The nuclear factor-κB pathway controls the progression of prostate cancer to androgen-independent growth. Cancer Res. 2008;68:6762–6769. doi: 10.1158/0008-5472.CAN-08-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessard L, Begin LR, Gleave ME, Mes-Masson AM, Saad F. Nuclear localisation of nuclear factor-κB transcription factors in prostate cancer: an immunohistochemical study. Br J Cancer. 2005;93:1019–1023. doi: 10.1038/sj.bjc.6602796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suh J, Payvandi F, Edelstein LC, Amenta PS, Zong WX, Gelinas C, Rabson AB. Mechanisms of constitutive NF-κB activation in human prostate cancer cells. Prostate. 2002;52:183–200. doi: 10.1002/pros.10082. [DOI] [PubMed] [Google Scholar]
- 13.Sweeney C, Li L, Shanmugam R, Bhat-Nakshatri P, Jayaprakasan V, Baldridge LA, Gardner T, Smith M, Nakshatri H, Cheng L. Nuclear factor-κB is constitutively activated in prostate cancer in vitro and is overexpressed in prostatic intraepithelial neoplasia and adenocarcinoma of the prostate. Clin Cancer Res. 2004;10:5501–5507. doi: 10.1158/1078-0432.CCR-0571-03. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Altuwaijri S, Deng F, Chen L, Lal P, Bhanot UK, Korets R, Wenske S, Lilja HG, Chang C, et al. NF-κB regulates androgen receptor expression and prostate cancer growth. Am J Pathol. 2009;175:489–499. doi: 10.2353/ajpath.2009.080727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ammirante M, Luo JL, Grivennikov S, Nedospasov S, Karin M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature. 2010;464:302–305. doi: 10.1038/nature08782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yemelyanov A, Gasparian A, Lindholm P, Dang L, Pierce JW, Kisseljov F, Karseladze A, Budunova I. Effects of IKK inhibitor PS1145 on NF-κB function, proliferation, apoptosis and invasion activity in prostate carcinoma cells. Oncogene. 2006;25:387–398. doi: 10.1038/sj.onc.1209066. [DOI] [PubMed] [Google Scholar]
- 17.May MJ, Marienfeld RB, Ghosh S. Characterization of the IκB-kinase NEMO binding domain. J Biol Chem. 2002;277:45992–46000. doi: 10.1074/jbc.M206494200. [DOI] [PubMed] [Google Scholar]
- 18.Marienfeld RB, Palkowitsch L, Ghosh S. Dimerization of the IκB kinase-binding domain of NEMO is required for tumor necrosis factor α-induced NF-κB activity. Mol Cell Biol. 2006;26:9209–9219. doi: 10.1128/MCB.00478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadiminty N, Lou W, Sun M, Chen J, Yue J, Kung HJ, Evans CP, Zhou Q, Gao AC. Aberrant activation of the androgen receptor by NF-κB2/p52 in prostate cancer cells. Cancer Res. 2010;70:3309–3319. doi: 10.1158/0008-5472.CAN-09-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross JS, Kallakury BV, Sheehan CE, Fisher HA, Kaufman RP Jr, Kaur P, Gray K, Stringer B. Expression of nuclear factor-κB and IκBα proteins in prostatic adenocarcinomas: correlation of nuclear factor-κB immunoreactivity with disease recurrence. Clin Cancer Res. 2004;10:2466–2472. doi: 10.1158/1078-0432.ccr-0543-3. [DOI] [PubMed] [Google Scholar]
- 21.Burke JR, Pattoli MA, Gregor KR, Brassil PJ, MacMaster JF, McIntyre KW, Yang X, Iotzova VS, Clarke W, Strnad J, et al. BMS-345541 is a highly selective inhibitor of IκB kinase that binds at an allosteric site of the enzyme and blocks NF-κB-dependent transcription in mice. J Biol Chem. 2003;278:1450–1456. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- 22.Nadiminty N, Chun JY, Lou W, Lin X, Gao AC. NF-κB2/p52 enhances androgen-independent growth of human LNCaP cells via protection from apoptotic cell death and cell cycle arrest induced by androgen-deprivation. Prostate. 2008;68:1725–1733. doi: 10.1002/pros.20839. [DOI] [PubMed] [Google Scholar]
- 23.Gioeli D, Ficarro SB, Kwiek JJ, Aaronson D, Hancock M, Catling AD, White FM, Christian RE, Settlage RE, Shabanowitz J, et al. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J Biol Chem. 2002;277:29304–29314. doi: 10.1074/jbc.M204131200. [DOI] [PubMed] [Google Scholar]
- 24.Rytinki MM, Kaikkonen S, Sutinen P, Palvimo JJ. Analysis of androgen receptor SUMOylation. Methods Mol Biol. 2011;776:183–197. doi: 10.1007/978-1-61779-243-4_12. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee S, Thomas M, Dadgar N, Lieberman AP, Iniguez-Lluhi JA. Small ubiquitin-like modifier (SUMO) modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J Biol Chem. 2009;284:21296–21306. doi: 10.1074/jbc.M109.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chymkowitch P, LeMay N, Charneau P, Compe E, Egly JM. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 2011;30:468–479. doi: 10.1038/emboj.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, Cheresh DA, Karin M. Nuclear cytokine-activated IKKα controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- 28.Blok LJ, de Ruiter PE, Brinkmann AO. Androgen receptor phosphorylation. Endocr Res. 1996;22:197–219. doi: 10.3109/07435809609030508. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Sadar MD. Amino-terminus domain of the androgen receptor as a molecular target to prevent the hormonal progression of prostate cancer. J Cell Biochem. 2006;98:36–53. doi: 10.1002/jcb.20802. [DOI] [PubMed] [Google Scholar]
- 30.Weitsman GE, Li L, Skliris GP, Davie JR, Ung K, Niu Y, Curtis-Snell L, Tomes L, Watson PH, Murphy LC. Estrogen receptor-α phosphorylated at Ser118 is present at the promoters of estrogen-regulated genes and is not altered due to HER-2 overexpression. Cancer Res. 2006;66:10162–10170. doi: 10.1158/0008-5472.CAN-05-4111. [DOI] [PubMed] [Google Scholar]
- 31.Schutz SV, Cronauer MV, Rinnab L. Inhibition of glycogen synthase kinase-3β promotes nuclear export of the androgen receptor through a CRM1-dependent mechanism in prostate cancer cell lines. J Cell Biochem. 2010;109:1192–1200. doi: 10.1002/jcb.22500. [DOI] [PubMed] [Google Scholar]
- 32.Rinnab L, Schutz SV, Diesch J, Schmid E, Kufer R, Hautmann RE, Spindler KD, Cronauer MV. Inhibition of glycogen synthase kinase-3 in androgen-responsive prostate cancer cell lines: are GSK inhibitors therapeutically useful? Neoplasia. 2008;10:624–634. doi: 10.1593/neo.08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadiminty N, Chun JY, Hu Y, Dutt S, Lin X, Gao AC. LIGHT, a member of the TNF superfamily, activates Stat3 mediated by NIK pathway. Biochem Biophys Res Commun. 2007;359:379–384. doi: 10.1016/j.bbrc.2007.05.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Stat3 activation of NF-{κ}B p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci USA. 2006;103:7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.