Abstract
Netrin-4 is a laminin-related secreted molecule originally found to have roles in neuronal axon migration. Recent studies have indicated that netrin-4 also participates in the development of nonneural tissues and modulates tumor cell proliferation and tumor metastasis. Here we have explored the functions and molecular mechanisms of netrin-4 in glioblastoma multiforme. The suppression of netrin-4 expression in glioblastoma cell lines significantly reduced cell proliferation and motility and increased serum deprivation-induced apoptosis. Using tandem affinity purification combined with protein identification by mass spectrometry, we found that integrin β4 interacts with netrin-4 and that it mediates mitogenic effects as well as AKT and mammalian target of rapamycin phosphorylation induced by netrin-4. Interestingly, netrin-4 acted as an inhibitor of cell proliferation in integrin β4-silenced glioblastoma cells, and high concentrations of netrin-4 reduced cell proliferation. The negative effects of netrin-4 on proliferation were mediated by UNC5B. Analysis of more than 400 primary tumors from The Cancer Genome Atlas repository revealed that the expression of netrin-4 is significantly downregulated in glioblastoma and that the reduced expression is linked to poor patient survival time. The expression of integrin β4 is increased in glioblastoma, and it predicts poor patient survival time. Current results illustrate a novel mechanism for glioma progression, where glioma cells reduce netrin-4 expression to decrease its inhibitory effects. In parallel, the expression of integrin β4 is upregulated to sensitize the cells to low concentrations of netrin-4 for maintaining cell proliferation.
Introduction
Netrins (NTNs) are laminin-related secreted molecules with roles in embryogenesis and tumor development. So far, five mammalian members have been identified in this family: netrin-1, -3, -4, -G1, and -G2 [1].
Netrin-1 (NTN1) has been defined as a neuronal guidance cue displaying both attractive and repulsive roles for neuronal cells and axons. Both netrin receptor deleted in colorectal cancer (DCC) and netrin receptor uncoordinated 5 (UNC5s) are major receptors mediating the attractive and repulsive effects of NTN1 [1,2]. Similar to the dual role in axon guidance, the biphasic function of NTN1 appears in the development of the vasculature, reflected by contrasting results in various studies [3–6]. NTN1 can also modulate the development of many other tissues such as the mammary gland, the pancreas, and the lung [7–9]. During tumor progression, NTN1 acts as a survival factor for numerous types of tumor cells during tumor progression through a “dependence-receptor” mechanism [10].
Another member of the NTN family, netrin-4 (NTN4), is expressed throughout the central nervous system (CNS) and brings beneficial effects for the neuronal development [11–13]. Recent studies have illustrated that NTN4 participates in the development of nonneural tissues by modulating the adhesion, migration, proliferation, and apoptosis of endothelial cells [5,14–17]. NTN4 can also act as a regulator of tumor cell proliferation, apoptosis, angiogenesis, and metastasis [15,18,19]. However, the results of the effects of NTN4 have been discordant in the regulation of tumor progression. NTN4 can also act as an inhibitor of both tumor growth and angiogenesis with relatively high concentrations of NTN4 [16,18,19]. NTN4 has also beneficial effects for tumor cells by promoting tumor cell proliferation, angiogenesis, and lymphangiogenesis at relatively low concentrations [5,15,19]. The expression of NTN4 is upregulated in the effusions or invading edge of solid tumor compared with corresponding tumor core [20,21], suggesting that NTN4 may have roles in tumor cell migration and invasion.
Several molecules have been identified to interact or form complex with NTN4, such as neogenin, UNC5B, UNC5D, laminin γ chain, and integrin α6β1, integrin α3β1, integrin α2β1 [12,16,17,22–24]. However, the roles of these interactions are still under discussion, and the biologic functions mediated by these molecules remain largely unclear.
Glioblastoma multiforme (GBM) is the most common malignant tumor of CNS [25]. NTN4 is strongly expressed by astrocytes [11] and astrocyte stem cells [12]. White matter-invading glioblastoma cells express more NTN4 than tumor cores do [20]. However, the functions and molecular mechanisms of NTN4 in glioblastoma need more elucidation. We have explored here the biologic functions of NTN4 in glioblastoma cell lines and analyzed the potential underlying molecular mechanisms for growth modulation.
Materials and Methods
Cell Lines and Reagents
293FT cells (Invitrogen Life Technology, Carlsbad, CA), Astrocytes (Lonza, Switzerland), U251MG cells (Health Science Research Resources Bank, Osaka, Japan), and U87MG and U373MG (American Type Culture Collection, Rockville, MD) were cultured according to the supplier's instructions. Cell migration and proliferation assays were performed by using recombinant NTN4 (R&D Systems, Minneapolis, MN) as modulator.
The following primary antibodies were used: anti-HA.11 from Covance, Princeton, NJ; anti-NTN4 from R&D Systems; anti-ITGB4 from Sigma-Aldrich, St Louis, MO; anti-phospho-AKT (Ser473), anti-phospho-p44/43MAPK (ERK1/2) (Thr202/Tyr204), and anti- phospho-mammalian target of rapamycin (mTOR; Ser2448) from Cell Signaling (Danvers, MA); and anti-BrdU and anti-β-tubulin from Santa Cruz Biotechnology (Santa Cruz, CA).
Bromodeoxyuridine Incorporation Assay
Cells were cultured on a 96-well plate (PerkinElmer, Waltham, MA) with the indicated medium. Subsequently, the medium was replaced to medium with 5-bromo-2-deoxyuridine (BrdU) and incubated for 70 (serum containing) or 120 minutes (serum starvation). After incubation, the BrdU labeling medium was removed, and the cells were washed with phosphate-buffered saline (PBS) twice for 5 minutes each. The cells were then fixed with 70% ethanol containing glycine at -20°C for 30 minutes. Next, the cells were washed three times with PBS and incubated in 2 M HCl at room temperature for 60 minutes. Subsequently, the cells were rinsed and treated with PBS containing 3% bovine serum albumin (BSA) at room temperature for 30 minutes and incubated with anti-BrdU antibodies. The primary antibody was detected by A594 Alexa Fluor secondary antibodies (Invitrogen). After Hoechst staining at 4°C for 15 minutes, cells were washed again. Finally, the images were captured and quantified by using ArrayScan 4.5 high-content-screening system (Cellomics, Pittsburg, PA).
Tandem Affinity Purification and Protein Identification by Mass Spectrometry
For tandem affinity purification (TAP-TAG) and mass spectrometry analysis, we separated and amplified NTN4 full-length sequence to five fragments based on functional domains (Figure W4B).
Using the lentivirus system, we successfully obtained U251MG cells expressing FLAG/HA-tagged fragments and full-length NTN4 complementary DNA (cDNA). Immunofluorescence and immunoblot analysis with anti-HA antibody were performed to confirm target sequence expression (Figure W4, B and C). U251MG cells expressing the indicated protein were lysed in RIPA buffer (50 mM Tris-HCl buffer pH 7.4, containing 150 mM NaCl, 1% NP-40, 0.1% SDS), and lysates were incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich) at 4°C overnight. Subsequently, Sepharose beads were washed with RIPA buffer for five times using micro bio-spin column (Bio-Rad, Hercules, CA). FLAG-containing proteins were eluted by incubating the Sepharose beads with 3x FLAG peptide (Sigma-Aldrich) for 30 minutes on ice. Eluates were incubated with anti-HA Affinity Gel (Sigma-Aldrich) at 4°C for 2 hours. Sepharose beads were washed for five times in spin column, and proteins were eluted with Laemmli sample buffer for 5 minutes on ice.
For identification, proteins were separated by SDS-PAGE and visualized by silver staining followed by in-gel digestion with trypsin and liquid chromatography-tandem mass spectrometry analysis of the resulting peptides as previously described [26]. The nonspecifically interacting proteins identified from control samples were excluded from the identification lists of interacting proteins for full-length NTN4 and fragments of it to create the final list of candidate interacting proteins (Supplemental Data Set).
Statistical Analysis
All numerical values represent mean ± SE or SD. Statistical significance was determined with the nonparametric Mann-Whitney U test.
Lentiviral transfection, quantitative real-time polymerase chain reaction (Q-RT-PCR), migration assay, apoptosis assay, immunofluorescence, and immunoblot analysis were carried out using standard protocols, and these are described in detail in Supplemental Materials and Methods. Bioinformatics analyses were performed as described previously [27,28], see Supplemental Materials and Methods for details.
Results
NTN4 Regulates Glioblastoma Cell Proliferation and Migration in a Concentration-Dependent Manner
NTN4 is highly expressed in astrocytes [11–13]. In vitro and in vivo studies have also revealed that NTN4 regulates tumor cell growth in various cancer types [15,18,19]. It is therefore possible that glioblastoma, the malignant tumor stage of astrocyte, responds to NTN4.
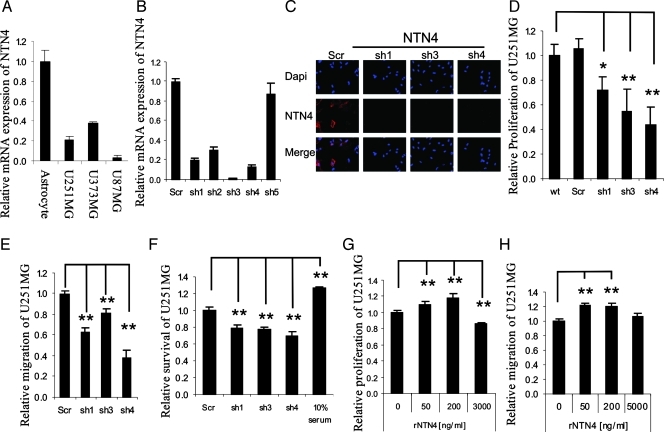
We first investigated the expression of NTN4 in astrocytes and three glioblastoma cell lines by Q-RT-PCR. We detected decent levels of NTN4 gene expression in three invasive glioblastoma cell lines: U251MG, U373MG, and U87MG. However, the expression level of NTN4 in glioblastoma cells was lower than in astrocytes (Figure 1A). We then analyzed the behavior of glioblastoma cell lines after suppression of NTN4 expression. By using the lentiviral short hairpin RNA (shRNA) system, we successfully decreased NTN4 expression levels in the three glioblastoma cell lines. Three efficient shRNAs of NTN4 were identified by both Q-RT-PCR (Figure 1B) and immunofluorescence (Figure 1C).
Figure 1.
NTN4 regulates glioblastoma cell proliferation and migration in a concentration-dependent bell-shaped manner. (A) Astrocytes and glioblastoma cell lines express NTN4. Using Q-RT-PCR, NTN4 expression levels were determined in astrocytes and three invasive glioblastoma cell lines: U87MG, U251MG, and U373MG. Glioblastoma cells have a lower expression of NTN4 than astrocytes in vitro. (B and C) U251MG cells were infected by lentivirus harboring NTN4 shRNAs. After puromycin selection for 72 hours, Q-RT-PCR was performed to detect the expression of NTN4. NTN4 shRNAs resulted in NTN4 mRNA reduction of approximately 80% for shRNA1, 95% for shRNA3, and 87% for shRNA4 (B). The silencing effects of these three shRNAs were also detected by immunofluorescence (C). (D–F) Cells expressing NTN4 shRNAs were used for proliferation assay, migration assay, and apoptosis assay. Mitogenic rate, migration distance, and survival rate of NTN4-silenced cells were compared with nontransfected cells (wt) or cells transfected with nontargeting shRNA (Scr). In the apoptosis assay, cells cultured in 10% serum-containing medium were used as positive control. Suppression of NTN4 gene expression in U251MG cells significantly reduced cell mitogenicity, migration, and increased apoptosis caused by serum deprivation. (G and H) U251MG cells were cultured in serum-free medium for 4 hours and then cultured in the presence of recombinant NTN4 with the indicated concentrations. Recombinant NTN4 was reapplied every 24 hours for 48 hours. Cells were then used for proliferation assay. NTN4 promoted U251MG cell proliferation at 50 and 200 ng/ml, but it inhibited cell proliferation at 3 µg/ml (G). U251MG cells were cultured in serum-free medium overnight and then used for the wound closure assays in the presence of recombinant NTN4 as in the indicated concentration. NTN4 promoted U251MG cell migration at 50 and 200 ng/ml. However, 5 µg/ml NTN4 had no significant effect on U251MG cell migration (H). Mean ± SE, n ≥ 3. **P < .01. *P < .05.
BrdU incorporation assay revealed that the suppression of NTN4 expression in U251MG cells inhibited cell proliferation (Figure 1D). Using wound closure assays, we determined that the reduction of NTN4 expression suppressed U251MG cell motility (Figure 1E). It has been found that NTN4 acts as a survival factor for lymphatic endothelial cells [15]. Therefore, we investigated the effects of reduced NTN4 expression on glioblastoma cell apoptosis by TUNEL assay. We found that inhibition of NTN4 expression significantly increased glioblastoma cell apoptosis caused by serum deprivation (Figure 1F). In addition, suppression of NTN4 expression also reduced U373MG and U87MG cell proliferation, inhibited U373MG cell motility, and increased U373MG cell apoptosis caused by serum deprivation (Figure W1). NTN4 may therefore play an important role in promoting glioblastoma cell proliferation and migration.
To further assess the role of NTN4 on the migration and proliferation of glioblastoma cells, NTN4 was overexpressed in U251MG cells. We used two overexpression methods and successfully obtained two NTN4-overexpressing U251MG cell populations (NTN4.pLVX and NTN4.pcDNA3). Q-RT-PCR was performed to confirm NTN4 gene overexpression at the mRNA level (Figure W2A). BrdU incorporation assays and wound closure assays revealed that stable NTN4 overexpression alters neither U251MG cell proliferation (Figure W2B) nor migration (Figure W2C). These results are in agreement with studies on prostate cancer, breast cancer, and colorectal carcinoma [15,16,18] but are not consistent with the effect of silencing NTN4 in glioblastoma cells.
The reason why we did not observe any effects on cell proliferation and migration with stable overexpression may arise from clonal selection and/or cell adaptation to increased NTN4 expression during selection process. Therefore, we assessed U251MG cell proliferation (Figure 1G) and migration (Figure 1H) by using increasing concentrations of exogenous NTN4. NTN4 promoted cell proliferation and migration concentration dependently with a maximum at 200 ng/ml. However, 5 µg/ml NTN4 did not display any effect on cell migration and 3 µg/ml significantly inhibited cell growth. These data imply that the effects of NTN4 on glioblastoma cells are concentration dependent.
NTN4/ITGB4 Interaction Promotes Glioblastoma Cell Proliferation
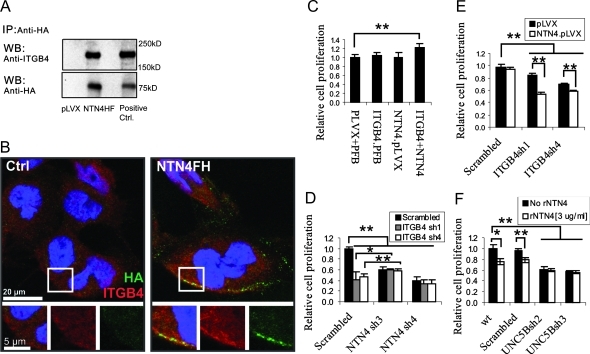
Similar to NTN1, the biphasic function of NTN4 on glioblastoma cells may arise from the binding to different receptors. To identify the receptors that mediate the biologic functions of NTN4, we first used Q-RT-PCR to determine which previously known receptors of the NTN family are expressed in the U251MG, U87MG, and U373MG cells. We found that these three glioblastoma cells express several receptors of NTN1 (Figure W3). Next, we applied TAP-TAG combined with protein identification by mass spectrometry by using NTN4 as a bait (Figure W4 and Supplemental Data Set). In the process, we reliably identified integrin β4 (ITGB4) as a partner interacting with NTN4 by immunoprecipitation (Figure 2A) and observed that NTN4 and ITGB4 colocalized at the edge of U251MG cell (Figure 2B).
Figure 2.
NTN4/ITGB4 interaction promotes glioblastoma cell proliferation. (A) U251 cells expressing empty vector or FLAG/HA-tagged NTN4 were cultured to 50% confluence and were subsequently lysed for immunoprecipitation. After immunoprecipitation with anti-HA agarose, ITGB4 was detected in immunoprecipitates of FLAG/HA-tagged NTN4. Immunoprecipitates of empty vector and cell lysates were set as negative control and positive control, respectively. (B) U251MG cells were cultured to 50% confluence and were incubated with conditioned medium from either control or FLAG/HA-tagged NTN4 (NTN4FH)-expressing cells as described in Supplemental Materials and Methods, and then used for immunofluorescence. Micrographs of ITGB4 (red) and HA (green) in U251MG were obtained using a confocal laser scanning microscope. Yellow indicates the colocalization of ITGB4 and NTN4. (C) U251 cells expressing the indicated gene or shRNAs were cultured in 96-well plates for 2 days and then used for the proliferation assay. Neither NTN4 overexpression nor ITGB4 overexpression alone promoted U251MG cell growth. Combined overexpression of NTN4 and ITGB4 increased cell proliferation by approximately 30%. (D) U251 cells expressing the indicated gene or shRNAs were used for proliferation assay. Suppression of ITGB4 expression in NTN4-silenced U251MG cells did not further reduce cell proliferation. Complete suppression of NTN4 expression, NTN4sh3, slightly increased the proliferation of ITGB4-silenced U251MG cells. (E) Suppression of ITGB4 expression leads to less mitogenic rate in NTN4-overexpressing U251MG cell than in control cells. (F) Suppression of UNC5B expression in U251MG cells reduced cell mitogenic ability. Exogenous addition (3 µg/ml) of NTN4 inhibited cell proliferation in control cells but not in UNC5B-silenced cells. Mean ± SE, n ≥ 3. **P < .01. *P < .05.
Recent studies have revealed that ITGB4 is involved in tumor progression [29], and many glioblastoma cell lines express ITGB4 [30]. All of U87MG, U251MG, and U373MG cells express ITGB4, but the expression levels of ITGB4 in these three cell lines are different. A previous report described that U373MG cells migrate and proliferate slightly faster than U87MG cells in vitro and leads to poorer survival than U87MG in vivo [31]. To investigate the function of ITGB4 and NTN4, we overexpressed them either separately or together (Figure W5A). We found that neither NTN4 overexpression nor ITGB4 overexpression alone altered U251MG cell proliferation. However, co-overexpression of ITGB4 and NTN4 increased glioblastoma cell growth slightly but significantly (Figure 2C).
We subsequently knocked down the expression of ITGB4 in the glioblastoma cell lines. Suppression of ITGB4 inhibited the proliferation of both U87MG and U373MG cell. Nontargeting shRNA was used as control (Figure W5, B–E). We then transduced the most efficient shRNAs of ITGB4 and NTN4 into U251MG cells in combination (Figure W5, F and G). BrdU incorporation assays revealed that suppression of ITGB4 in U251MG cells inhibited cell growth but did not further reduce the proliferation rate of NTN4-silenced cells. These data imply that ITGB4 mediates NTN4's effects in the promotion of cell proliferation. Interestingly, an almost complete suppression of the expression of NTN4 (NTN4sh3) even slightly increased the mitogenic rate of ITGB4-suppressed cells (Figure 2D). One explanation for this is that, after silencing ITGB4, the remaining NTN4 interacts with other potential lower-affinity receptors, which induce negative effects for proliferation. To obtain support for this hypothesis, we suppressed ITGB4 expression in both control and NTN4-overexpressing U251MG cells (Figure W5H). BrdU incorporation assay showed that overexpression of NTN4 in ITGB4-suppressed U251MG cells led to a lower proliferation rate compared to cells with normal ITGB4 expression level (Figure 2E). This observation is in accordance with the hypothesis that NTN4 may negatively regulate cell proliferation when not bound to ITGB4.
Because the complex of NEO1 and UNC5B has been observed to mediate the angiogenesis inhibition of NTN4 [16,32], we investigated the role of UNC5B in cell growth inhibition of NTN4 by using UNC5B shRNAs (Figure W5I). Suppression of UNC5B expression decreased U251MG cell proliferation. However, higher concentrations of NTN4 were not able to further inhibit glioblastoma cell proliferation (Figure 2F).
NTN1 binds also to ITGB4 [9]. Therefore, we asked whether NTN4 competes with NTN1 for binding to ITGB4. By using the overexpression of either tagged or native NTNs, we revealed that NTN1 and NTN4 do not compete with each other in binding to ITGB4 (Figure W6). Similarly, we found that both NTN1 and NTN4 promote cell proliferation, but neither NTN1 nor NTN4 can compensate each other for the growth-promoting effects. Furthermore, combined overexpression of NTN1 and NTN4 did not have any effect on cell proliferation, and silencing of the expression of both NTN1 and NTN4 had an additive effect on cell proliferation (Figure W7).
NTN4 Expression Is Downregulated in Clinical Glioblastoma Specimen and Associates with Patient Survival
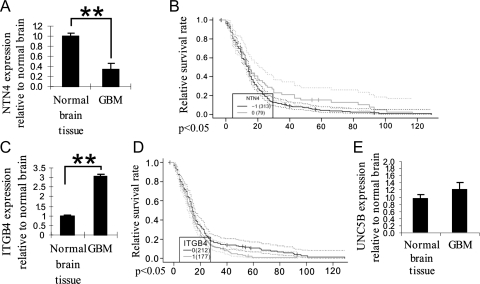
In vitro data suggest that NTN4 may act as a stimulator or an inhibitor of glioblastoma cell proliferation through binding to different receptors. To investigate the role of NTN4 and ITGB4 in glioblastoma tissues, we analyzed the expression levels of primary tumors from The Cancer Genome Atlas - Glioblastoma multiforme (TCGA-GBM) repository [27,28,33]. NTN4 is expressed less in grade 4 glioblastoma than in normal brain tissue (Figure 3A). In addition, we analyzed NTN4 expression in relation to the survival time of the patients with grade 4 glioblastoma. We found that low expression of NTN4 predicts poor patient survival time (Figure 3B). To confirm these results, we used the Oncomine database and two published reports [34,35]. The expression of NTN4 decreased after glioma progression from grade 2 to grade 4 (Figure W8A). However, most glioma cell lines express NTN4 (Figure W8B). These data suggest that proper concentration of NTN4 is beneficial for glioma cells, whereas high concentrations of NTN4 may have negative effects on glioma progression.
Figure 3.
NTN4 expression is downregulated in glioblastoma specimens and associates with reduced patient survival. (A) On the basis of 425 primary GBM tumors from TCGA [27,33], bioinformatics analysis revealed that the expression of NTN4 was reduced to 33% in grade 4 glioblastoma, compared with normal brain tissue. (B) Kaplan-Meier survival analysis showed that the low expression of NTN4 predicts poor patient survival. Black line represents all glioblastoma samples with low expression of NTN4 compared with normal tissue (fold change < 0.5), and gray line represents a group of glioblastoma samples which have similar expression of NTN4 to the normal brain tissue. Dashed lines represent 0.95 confidence intervals for the curves with the corresponding color. P < .043. (C) The expression of ITGB4 increased 2.9-fold in grade 4 glioblastoma, compared with normal brain tissue. (D) Kaplan-Meier survival analysis showed that the upregulated expression of ITGB4 predicts poor patient survival. Gray line represents all glioblastoma samples with high expression of ITGB4 compared with normal tissue (fold change > 3), and black line represents a group of glioblastoma samples that have similar expression of ITGB4 to the normal brain tissue. P < .029. (E) The expression of UNC5B does not significantly change in glioblastoma compared with the normal brain tissue.
Interestingly, the expression of ITGB4 is significantly increased in grade 4 glioblastoma (Figure 3C), and the up-regulation of ITGB4 expression predicts poor patient survival time (Figure 3D). However, the expression of UNC5B has no significant change in GBM specimens compared with normal brain tissue (Figure 3E).
NTN4/ITGB4 Interaction Activates AKT/mTOR Signaling Pathways
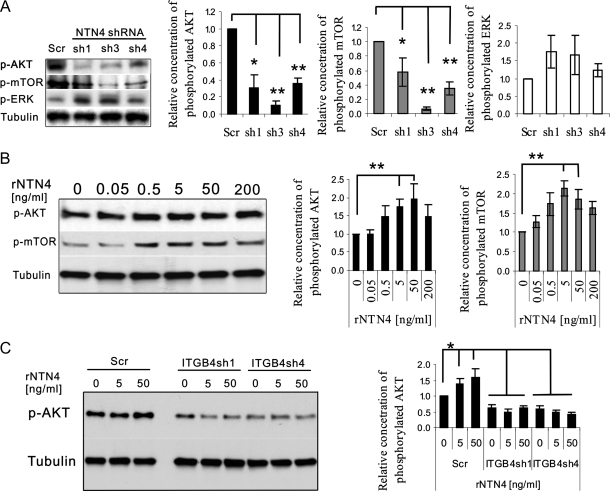
AKT and ERK1/2 phosphorylation have been linked to ITGB4-mediated cell proliferation [36,37], and a recent study has shown that NTN4 activates S6K [15]. Therefore, we assessed the effect of NTN4 on the activation of the MAP kinase, AKT, and mTOR signaling pathways.
Suppression of NTN4 expression in U251MG significantly reduced AKT and mTOR phosphorylation, whereas it slightly increased ERK1/2 phosphorylation (Figure 4A). Thirty minutes after exogenous addition of NTN4, U251MG cells displayed concentration-dependent activation of AKT and mTOR (Figure 4B). This suggested that NTN4 promotes glioblastoma cell growth by stimulating the phosphorylation of AKT-mTOR signaling pathway.
Figure 4.
NTN4/ITGB4 interaction activates AKT/mTOR signaling pathways. (A) U251 cells expressing NTN4 shRNAs were cultured in serum-free medium for 12 hours, followed by immunoblot analysis as indicated. U251 cells expressing NTN4 shRNAs displayed reduced phosphorylation of AKT and mTOR but slightly increased phosphorylation of ERK. Chart illustrates the relative phosphorylation levels of AKT, mTOR, and ERK. (B) NTN4 concentration-dependently increased AKT and mTOR phosphorylation. U251MG cells were cultured in serum free medium for 12 hours and then incubated with increasing concentrations of exogenous recombinant NTN4 for 30 minutes, followed by immunoblot analysis as indicated. Concentrations of 5 and 50 ng/ml NTN4 significantly increased AKT and mTOR activation in U251MG wild-type cells and scrambled transfected control cells. Chart illustrates the relative phosphorylation levels of AKT and mTOR. (C) Concentrations of 5 and 50 ng/ml NTN4 were not able to stimulate AKT phosphorylation in ITGB4-silenced U251MG cell. Chart illustrates the relative phosphorylation levels of AKT. β-Tubulin was used as loading control. Mean ± SE, n ≥ 3. **P < .01. *P < .05.
ITGB4 has been shown to induce AKT-mTOR signaling [38,39], and mTOR plays a crucial role in cell proliferation [40,41]. Insulin receptor substrates participate in the ITGB4-AKT signaling pathway [37], and these molecules also appeared in the list of interacting candidates for NTN4 (Supplemental Data Set). Therefore, we investigated the role of NTN4 in ITGB4 mediated AKT-mTOR signaling pathway. Addition of recombinant NTN4 increased AKT phosphorylation in control cells. However, it was not able to activate AKT in ITGB4-suppressed U251MG cells. This indicates that ITGB4 mediates NTN4-induced AKT-mTOR phosphorylation (Figure 4C).
Discussion
NTN4 was discovered a decade ago as an axon guidance cue [11,13]. Recent studies have described that NTN4 regulates endothelial cell adhesion, migration, proliferation, and apoptosis and that its effects correlate with its concentration [5,15,16,42]. In cancer cells, NTN4 can act as a modulator of tumor growth, angiogenesis, and metastasis [15,18,19,21]. As a functional molecule in CNS, high expression of NTN4 has been found in astrocytes and at the invading edge of glioblastoma specimens [12,13,20]. However, the functions and molecular mechanisms of NTN4 in glioblastoma are not clear yet.
So far, many studies support the hypothesis that NTNs, especially NTN1, have biphasic functions during development [1,2,6]. In the present study, we provide evidence that NTN4 plays a dual role in the modulation of glioblastoma cell proliferation. The mitogenic facilitation of glioblastoma cells may convert to inhibition of proliferation at higher NTN4 concentrations in a bell-shaped manner. This is consistent with the result that NTN4 is downregulated in glioblastoma specimen and predicts patient survival. It is also consistent with the dual role of NTN4 in other tumor cell lines [19].
The receptors and downstream signaling pathway of NTN4 mediating these functions have been largely unknown. ITGB4, the β-subunit of integrin heterodimers, is predominantly expressed in epithelial cells. It is upregulated during tumor progression and pathologic angiogenesis [43]. In the CNS, ITGB4 is expressed strongly in astrocytes, and it is upregulated during glioma progression [44,45], indicating that ITGB4 plays an integral role in glioblastoma. Exon-array analysis with more than 400 GBM primary tumors revealed that the expression of ITGB4 is upregulated in grade 4 glioblastomas, and it predicts poor patient survival whereas it negatively associates with NTN4 expression. We discovered that ITGB4 binds to NTN4 and mediates the positive effects of NTN4 on glioblastoma cell proliferation. Interestingly, suppression of NTN4 expression in ITGB4-silenced cells slightly increases cell mitogenicity, and NTN4 overexpression leads to mitogenic inhibition in ITGB4-silenced glioblastoma cells. These results suggest that there are other mechanisms mediating the negative effects of NTN4. So far, the complex of NEO1 and UNC5B has been identified to participate in the antiangiogenic activity of NTN4 [16,18,32]. Our results indicate that UNC5B mediates the negative effects of NTN4 on glioblastoma cell proliferation.
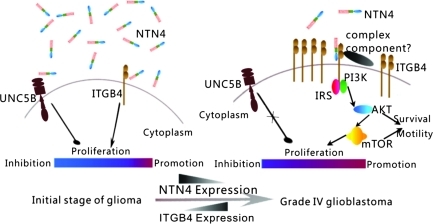
Although NTN4 and ITGB4 interaction induces glioblastoma cell proliferation, their expression levels in glioblastomas correlate negatively. In addition, the expression levels of NTN4 and ITGB4 are related to patient survival positively and negatively, accordingly. Regarding the dual role of NTN4 in glioblastoma cells and its potential inhibitory effects on angiogenesis at high concentrations [16,18], we propose a mechanism for NTN4 and ITGB4 in glioma progression. At the initial stage of glioma progression, the concentration of NTN4 in the extracellular environment is relatively high. Glioma cells express low levels of ITGB4, which is sufficient to respond to high concentrations of NTN4. Meanwhile, high concentrations of NTN4 have the ability to interact with UNC5B. Therefore, NTN4 may reduce cell proliferation through UNC5B and maintain the balance of glial cell proliferation. During tumor progression, glioma cells reduce their NTN4 expression. Because of the unaltered expression of UNC5B, decreased NTN4 expression endows glioma cells with at least two benefits: First, it helps glioma cells to escape from NTN4 induced growth reduction. Second, glioma tumor is relieved from potential angiogenesis inhibition induced by high concentration of NTN4. In parallel, ITGB4 expression increases. In this way, excessive ITGB4 may make glioblastoma cell hyperresponsive to low concentrations of NTN4 and maintain glioma cell proliferation (Figure 5).
Figure 5.
Interplay mechanism between NTN4 and ITGB4 during glioma progression. At the initial stage of glioma progression, NTN4 concentration stays relatively high in extracellular environment. High concentrations of NTN4 have an ability to reduce cell proliferation through UNC5B. During progression, glioma cells reduce NTN4 expression to prevent the negative effects on glioma cell growth induced by NTN4. In parallel, ITGB4 expression increases. In this way, excessive ITGB4 makes glioblastoma cells hyperresponsive to low concentrations of NTN4 for keeping AKT/mTOR phosphorylation and glioma cell proliferation.
The complicated functions of NTN4 in glioblastoma cells indicate that NTN4 may be associated with multiple signaling pathways. In cancer cells, ITGB4 interacts with several growth factor receptors including ErbB2, epidermal growth factor receptor, and Met [46–48]. These interactions lead to enhanced receptor tyrosine kinase signaling and associate ITGB4 with tumor progression and invasion. ITGB4 promotes tumor progression through three signaling pathways: Ras-ERK pathway, PI3K-AKT pathway, and NFAT pathway [36,37,49]. Meanwhile, it has also been found that ITGB4 activates the AKT-mTOR signaling pathway, and insulin receptor substrates act as components in ITGB4-transduced AKT phosphorylation [37–39]. We have identified insulin receptor substrates in the interacting candidate list of NTN4. These results indicate that NTN4 may act as a stimulator in ITGB4 activation. Furthermore, a recent study reported that NTN4 activates S6K to stimulate protein synthesis [15]. These observations suggest that NTN4 activates the ITGB4-transduced AKT-mTOR signaling pathway. In the current work, we provide evidence that NTN4 induces phosphorylation of AKT and mTOR through ITGB4. As mTOR has been found to associate with glioblastoma cell proliferation, apoptosis, and transformation [50–52], it is likely that NTN4 promotes glioblastoma cell proliferation through ITGB4-transduced PI3K-AKT-mTOR signaling pathway.
In summary, the complicated functions of NTN4 in GBM cells are mediated by different receptors and multiple signaling pathways. The observation of ITGB4 and NTN4 interaction provides a novel pathway for promotion of tumor growth. This suggests that blocking ITGB4 may be a promising therapy method for treating glioblastoma and that NTN4/ITGB4 interaction possibly provides a way to modulate ITGB4 signaling.
Supplemental Materials and Methods
Total RNA Extraction, Reverse Transcription, and Real-time Reverse Transcription-PCR
Total cellular RNA was isolated and purified using RNeasy Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was carried out with random hexamer primers (Invitrogen) and SuperScript III reverse transcription (Invitrogen) using 1.0 µg of total RNA according to the manufacturer's instructions. The cDNA products were amplified by TaqMan Assays-on-Demand gene expression products (Applied Biosystems, Foster City, CA), TaqMan Universal PCR Master Mix (Applied Biosystems), and CFX96 Real-time PCR Detection System (Bio-Rad). Levels of gene expression were determined by ΔΔCT method, with the results being expressed as mRNA expression levels normalized to the levels of a gene with a constant expression (GAPDH).
Vectors and Expression Constructs
pRK5-FLAG /HA (pRK5-FH) plasmid was provided by Dr Giancotti's laboratory, Memorial Sloan-Kettering Cancer Center [1]. pRK5-FH contains EcoRI and BamHI restriction sites for cloning the target gene and C-terminal Flag and HA tag.
The indicated fragments or full-length gene were amplified and cloned to pRK5-FH. In constructs missing the signal sequence, CD33 signal sequence coding cDNA was cloned 5′ to the coding region of each construct. Next, by using a pair of primers recognizing pRK5 plasmid DNA sequence upstream of target gene and downstream of FLAGHA, we amplified all the constructs with FLAG/HA tag from pRK5 plasmid, and then cloned them to pLVX-puro (Clontech) plasmid for lentiviral expression. Full-length human NTN1 and NTN4 were cloned to both pLVX-puro and pcDNA3 (Invitrogen) plasmid for subsequent assays. All constructs were confirmed by DNA sequencing. Details on the cloning are available on request.
Transfection of Cells
Cells were cultured on six-well plates to 50% to 80% confluence and transfected using 2 µg of plasmid DNA and 6 µl of FuGENE 6 transfection reagents for each transfection according to the manufacturer's instructions (Boehringer Mannheim, Mannheim, Germany). Stably transfected cells were selected with ∼0.8 mg/ml G418 (Gibco/Invitrogen) in complete medium. The expression levels of each indicated gene in transfected cells were analyzed by Q-RT-PCR.
Lentiviral Silencing/Overexpression of Gene Expression
Plasmids expressing shRNAs targeted against the indicated gene were obtained from the RNAi Consortium through Open Biosystems (hairpin-pLKO.1 vector). Five different constructs for each gene were tested. A nontargeting scrambled construct was used as control for nonspecific effects (scrambled-pLKO.1 vector). Plasmids harboring the indicated gene were constructed as described above. pLVX-puro empty vector was used as a control.
To produce recombinant lentivirus particles, 293FT producer cells were cotransfected with the packaging/envelope plasmids (pCMVdr8.74 and pMD2-VSVG; Addgene, Cambridge, MA) and target plasmids by using the Lipofectamine (Invitrogen) transfectionmethod. Normal complete culture medium (high-glucose Dulbecco modified Eagle medium [DMEM]) was changed to the 293FT cells 24 hours after transfection. Subsequently, 48 hours later, the viral supernatants were harvested and filtered through a 0.45-µm filter. The titer of the virus was investigated. After a 24-hour infection with virus, the supernatants were replaced with complete medium (DMEM) for the subsequent assays. For stable shRNA expression, the infected U251MG cells were subjected to selection with 2 µg/ml puromycin for 72 hours. The efficiency of the transduction was measured by monitoring the indicated gene expression.
Scratch-Induced In Vitro Wound Closure Assay
Cells were plated in 24-well tissue culture plate at a concentration of 30,000 cells and maintained under the indicated conditions for 3 days. After the cell cultures reached ∼90% confluence, the tip of a 1000-µl micropipette was used to scratch the cells, creating linear and cross-shaped scratches. Cell layers were washed twice with PBS to remove floating cells and medium. The medium was replaced by DMEM containing 0.5% serum for an additional 12-hour incubation. Cell migration was photographed at the edge where the scrape was introduced at the time point of 0 and 8 hours using Axiovert 200 inverted epifluorescence microscope (Carl Zeiss). The migration rate was analyzed with the ImageJ program (National Institutes of Health, Bethesda, MD).
Serum Deprivation-Induced Apoptosis Assay
Cells were cultured on glass coverslips overnight and then changed the medium to serum free DMEM and cultured for 72 hours. When cells started to float up, the coverslips were washed with PBS and fixed with 4% paraformaldehyde in PBS for 20 minutes. The cells were then subject to TUNEL labeling by using In Situ Cell Death Detection Kit (Roche, Switzerland) according to the manufacturer's instructions.
After several washing steps, the coverslips were washed once inwater and then mounted with VECTASHIELD antifading reagent (Vector Laboratories, Burlingame, CA). Nuclei were visualized by 4′,6-diamidino-2-phenylindole staining. The images were captured using the 20x objective of the Axioplan microscope (Carl Zeiss, Inc, Thornwood, NY), acquired with AxioCamHRc camera (Carl Zeiss) and AxioVision3.1 software (Carl Zeiss).
Immunofluorescence
Cells were cultured on glass coverslips for the indicated times. For observing the colocalization of ITGB4 and NTN4 in U251MG cell, U251MG cells were incubated with the conditioned medium from either control or HA-tagged NTN4-expressing cells on ice for 1 hour. Cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 20 minutes and then incubated in permeabilization buffer (0.1% Triton X-100 in PBS) for 15 minutes on ice. Subsequently, the cells were washed three times with PBS for 5 minutes each and treated with Dulbecco PBS containing 3% BSA at room temperature for 30 minutes. The cells were rinsed and incubated with primary antibody diluted in Dulbecco PBS containing 3% BSA at room temperature for 1 hour. After washing with PBS, the cells were incubated with Alexa Fluor secondary antibodies (Invitrogen) diluted in Dulbecco PBS containing 3% BSA at room temperature for 30 minutes. After several washing steps, the coverslips were washed once in water and then mounted with Mowiol-mounting medium (Calbiochem/Merck, Darmstadt, Germany) containing 6 mg/ml DABCO (Sigma) as antifading reagent. Nuclei were visualized by Hoechst (Sigma) staining. The images were captured using the Axioplan microscope (Carl Zeiss), laser scanning confocal microscope (Zeiss LSM 510 Meta), and acquired with AxioCamHRc camera (Carl Zeiss), AxioVision3.1 software (Carl Zeiss), and LSM software at the Biomedicum Imaging Unit of the University of Helsinki.
SDS-PAGE and Immunoblot Analysis
Cells were lysed in RIPA buffer supplemented with protease inhibitors (Roche). SDS-PAGE was performed using commercial 4% to 20% gradient Tris-HCl polyacrylamide gels (Lonza). Electrophoretically separated proteins were transferred onto nitrocellulose membranes (PROTRAN; Schleicher and Schuell, Keene, NH) by semidry blot analysis system (Bio-Rad). The membranes were subsequently washed three times with 0.05% Tween-20/Tris-buffered saline and incubated with the primary antibodies in 5% BSA/0.05% Tween-20/Tris-buffered saline at 4°C overnight. Membranes were washed twice for 10 minutes each. HRP-conjugated secondary antibodies (Santa Cruz Biotechnology) were then diluted in 5% BSA/0.05% Tween-20/TBS and incubated with the membrane at 4°C for 1 hour. After several washing steps, the detection was performed using enhanced chemiluminescence reagents (GE Healthcare, United Kingdom). The images were obtained using an x-ray film (Fujifilm, Tokyo, Japan) and Kodak X-OMAT 2000 equipment (Kodak, Rochester, NY).
Phospho-specific Western blot analysis was performed following the manufacturer's instructions (Cell Signaling Technology, Danvers, MA). Protein concentrations were measured using a bicinchoninic acid kit (Thermo Scientific, Rockford, IL). Equal amounts of total protein were loaded per lane.
Band intensity quantification was carried out using ImageJ software.
Bioinformatics Analysis
Exon array data for 425 primary GBMsamples and 10 normal brain samples were downloaded from the TCGA repository and preprocessed at gene level by Multiple Exon Array Preprocessing (MEAP) algorithm [2]. All data analyses, such as statistical tests and Kaplan-Meier survival estimates, were done with the Anduril framework [3]. In the survival analyses, we used 393 samples that had survival end time in the TCGA GBM repository. Fold change (FC) values were calculated by taking the median and used in grouping for Kaplan-Meier survival analysis, where patients were divided into groups denoted by “-1” (underexpression, FC < 0.5), “1” (overexpression, FC > 3), and “0” (stable expression). Survival P values were calculated with the log rank test, and the threshold used in our analysis was P < .05.
Supplemental Data Set
The peptide data obtained with mass spectrometry were used to identify proteins with Mascot search engine. The searches were done against the SwissProt database. The identification lists for negative control, full-length NTN4, and each of the fragments are shown.
Acknowledgments
The authors thank Sami Starast and Anne Remes for excellent technical assistance. Anders Paetau, Department of Pathology, University of Helsinki, is acknowledged for the critical comments on the article. Filippo Giancotti, Memorial Sloan-Kettering Cancer Center, is acknowledged for the discussions during the postdoctoral fellowship of M.H. in his laboratory. The microscopic analyses were carried out at the Biomedicum Imaging Unit, University of Helsinki.
Abbreviations
- AKT
serine/threonine-protein kinase akt (protein kinase B)
- DCC
netrin receptor deleted in colorectal cancer
- GBM
glioblastoma multiforme
- ITGB4
integrin β4
- mTOR
mammalian target of rapamycin
- NTN
netrin
- Q-RT-PCR
quantitative real-time polymerase chain reaction
- TAP-TAG
tandem affinity purification
- TCGA
The Cancer Genome Atlas
- UNC5
netrin receptor uncoordinated 5
Footnotes
This research was supported by Finnish Cultural Foundation, the Academy of Finland, the Finnish Cancer Foundation, the Sigrid Juselius Foundation, Finska Lākaresällskapet, the Finnish Society of Sciences and Letters, Biocentrum Helsinki, Helsinki University Hospital Fund and the University of Helsinki.
This article refers to supplementary materials, which are designated by Figures W1 to W8 and Supplemental Data Set and are available online at www.neoplasia.com.
References
- 1.Cirulli V, Yebra M. Netrins: beyond the brain. Nat Rev Mol Cell Biol. 2007;8:296–306. doi: 10.1038/nrm2142. [DOI] [PubMed] [Google Scholar]
- 2.Hong K, Hinck L, Nishiyama M, Poo MM, Tessier-Lavigne M, Stein E. A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell. 1999;97:927–941. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- 3.Larrivee B, Freitas C, Trombe M, Lv X, Delafarge B, Yuan L, Bouvree K, Breant C, Del Toro R, Brechot N, et al. Activation of the UNC5B receptor by Netrin-1 inhibits sprouting angiogenesis. Genes Dev. 2007;21:2433–2447. doi: 10.1101/gad.437807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park KW, Crouse D, Lee M, Karnik SK, Sorensen LK, Murphy KJ, Kuo CJ, Li DY. The axonal attractant Netrin-1 is an angiogenic factor. Proc Natl Acad Sci USA. 2004;101:16210–16215. doi: 10.1073/pnas.0405984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson BD, Ii M, Park KW, Suli A, Sorensen LK, Larrieu-Lahargue F, Urness LD, Suh W, Asai J, Kock GA, et al. Netrins promote developmental and therapeutic angiogenesis. Science. 2006;313:640–644. doi: 10.1126/science.1124704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Zou L, Wang Y, Xu KS, Zhang JX, Zhang JH. Axon guidance cue Netrin-1 has dual function in angiogenesis. Cancer Biol Ther. 2007;6:743–748. doi: 10.4161/cbt.6.5.3976. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasan K, Strickland P, Valdes A, Shin GC, Hinck L. Netrin-1/neogenin interaction stabilizes multipotent progenitor cap cells during mammary gland morphogenesis. Dev Cell. 2003;4:371–382. doi: 10.1016/s1534-5807(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Stein E, Oliver T, Li Y, Brunken WJ, Koch M, Tessier-Lavigne M, Hogan BL. Novel role for Netrins in regulating epithelial behavior during lung branching morphogenesis. Curr Biol. 2004;14:897–905. doi: 10.1016/j.cub.2004.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yebra M, Montgomery AM, Diaferia GR, Kaido T, Silletti S, Perez B, Just ML, Hildbrand S, Hurford R, Florkiewicz E, et al. Recognition of the neural chemoattractant Netrin-1 by integrins α6β4 and α3α1 regulates epithelial cell adhesion and migration. Dev Cell. 2003;5:695–707. doi: 10.1016/s1534-5807(03)00330-7. [DOI] [PubMed] [Google Scholar]
- 10.Mehlen P, Delloye-Bourgeois C, Chedotal A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nat Rev Cancer. 2011;11:188–197. doi: 10.1038/nrc3005. [DOI] [PubMed] [Google Scholar]
- 11.Koch M, Murrell JR, Hunter DD, Olson PF, Jin W, Keene DR, Brunken WJ, Burgeson RE. A novel member of the netrin family, β-netrin, shares homology with the β chain of laminin: identification, expression, and functional characterization. J Cell Biol. 2000;151:221–234. doi: 10.1083/jcb.151.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staquicini FI, Dias-Neto E, Li J, Snyder EY, Sidman RL, Pasqualini R, Arap W. Discovery of a functional protein complex of netrin-4, laminin γ1 chain, and integrin α6β1 in mouse neural stem cells. Proc Natl Acad Sci USA. 2009;106:2903–2908. doi: 10.1073/pnas.0813286106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Y, Sanes JR, Miner JH. Identification and expression of mouse netrin-4. Mech Dev. 2000;96:115–119. doi: 10.1016/s0925-4773(00)00369-5. [DOI] [PubMed] [Google Scholar]
- 14.Hoang S, Liauw J, Choi M, Guzman RG, Steinberg GK. Netrin-4 enhances angiogenesis and neurologic outcome after cerebral ischemia. J Cereb Blood Flow Metab. 2009;29:385–397. doi: 10.1038/jcbfm.2008.128. [DOI] [PubMed] [Google Scholar]
- 15.Larrieu-Lahargue F, Welm AL, Thomas KR, Li DY. Netrin-4 induces lymphangiogenesis in vivo. Blood. 2010;115:5418–5426. doi: 10.1182/blood-2009-11-252338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lejmi E, Leconte L, Pedron-Mazoyer S, Ropert S, Raoul W, Lavalette S, Bouras I, Feron JG, Maitre-Boube M, Assayag F, et al. Netrin-4 inhibits angiogenesis via binding to neogenin and recruitment of Unc5B. Proc Natl Acad Sci USA. 2008;105:12491–12496. doi: 10.1073/pnas.0804008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneiders FI, Maertens B, Bose K, Li Y, Brunken WJ, Paulsson M, Smyth N, Koch M. Binding of netrin-4 to laminin short arms regulates basement membrane assembly. J Biol Chem. 2007;282:23750–23758. doi: 10.1074/jbc.M703137200. [DOI] [PubMed] [Google Scholar]
- 18.Eveno C, Broqueres-You D, Feron JG, Rampanou A, Tijeras-Raballand A, Ropert S, Leconte L, Levy BI, Pocard M. Netrin-4 delays colorectal cancer carcinomatosis by inhibiting tumor angiogenesis. Am J Pathol. 2011;178(4):1861–1869. doi: 10.1016/j.ajpath.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nacht M, St Martin TB, Byrne A, Klinger KW, Teicher BA, Madden SL, Jiang Y. Netrin-4 regulates angiogenic responses and tumor cell growth. Exp Cell Res. 2009;315:784–794. doi: 10.1016/j.yexcr.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Hoelzinger DB, Mariani L, Weis J, Woyke T, Berens TJ, McDonough WS, Sloan A, Coons SW, Berens ME. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005;7:7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y, Leszczynska M, Konstantinovsky S, Trope CG, Reich R, Davidson B. Netrin-4 is upregulated in breast carcinoma effusions compared to corresponding solid tumors. Diagn Cytopathol. 2011;38(8):562–566. doi: 10.1002/dc.21424. [DOI] [PubMed] [Google Scholar]
- 22.Yebra M, Diaferia GR, Montgomery AM, Kaido T, Brunken WJ, Koch M, Hardiman G, Crisa L, Cirulli V. Endothelium-derived Netrin-4 supports pancreatic epithelial cell adhesion and differentiation through integrins α2β1 and α3β1. PLoS One. 2011;6:e22750. doi: 10.1371/journal.pone.0022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larrieu-Lahargue F, Welm AL, Thomas KR, Li DY. Netrin-4 activates endothelial integrin α6β1. Circ Res. 2011;109:770–774. doi: 10.1161/CIRCRESAHA.111.247239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takemoto M, Hattori Y, Zhao H, Sato H, Tamada A, Sasaki S, Nakajima K, Yamamoto N. Laminar and areal expression of Unc5d and its role in cortical cell survival. Cereb Cortex. 2011;21(8):1925–1934. doi: 10.1093/cercor/bhq265. [DOI] [PubMed] [Google Scholar]
- 25.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 26.Öhman T, Lietzen N, Välimäki E, Melchjorsen J, Matikainen S, Nyman TA. Cytosolic RNA recognition pathway activates 14-3-3 protein mediated signaling and caspase-dependent disruption of cytokeratin network in human keratinocytes. J Proteome Res. 2010;9:1549–1564. doi: 10.1021/pr901040u. [DOI] [PubMed] [Google Scholar]
- 27.Chen P, Lepikhova T, Hu Y, Monni O, Hautaniemi S. Comprehensive exon array data processing method for quantitative analysis of alternative spliced variants. Nucleic Acids Res. 2011;39(18):e123. doi: 10.1093/nar/gkr513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovaska K, Laakso M, Haapa-Paananen S, Louhimo R, Chen P, Aittomaki V, Valo E, Nunez-Fontarnau J, Rantanen V, Karinen S, et al. Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme. Genome Med. 2010;2:65. doi: 10.1186/gm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo W, Pylayeva Y, Pepe A, Yoshioka T, Muller WJ, Inghirami G, Giancotti FG. β4 Integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 30.Kawataki T, Yamane T, Naganuma H, Rousselle P, Anduren I, Tryggvason K, Patarroyo M. Laminin isoforms and their integrin receptors in glioma cell migration and invasiveness: evidence for a role of α5-laminin(s) and α3β1 integrin. Exp Cell Res. 2007;313:3819–3831. doi: 10.1016/j.yexcr.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 31.Belot N, Rorive S, Doyen I, Lefranc F, Bruyneel E, Dedecker R, Micik S, Brotchi J, Decaestecker C, Salmon I, et al. Molecular characterization of cell substratum attachments in human glial tumors relates to prognostic features. Glia. 2001;36:375–390. doi: 10.1002/glia.1124. [DOI] [PubMed] [Google Scholar]
- 32.Hata K, Kaibuchi K, Inagaki S, Yamashita T. Unc5B associates with LARG to mediate the action of repulsive guidance molecule. J Cell Biol. 2009;184:737–750. doi: 10.1083/jcb.200807029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network, author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 35.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Giancotti FG. Integrin β4 signaling promotes tumor angiogenesis. Cancer Cell. 2004;6:471–483. doi: 10.1016/j.ccr.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 37.Shaw LM. Identification of insulin receptor substrate 1 (IRS-1) and IRS-2 as signaling intermediates in the α6β4 integrin-dependent activation of phosphoinositide 3-OH kinase and promotion of invasion. Mol Cell Biol. 2001;21:5082–5093. doi: 10.1128/MCB.21.15.5082-5093.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bon G, Folgiero V, Bossi G, Felicioni L, Marchetti A, Sacchi A, Falcioni R. Loss of β4 integrin subunit reduces the tumorigenicity of MCF7 mammary cells and causes apoptosis upon hormone deprivation. Clin Cancer Res. 2006;12:3280–3287. doi: 10.1158/1078-0432.CCR-05-2223. [DOI] [PubMed] [Google Scholar]
- 39.Chung J, Bachelder RE, Lipscomb EA, Shaw LM, Mercurio AM. Integrin (α6β4) regulation of eIF-4E activity and VEGF translation: a survival mechanism for carcinoma cells. J Cell Biol. 2002;158:165–174. doi: 10.1083/jcb.200112015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah OJ, Hunter T. Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 42.Yang YH, Szabat M, Bragagnini C, Kott K, Helgason CD, Hoffman BG, Johnson JD. Paracrine signalling loops in adult human and mouse pancreatic islets: netrins modulate β cell apoptosis signalling via dependence receptors. Diabetologia. 2011;54:828–842. doi: 10.1007/s00125-010-2012-5. [DOI] [PubMed] [Google Scholar]
- 43.Giancotti FG. Targeting integrin β4 for cancer and anti-angiogenic therapy. Trends Pharmacol Sci. 2007;28:506–511. doi: 10.1016/j.tips.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Previtali S, Quattrini A, Nemni R, Truci G, Ducati A, Wrabetz L, Canal N. α6β4 and α6β1 integrins in astrocytomas and other CNS tumors. J Neuropathol Exp Neurol. 1996;55:456–465. doi: 10.1097/00005072-199604000-00008. [DOI] [PubMed] [Google Scholar]
- 45.Shaw CE, Milner R, Compston AS, ffrench-Constant C. Analysis of integrin expression on oligodendrocytes during axo-glial interaction by using rat-mouse xenocultures. J Neurosci. 1996;16:1163–1172. doi: 10.1523/JNEUROSCI.16-03-01163.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gambaletta D, Marchetti A, Benedetti L, Mercurio AM, Sacchi A, Falcioni R. Cooperative signaling between α6β4 integrin and ErbB-2 receptor is required to promote phosphatidylinositol 3-kinase-dependent invasion. J Biol Chem. 2000;275:10604–10610. doi: 10.1074/jbc.275.14.10604. [DOI] [PubMed] [Google Scholar]
- 47.Mariotti A, Kedeshian PA, Dans M, Curatola AM, Gagnoux-Palacios L, Giancotti FG. EGF-R signaling through Fyn kinase disrupts the function of integrin α6β4 at hemidesmosomes: role in epithelial cell migration and carcinoma invasion. J Cell Biol. 2001;155:447–458. doi: 10.1083/jcb.200105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santoro MM, Gaudino G, Marchisio PC. The MSP receptor regulates α6β4 and α3β1 integrins via 14-3-3 proteins in keratinocyte migration. Dev Cell. 2003;5:257–271. doi: 10.1016/s1534-5807(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 49.Jauliac S, Lopez-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- 50.Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23:2411–2422. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura JL, Garcia E, Pieper RO. S6K1 plays a key role in glial transformation. Cancer Res. 2008;68:6516–6523. doi: 10.1158/0008-5472.CAN-07-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao RD, Mladek AC, Lamont JD, Goble JM, Erlichman C, James CD, Sarkaria JN. Disruption of parallel and converging signaling pathways contributes to the synergistic antitumor effects of simultaneous mTOR and EGFR inhibition in GBM cells. Neoplasia. 2005;7:921–929. doi: 10.1593/neo.05361. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Li W, You L, Cooper J, Schiavon G, Pepe-Caprio A, Zhou L, Ishii R, Giovannini M, Hanemann CO, Long SB, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen P, Lepikhova T, Hu Y, Monni O, Hautaniemi S. Comprehensive exon array data processing method for quantitative analysis of alternative spliced variants. Nucleic Acids Res. 2011;39(18):e123. doi: 10.1093/nar/gkr513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ovaska K, Laakso M, Haapa-Paananen S, Louhimo R, Chen P, Aittomäki V, Valo E, Nûñez-Fontarnau J, Rantanen V, Karinen S, et al. Large-scale data integration framework provides a comprehensive view on glioblastomamultiforme. Genome Med. 2010;2:65. doi: 10.1186/gm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.