Abstract
Drosha is a key enzyme in microRNA biogenesis, generating the precursor miRNA (pre-miRNA) by excising the stem-loop embedded in the primary transcripts (pri-miRNA). The specificity for the pri-miRNAs and determination of the cleavage site are provided by its binding partner DGCR8, which is necessary for efficient processing. The crucial Drosha domains for pri-miRNA cleavage are the middle part, the two enzymatic RNase III domains (RIIID), and the dsRNA binding domain (dsRBD) in the C-terminus. Here, we identify alternatively spliced transcripts in human melanoma and NT2 cell lines, encoding C-terminally truncated Drosha proteins lacking part of the RIIIDb and the entire dsRBD. Proteins generated from these alternative splice variants fail to bind to DGCR8 but still interact with Ewing sarcoma protein (EWS). In vitro as well as in vivo, the Drosha splice variants are deficient in pri-miRNA processing. However, the aberrant transcripts in melanoma cells do not consistently reduce mature miRNA levels compared with melanoma cell lines lacking those splice variants, possibly owing to their limited abundance. Our findings show that alternative processing-deficient Drosha splice variants exist in melanoma cells. In elevated amounts, these alternatively spliced transcripts could provide one potential mechanism accounting for the deregulation of miRNAs in cancer cells. On the basis of our results, the search for alternative inactive splice variants might be fruitful in different tumor entities to unravel the molecular basis of the previously observed decreased microRNA processing efficiency in cancer.
Introduction
MicroRNAs (miRNAs) are single-stranded noncoding RNAs of approximately 22 nucleotides (nt) involved in posttranscriptional gene regulation. Their mode of action is degradation or translational repression of target messenger RNAs (mRNAs) harboring complementary sequences [1,2]. Transcripts modulated by miRNAs are involved in a wide range of cellular functions. Consequently, dysregulation of miRNA biogenesis, and thus alteration in miRNA expression levels, contributes to multiple diseases, including various forms of cancer.
In cancer cells, aberrant expression of mature miRNAs has been frequently observed [3–9]. Deviations from the normal miRNA expression pattern could be caused, for example, by genomic deletion or mutation, epigenetic silencing, or aberrant transcription. The abundance of miRNA stabilizing factors could also potentially alter miRNA activity in cancer [10,11]. In some instances, for example, for mir-143 and mir-145, the precursor miRNAs (pri- and pre-miRNAs) do not differ in their quantities between normal and cancer cells, although mature miRNA levels are decreased [12]. Thus, a deficit in miRNA processing could be another alternative mechanism for a decrease in mature miRNA expression. Indeed, in some tumors, expression of key players of the miRNA biogenesis pathway is reduced or abrogated by genomic deletion and correlates with survival and mature miRNA levels [13–16]. Moreover, impairment of the miRNA biogenesis pathway by knockdown of pivotal processing factors favors tumor formation [17].
The maturation of miRNAs is a highly complex and regulated process, which is characterized by two cleavage reactions mediated by RNase III enzymes [18]. Hairpin structures of long primary transcripts (pri-miRNA) are first cleaved close to the base of the stem by the Microprocessor complex, consisting of Drosha and its binding partner DGCR8. This cropping liberates an approximately 60- to 70-nt-long hairpin structure, termed precursor miRNA (pre-miRNA) [19–22]. Whereas the N-terminus of Drosha is required for nuclear localization [23], the middle part of Drosha as well as the C-terminal part, harboring the two RNase III domains, RIIIDa and RIIIDb, and the dsRNA binding domain (dsRBD) are essential for pri-miRNA processing [22]. The two enzymatic domains form a processing center with two catalytic sites of RIIIDa and RIIIDb cleaving the 3′-arm and the complementary 5′-arm of the hairpin, respectively [22]. For the conversion of pri-miRNAs into pre-miRNAs, the interaction of Drosha with its cofactor DGCR8 is essential [21,22]. DGCR8 confers substrate specificity by recognizing the structural characteristics of the pri-miRNA and acts as a “molecular ruler” by determining the cleavage site within the pri-miRNA [21,24]. The basal segments, the stem as well as the terminal loop, are structural characteristics that play a role in cleavage site determination [24–26].
Although Drosha-mediated cleavage represents a critical processing step, it is not compulsory for the generation of all pre-miRNAs. Short introns that form hairpin structures resembling pre-miRNAs, termed mirtrons, can be spliced and debranched into pre-miRNA mimics, thereby omitting Drosha processing. The debranched hairpins are then exported, and further processed by the canonical miRNA biogenesis pathway [27–30].
After the initial cropping by the Microprocessor, the processing intermediate is exported from the nucleus by Exportin-5 in a Ran-dependent manner [25,31–33]. In the cytoplasm, Dicer removes the terminal loop in a second cleavage reaction, thereby producing the miRNA duplex of 21 to 24 nt length [34–38]. After incorporation of one selected single strand into the effector complex RISC (RNA-induced silencing complex) with an Argonaute protein [39,40], the mature miRNA causes silencing of its targets by base pairing mostly with the 3′-UTR mediating mRNA cleavage, translational repression, or mRNA degradation [1,2].
Here, we identify novel endogenous alternative splice variants of the important microRNA processing factor Drosha in melanoma cell lines and NT2 embryonal carcinoma cells. These transcripts encode proteins lacking critical functional domains, namely part of the 3′-end proximal RNase III domain and the dsRBD. These C-terminally truncated Drosha variants do not bind DGCR8 and hence also lack pri-miRNA processing activity. Although processing efficiency is impaired, the splice variants do not have a consistent major effect on the mature miRNA repertoire of the melanoma cells most likely because of the minor pool of alternative splice forms. However, the discovery of the processing-deficient Drosha variants can lead the way to a thorough search in other tumor entities where they could be more abundant and validated as one mechanism of miRNA deregulation in cancer.
Materials and Methods
Cell Lines
Cell lines have been established from skin or lymph node metastasis and were cultured as described elsewhere [41]. Patients were experiencing histologically confirmed melanoma of the skin, mucosa, or unknown primary and were classified as stage III or IV according to American Joint Committee on Cancer.
Plasmids
Wild-type Drosha was cloned into pcDNA3.1D/V5-His-TOPO as described previously [42]. The N-terminal Flag-tag was removed and replaced by a YFP-tag using HindIII and BamHI restriction sites. The BamHI site and the Drosha start codon were separated by the nucleotides acc. At the end of the Drosha coding sequence, a stop codon was introduced. RIIIDb splice variants were created by deleting exon 31, exons 31 and 32, or exons 30 to 32 from pcDNA3.1D/TOPO-YFP-Drosha-V5-His. The Drosha mutant E1045Q E1222Q was generated by site-directed mutagenesis of pcDNA3.1D/TOPO-YFP-Drosha-V5-His. HA-DGCR8 was created by deleting the Flag-tag of pFLAG/HA-DGCR8, which was kindly provided by Thomas Tuschl (Rockefeller University). The expression plasmid-encoding pri-mir-145 was described previously [43]. All constructs were verified by sequencing. Primer sequences are listed in Table W1. Plasmid sequences are available on request.
Reverse Transcription and Quantitative Polymerase Chain Reaction
Total or fractionated RNA was extracted using TRIzol (Invitrogen/Life Technologies, Carlsbad, CA) and treated with DNaseI (Roche, Mannheim, Germany). RNA was reverse transcribed using RevertAid H Minus Reverse Transcriptase (Fermentas, St Leon-Rot, Germany). To test mature miRNA levels, gene-specific RT was performed using stem-loop primers shown in Table W1 according to Chen et al. [44]. For the identification of Drosha splice variants, complementary DNA (cDNA) of different cell lines was used as a template to amplify exons 29 to 35 of Drosha. Polymerase chain reaction (PCR) products were visualized on a 1.2% agarose gel using ethidium bromide. Bands that ran lower than the expected wild-type band were purified from Ma-Mel-71, Ma-Mel-75, and NT2; cloned into pCRII-TOPO (Invitrogen) by TOPO-TA cloning; and sequenced.
Quantitative PCR was performed on an ABI StepOne Plus using 2x Power SybrGreen MasterMix (Applied Biosystems/Life Technologies). Primers are shown in Table W1.
Cellular Fractionation
Nuclear and cytoplasmic fractionation was basically performed according to [45] with minor changes. In brief, Ma-Mel-71 cells were scraped off with phosphate-buffered saline and pelleted at 300g for 5 minutes at 4°C. Pellets were resuspended in 1 ml of RSB (10 mM Tris, pH7.4,10 mM NaCl, 3 mM MgCl2), incubated for 3 minutes on ice and centrifuged at 1000g for 5 minutes at 4°C. The cell pellet was resuspended with four times its volume of RSBG40 (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 10% glycerol, 0.5% Nonidet P-40, 0.5 mM dithiothreitol), and 100 U/ml RNasin (Fermentas), and 10 mM ribonucleosid vanadyl complex (NEB) were used as RNA inhibitor. After incubating for 3 minutes on ice and centrifuging at 4500g for 3 minutes at 4°C, the supernatant was saved as the cytoplasmic fraction, and the nuclear pellet was resuspended in RSBG40 containing one-tenth volume of detergent (3.3% [wt/wt] sodium deoxycholate and 6.6% [vol/vol] Tween-20) and incubated for 5 minutes on ice. Nuclei were pelleted again at 4500g for 3 minutes at 4°C, washed with RSBG40, and collected at 9300g for 5 minutes. RNA from nuclear and cytoplasmic fraction was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions.
Cycloheximide Treatment
Ma-Mel-71 human melanoma cells were grown to 80% confluence and treated either with the solvent dimethyl sulfoxide or with 50 µg/ml cycloheximide (Sigma, St Louis, MO) for 20 minutes at 37°C as described previously [46]. The cells were then washed with phosphate-buffered saline and lysed in TRIzol (Invitrogen) for total RNA isolation, DNase I treatment, reverse transcription, and quantitative PCR as described previously.
Immunoprecipitation
HEK293 cells were transfected in 10-cm dishes with a total of 24 µg of N-terminally YFP-tagged Drosha, splice variants thereof, or empty vector with or without cotransfection of HA-DGCR8 using polyethylenimine. At 48 hours after transfection, cells were lysed in 500 µlofcoldlysis buffer(50 mM Tris-HCl, pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1x complete EDTA-free protease inhibitor cocktail [Roche]) for 20 minutes at 4°C under gentle shaking followed by centrifugation at 14,000g for 15 minutes at 4°C. A ratio of 1:20 of the supernatant was saved as input sample. The remaining lysates were incubated for 2 hours with 30 µl of slurry of the GFP-nanotrap [47] with constant rotation. Beads were pelleted at 400g for 2 minutes at 4°C and washed three times with cold lysis buffer and eluted with 100 µl of sample buffer (250 mM Tris-HCl, pH 6.8, 9.2% SDS, 40% glycerol, bromophenol blue) boiling for 2 minutes. Samples were analyzed by 7.5% SDS-PAGE and Western blot using the following primary antibodies: anti-Drosha (1:500, Drosha [D28B1] XP no. 3364; Cell Signaling Technology, Danvers, MA), anti-DGCR8 (1:300, 10996-1-AP; ProteinTechGroup, Inc, Chicago, IL), anti-EWS(1:1000, EWS [G-5], sc-28327; Santa Cruz Biotechnology, Inc, Santa Cruz, CA), antitubulin (1:1000, α/β-tubulin antibody no. 2148; Cell Signaling Technology), and anti-HA (1:500, Mono HA.11 MMS-101P; Covance, Münster, Germany).
In Vivo Processing Assay
HEK293 cells were reverse transfected with the Drosha-siRNA: 5′-GCAUGCAAGCGCGGAGUAU(dTdT) or scrambled small interfering RNA (siRNA) using Lipofectamine RNAiMAX in a six-well plate. After 24 hours, the cells were transfected each with 1.5 µg of plasmids encoding YFP-Drosha, HA-DGCR8, and pri-mir-145 to challenge the pri-miRNA processing factors. At 48 hours after plasmid transfection, RNA was isolated using TRIzol (Invitrogen), digested with DNaseI (Roche), and reverse transcribed. The samples were analyzed for pri-miRNA and endogenous Drosha levels by quantitative PCR.
In Vitro Processing Assay
HEK293 cells were reverse transfected with the Drosha-siRNA: 5′-GCAUGCAAGCGCGGAGUAU(dTdT) or scrambled siRNA using Lipofectamine RNAiMAX in a six-well plate. After 24 hours, the cells were transfected each with 1.5 µg of plasmids encoding YFP-Drosha and HA-DGCR8. Whole-cell extract was prepared in a buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM KCl by sonication, followed by centrifugation. The processing reaction (total, 35 µl) contained 40 µg of total protein, 0.5 mM ATP, 20 mM creatine phosphate, 4 mM MgCl2, 20 U RiboLock RNase Inhibitor (Fermentas), and 104 to 105 counts per minute of the [32P]-labeled transcript containing pri-15b∼16-2. The assay was incubated for 30 minutes at 37°C. The T7 template for the primary transcript was amplified by PCR (Table W1). The labeled transcript was gel purified, precipitated, and allowed to fold for 10 minutes at 60°C before usage (as described in Allegra and Mertens [48]).
Array
MiRNA expression profiling for melanoma cell lines was performed on Illumina DASL (cDNA-mediated Annealing, Selection, Extension, and Ligation) platform using Human miRNA Expression Profiling V2 Panel (Illumina, San Diego, CA). Briefly, 500 ng of total RNA was polyadenylated, reverse transcribed using a biotin-labeled oligo-dT primer with a universal sequence at the 5′-end, attached to a solid phase, and annealed to miRNA-specific oligonucleotides that comprised three parts: another 5′ universal PCR priming sequence, an address sequence for capturing the product on the array, and the 3′ miRNA-specific sequence. The miRNA-specific oligonucleotides were extended, eluted, and subjected to PCR fluorescent universal primers. Then, single-stranded PCR products were prepared and hybridized to capture the probes immobilized on beads in the arrays. Arrays were scanned by the BeadArray Reader and intensity per bead type (miRNA) was extracted. Data were quantile normalized using BeadStudio Data Analysis Software (Illumina).
From the normalized array data, the average signal intensities for all 858 validated mature miRNAs (as annotated in the microRNA registry) and for the mirtron-derived mature miRNAs miR-877 and miR-1224–1238 were calculated. The ratio of the Drosha-dependent mature miRNAs divided by the mirtron-derived, Drosha-independent mature miRNAs was used as a normalized measure for genome-wide Drosha-dependent processing in the cell lines. For the analysis, six cell lines with consistently detected splice variants in reverse transcription (RT)-PCR and quantitative PCR (Ma-Mel-71, Ma-Mel-128, Ma-Mel-20, Ma-Mel-36, Ma-Mel-75, and Ma-Mel-12) were compared to five cell lines with no significant signs of splice variant expression (Ma-Mel-53, Ma-Mel-73, Ma-Mel-26a, Ma-Mel-122, and Ma-Mel-8a).
Results
Drosha RIIIDb Splice Forms Exist in Several Cancer Cell Lines
To examine whether aberrant splicing of key miRNA processing factors could be a potential cause of decreased mature miRNA levels in cancer cells, we searched for functionally inactive Drosha splice variants in different sequence databases. We hypothesized that functional inactivity of Drosha would most likely require a loss of either the RNase or dsRNA binding activity, hence the loss of the RIIID or dsRBD domain. Therefore, we blasted the C-terminal region of Drosha including these three domains (NM_013235.4, nt 2985-4361) against several nucleotide databases to determine whether expressed sequence tags with aberrant splicing patterns had ever been identified.
This approach led to the identification of two Drosha transcripts lacking different parts of the functional region in melanoma (GenBank BC054003.1) and teratocarcinoma (AK308093).
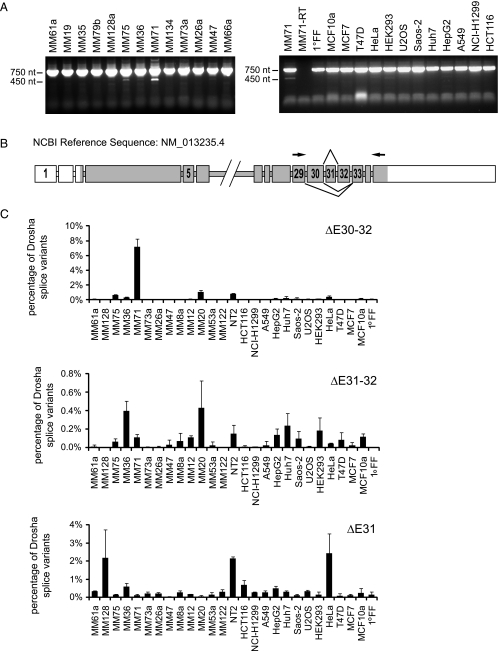
To validate whether these aberrant forms exist in tumor cells, we amplified Drosha's C-terminus encoding the RIIIDb and the dsRBD from cDNA of different cancer cell lines of diverse origins, several melanoma cell lines and NT2 cells. Amplification of the wild-type isoform with primers spanning exon 29 to 35 should yield a product of 759 base pairs (bp) and the alternatively spliced isoforms of BC054003.1 and AK308093 211 and 387 bp, respectively. The wild-type product was detected in every cell line tested (Figure 1A), but amplicons, which would reflect splicing events as in BC054003.1 and AK308093, were not produced, at least not in detectable amounts. However, additional weaker PCR products of unexpected size (approximately 450 bp) were observed in NT2 cells and the melanoma cell lines Ma-Mel-71 and Ma-Mel-75 (Figure 1A). Gel purification of these products, cloning, and subsequent sequence analysis revealed that these transcripts lack exon 31 (identified from NT2), exons 31 and 32 (identified from NT2 and Ma-Mel-71), or exon 30 to 32 (identified from Ma-Mel-71, Ma-Mel-75, and MaMel-20), which code for a part of the RIIIDb (Figure 1B).
Figure 1.
Identification of alternative Drosha splice variants in melanoma cells. (A) Identification of alternative Drosha splice variants by reverse transcription. Drosha cDNA was amplified from exon 29 to 35 by reverse transcription in several cancer cell lines (MM = Ma-Mel). PCR products were visualized on a 1.2% agarose gel using ethidum bromide. The upper band is derived from wild-type Drosha. The lower bands are derived from splice variants lacking exons in the amplified region. (B) Schematic representation of the Drosha gene and derived alternative spliced mRNAs. Exon skipping changes the reading frame resulting in premature stop codons. Protein coding exons are shown in dark gray, noncoding exons in white. Arrowheads indicate positions of primers used to amplify exon 29 to 35. Introns are not drawn to scale. (C) Expression levels of splice variants lacking exons 31, 31 and 32, and 30 to 32 in different cancer cell lines quantified by quantitative RT-PCR in relation to total Drosha levels. Total Drosha levels were obtained by totaling the 2-ΔCt values normalized to RPLP0. The sum was set as 100%. One representative data set is shown; error bars, SEM.
To quantitate the levels of these three alternatively spliced transcripts, we performed quantitative real-time PCR using a reverse primer in exon 35 and a forward primer specific for the individual splice forms. Splice variants ΔE31 and ΔE31–32 contribute to less than 3% of total Drosha levels in all cell lines tested, whereas ΔE30–32 accounts for approximately 10% of all Drosha transcripts in Ma-Mel-71 cells (Figures 1C and 2E). These cells also have already a comparably low level of wild-type Drosha (Figure W1).
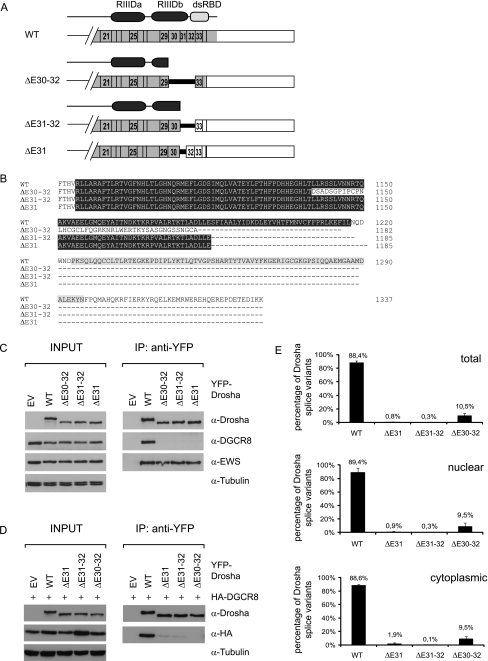
Figure 2.
Drosha splice variants do not bind DGCR8. (A) Schematic overview of Drosha splice variants and their protein products. The two conserved RNase III domains and the double-stranded RNA binding domain are indicated (RIIIDa, RIIIDb, dsRBD) according to the SMART program (http://smart.embl-heidelberg.de/). (B) Sequence alignment of Drosha wild-type and alternative splice variants. Amino acids coding for the RIIIDb are highlighted in dark gray, amino acids coding for the dsRBD in light gray. (C, D) Novel alternative splice variants do not bind DGCR8. YFP-tagged WT and alternatively spliced Drosha were overexpressed in HEK293 cells without (C) or with (D) coexpression of HA-DGCR8. Cells were lysed 48 hours after transfection, and Drosha was pulled down using the GFP-nanotrap. The eluate was analyzed by SDS-PAGE and Western blot analysis using anti-Drosha, anti-DGCR8, anti-HA, anti-EWS, and anti-tubulin antibodies. Tubulin served as loading control. EV indicates empty vector; EWS, Ewing sarcoma protein; WT, wild-type. (E) Ma-Mel-71 shows the same percentage of Drosha ΔE30–32 splice variants expression in total, cytoplasmic, and nuclear fractions as determined by quantitative RT-PCR. Total Drosha levels were obtained by totaling the 2-ΔCt values normalized to RPLP0. The sum was set as 100%. n = 3. Error bars, SEM.
Drosha RIIIDb Splice Variants Yield C-terminally Truncated Proteins, Which Do Not Bind to DGCR8
All of the three aberrant Drosha splice variants contain premature stop codons, encoding C-terminally truncated Drosha proteins. The lack of E31 or E31–32 creates a stop codon of the last two nucleotides of exon 30 and the first nucleotide of E32 or E33, respectively. Thus, ΔE31 and ΔE31–32 create the same protein lacking part of the RIIIDb and the dsRBD, ending with Glu1222 (E110b), the residue responsible for cutting the 5′ arm of the pri-miRNA hairpin [22]. Deletion of E30–32 creates a frameshift leading to a sequence of 44 amino acids different from the wild-type before creation of a premature stop codon. The encoded protein lacks more than half of the RIIIDb and the dsRBD (Figure 2, A and B).
Processing of pri-miRNA into pre-miRNA not only requires the RNase III Drosha but also necessitates the cofactor DGCR8 [21,22], which interacts with the middle region as well as the RIIIDs of Drosha [22]. Because the alternative splice variants lack part of RIIIDb, we tested whether they are still able to bind DGCR8. To this end, we transfected HEK293 cells with plasmids encoding N-terminally YFP-tagged Drosha proteins with wild-type or alternatively spliced sequences. The expression levels of the spliced forms are similar to that of wild-type protein (Figure 2C, left panel). On YFP pull-down, endogenous (Figure 2C) or ectopic (Figure 2D) DGCR8 coprecipitated with wild-type Drosha but not with the three alternatively spliced isoforms. On the contrary, another interaction partner of Drosha, the Ewing sarcoma protein [21], was still associated with the RIIIDb splice variants to the same extent as with the wild-type (Figure 2C). Hence, lack of the dsRBD and part of the RIIIDb results in loss of DGCR8 binding but has no impact on interaction with EWS.
Drosha RIIIDb Splice Variants Exist in the Cytoplasm
Alternative splicing of exon E31, E31–32 or E30–32 generates premature termination codons, which could potentially make the variant transcripts prone to nonsense-mediated decay (NMD). To find out, whether Drosha RIIIDb alternatively spliced transcripts are degraded by NMD or whether they are stable, we examined mRNA levels of wild-type Drosha and the splice variants in nuclear and cytoplasmic fractions of Ma-Mel-71 cells. To verify the purity of the fractions, we monitored pre-45S rRNA and the tRNALys, which showed a clear nuclear and cytoplasmic enrichment, respectively (data not shown). The distribution of wild-type and alternatively spliced Drosha transcripts was consistent in total, nuclear, and cytoplasmic fractions (Figure 2E). Hence, the cytoplasmic stability of wild-type and alternatively spliced Drosha transcripts seems to be comparable, suggesting that the novel splice forms are not degraded by NMD. To formally prove that Drosha ΔE30–32 is not subject to NMD, we treated Ma-Mel-71 cells with cycloheximide, which inhibits NMD by blocking translation [46]. Quantification of wild-type and alternatively spliced Drosha revealed that cycloheximide treatment did not increase the expression of the alternatively spliced isoforms indicating that the isoform is not subject to NMD (Figure W2).
Drosha RIIIDb Splice Variants Are Deficient in pri-miRNA Processing Activity
Drosha processing requires DGCR8 binding and two intact RIIIDs [21,22]. To determine whether Drosha proteins generated from RIIIDb alternative transcripts are catalytically inactive, we tested their ability to process pri-miRNA in vitro and in vivo.
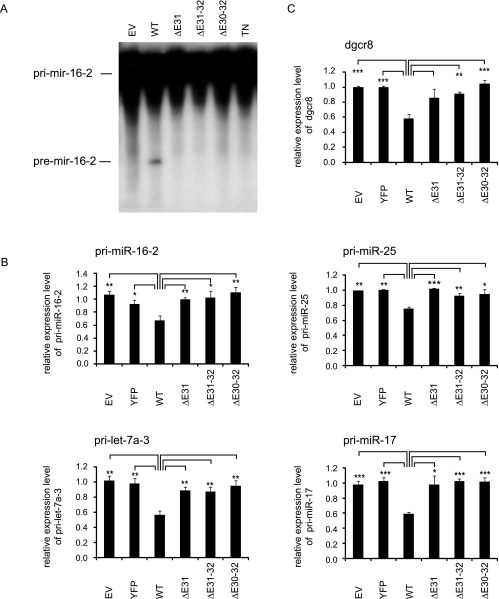
For the in vitro processing assay, lysates of cells overexpressing DGCR8 and wild-type or mutant Drosha constructs were incubated with radioactively labeled pri-miR-16-2. As negative controls, we used cells transfected with empty vector or a transdominant-negative, processing-deficient Drosha mutant (TN-Drosha) containing point mutations in both RNase III domains (E1045Q E1222Q) [49]. Only lysate containing wild-type Drosha was capable of generating pre-mir-16-2, whereas the three Drosha splice variants as well as empty vector and TN-Drosha were unable to execute pri-miRNA processing (Figure 3A).
Figure 3.
Drosha splice variants are defective in pri-miRNA processing activity. HEK293 cells depleted of endogenous Drosha using an siRNA against the 3′UTR of Drosha were transfected with HA-DGCR8 and siRNA-resistant Drosha constructs as indicated. EV indicates empty vector; WT, wild-type. (A) The cell lysates were incubated with labeled pri-mir-16-2 to assay pri-miRNA processing activity in vitro. (B, C) Quantification of several pri-miRNA levels (B) and endogenous DGCR8 mRNA levels (C) by quantitative RT-PCR normalized to cyclophilin mRNA levels. n = 3. Error bars, SEM. Asterisks denote statistically significant differences: * P < .05, ** P < .01, *** P < .001 (unpaired t test).
To confirm this result in vivo, we determined pri-miRNA levels by quantitative PCR on endogenous Drosha knockdown and subsequent overexpression of DGCR8 and wild-type or alternatively spliced Drosha. The siRNA was designed to target the 3′-UTR of the endogenous Drosha mRNA, so that only the endogenous not the ectopically expressed Drosha was silenced. Knockdown efficiency was monitored both at the mRNA and protein levels compared with control siRNA-treated cells by quantitative PCR and Western blot. Endogenous Drosha mRNA levels dropped to less than 60% (Figure W3A), and a much more potent down-regulation was observed at protein level (Figure W3B). Ectopic Drosha constructs were not affected by the siRNA (Figure W3B). On Drosha knockdown, we observed that pri-miRNAs accumulated (data not shown).
Expression of wild-type Drosha significantly diminished pri-miRNA levels compared with cells transfected with empty vector or YFP. In accordance with the in vitro assay results, the Drosha splice variants did not exhibit processing activity because no change in pri-miRNA levels was observed (Figure 3B). Also, the DGCR8 mRNA, which is also a substrate for Drosha cleavage [50–52], was only processed by wild-type Drosha but not by the splice variants (Figure 3C).
Upon overexpression of TN-Drosha, pri-miRNA levels accumulated (Figure W3C) as described previously [50]. To take into consideration that Drosha splice variants could be marginally less expressed than wild-type Drosha, we transfected cells with only half of the amount of the wild-type Drosha construct (WT 12). This resulted in a comparable expression of Drosha wild-type and splice variants at the protein level (Figure W3B). Nevertheless, pri-miRNA processing efficiency closely resembles that of cells transfected with the full amount of wild-type Drosha (Figure W3C). Thus, the difference in pri-miRNA expression (Figure 3, B and C) was due to a difference in processing activity in vivo, but not due to a potential difference in expression of the splice variants compared with wild-type Drosha.
Melanoma Cell Lines with Alternatively Spliced Drosha Variants Do Not Show Generally Decreased Mature MiRNA Levels
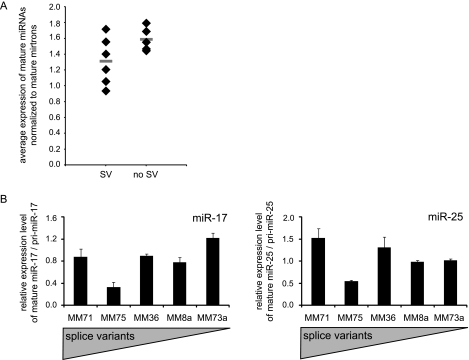
To test whether the Drosha RIIIDb splice variants affected mature miRNA levels, we determined the expression of 858 mature miRNAs using a microarray platform in eleven melanoma cell lines which consistently showed Drosha splice variants (n = 6) or no sign of splice variants (n = 5) in RT-PCR and quantitative PCR. For normalization purposes, we used average levels of mature mirtrons, which are derived from Drosha-independent substrates as they are generated by splicing [27,28]. Indeed, the average mature miRNA levels of cell lines expressing Drosha splice variants were lower than in the control cell lines, but this strong trend was not significant (P = .096, t test). On the contrary, the cell line with the highest level of splice variants, Ma-Mel-71, exhibited the highest mature miRNA levels within the group with splice variants (Figure 4A). Thus, these results were ambivalent with respect to the impact of Drosha splice variants on mature miRNA expression: Whereas a trend indicates decreased mature miRNA abundance coinciding with low-abundance Drosha splice variants, the cell line with the by-far highest level of processing deficient splicing variants did not follow this pattern.
Figure 4.
Cell lines with low levels of Drosha splice variants do not show consistently reduced miRNA levels. (A) Average mature miRNA expression levels from microarrays normalized to average mature mirtron levels. Cell lines with alternative splicing events at the C-terminus (SV) versus cell lines without detectable splicing events within exon 29 to 35 (no SV). The horizontal bars represent average values in the respective groups. (B) Expression levels of mature miRNAs, miR-25 and mir-17, normalized to the respective pri-miRNAs in different melanoma cell lines. The average miRNA level of Ma-Mel-8a and Ma-Mel-73a was set as 1. Bars indicate average expression; error bars, SEM (n =3).
Further, we investigated the levels of individual mature miRNAs, miR-17 and miR-25, relative to their respective pri-miRNA expression in cell lines expressing RIIIDb splice variants (Ma-Mel-71, Ma-Mel-75, and Ma-Mel-36) and cell lines with no detectable Drosha splice variants (Ma-Mel-8a and Ma-Mel-73a) by quantitative PCR. For both miRNAs, no significant correlation between the amount of splice variants and mature miRNA levels could be observed (Figure 4B).
Overall, these results suggest that, although alternatively spliced Drosha variants are pri-miRNA processing deficient, the amount in the tested cell lines might not be sufficient to consistently affect mature miRNA production.
Discussion
At least 30% of all human genes are predicted to be controlled by miRNAs [53]. Because many targets are involved in key cellular processes, deregulation of miRNA biogenesis and finally aberrant miRNA levels are associated with a variety of diseases, particularly cancer. Although several miRNAs are overexpressed in cancer cells, most miRNAs shows reduced expression in human tumors [3,4,8]; reviewed in references [54–56]. This phenotype could be attributed to altered levels of functional processing factors, which we hypothesized could be caused by aberrant alternative splicing. Our analyses focused on the major nuclear pri-miRNA processing enzyme Drosha, which is essential for the production of most mature miRNAs except the mirtrons. Moreover, other key factors of the miRNA biogenesis pathway might be subject to alternative splicing, as was previously shown for Dicer in neuroblastoma cell lines [57].
In this study, we identify novel alternative splice variants of the human Drosha gene in melanoma cell lines, which differ in their coding sequence and encode functionally inactive Drosha proteins. Skipping of exon E31, E31–32, or E30–32 changes the reading frame, leading to a premature stop codon.
Transcripts containing a stop codon that prematurely terminates translation can be eliminated by a quality mechanism, termed nonsense-mediated decay, to avoid the generation of truncated proteins that could have dominant-negative or otherwise detrimental effects. This process itself is interestingly also regulated by microRNAs [58,59]. Stop codons are sensed as premature and elicit NMD, for example, when they are followed by an exon-exon junction located more than 50 to 55 nt downstream [60]. Drosha ΔE31 and Drosha ΔE31–32 splice variants fulfill this criterion, as the premature stop codon is followed by three or two exon-exon junctions, respectively, more than 55 nt downstream, and hence, they might be targeted for degradation by the NMD machinery. In contrast, removal of E30–32 creates a premature stop codon only at the last but one exon, exon 34, 5 nt upstream of the last exonexon boundary. The most abundant transcript variant, Drosha ΔE30–32, should thus escape NMD and be translated into a truncated Drosha protein. The fact that the splice variants exist in cytoplasmic fractions of Ma-Mel-71 cells to the same extent as in total lysates argues in favor of transcripts that are stable and translated rather than rapidly degraded by the cytoplasmic NMD machinery. In addition, expression of the truncated variants from plasmids leads to approximately the same level of protein as expression of wild-type Drosha. Hence, the truncation does not compromise the structure in a way that leads to degradation of the protein. Lastly, inhibiting NMD using cycloheximide in Ma-Mel-71 cells did not alter the abundance of Drosha ΔE30–32 proving that it is not subject to NMD. Summing up, at least the most abundant splice variant Drosha ΔE30–32 is most likely translated into a truncated Drosha protein.
The encoded truncated proteins are lacking the dsRBD and part of the RIIIDb. Previous studies have shown that the dsRBD as well as two intact RIIIDs are required for cropping of the pri-miRNA [22]. The two enzymatic RIIIDs form a single processing center with two catalytic sites by intramolecular dimerization [22]. In addition, the interaction of DGCR8 with Drosha is needed for efficient cleavage [21,22] because Drosha alone does not exhibit pri-miRNA-binding activity [61]. We demonstrated that Drosha RIIID splice variants do not bind DGCR8, showing that two complete RIIIDs are required for DGCR8 binding. Consistently, Drosha splice forms are deficient in pri-miRNA processing in vitro as well as in vivo.
Although the ectopic isoforms did not exhibit DGCR8 binding or pri-miRNA processing activity, there was no consistent impact of the endogenous alternatively spliced Drosha transcripts in melanoma cell lines on mature miRNA production. Expression profiling indicates decreased miRNA expression associated with the Drosha splice variants, but paradoxically, Ma-Mel-71 cells, which exhibit the highest amounts of RIIID spliced transcripts among the cell lines tested, show high mature-to-pri-miRNA ratios, that is, good pri-miRNA processing. It could be possible that another factor involved in miRNA biogenesis might be altered in Ma-Mel-71 cells, which could overshadow the effect of the Drosha splice variants.
Given the importance of Drosha as a key player in miRNA biogenesis, it is most likely that sufficient amounts of alternatively spliced Drosha transcripts show a more pronounced effect on miRNA maturation. This could be envisioned in two ways: either the truncated transcripts are translated into nonfunctional proteins that execute dominant-negative functions or, in case the transcripts are degraded, wild-type RNA levels would be reduced resulting in limiting amounts of wild-type Drosha transcripts and thus less functional Drosha protein. As shown before, the artificial decrease in Drosha expression can lead to decreased miRNA activity and enhanced tumorigenesis [17]. In addition, a decrease of Drosha mRNA and protein in ovarian cancer [62] as well as neuroblastoma has been associated with a global miRNA down-regulation [63] corroborating the significance of Drosha regulation in cancer. Thus, our data suggest that the search for more strongly expressed Drosha splice variants might be fruitful in other tumor entities and might provide one way to solve the many open questions about miRNA deregulation and processing defects in human cancer.
In summary, we have discovered rare splice variants of the important miRNA biogenesis factor Drosha for the first time. In melanoma, these splice variants lack parts of one RNase domain as well as the dsRBD. They thus do not bind DGCR8 and are completely deficient in miRNA processing in vivo and in vitro. However, the variants lack a consistent effect on endogenous miRNA expression in melanoma cell lines. Thus, this study shows that processing-deficient splice variants of important processing factors exist and could impact the miRNA expression patterns found in human tumors. For the future, it will be of great interest to search for tumor-enriched processing-deficient splice variants in the miRNA biogenesis pathway to understand the mechanisms underlying the frequently observed deregulation of mature miRNAs in cancer beyond the transcriptional level.
Supplementary Material
Acknowledgments
The authors thank all members of the Diederichs laboratory for helpful discussions and Daniel Mertens and Danilo Allegra (University of Ulm) for support with the in vitro processing assays. The authors also thank Tom Tuschl (Rockefeller University) for providing expression plasmids.
Abbreviations
- RIIID
RNase III domain
- dsRBD
dsRNA binding domain
- miRNA
microRNA
Footnotes
Our research is supported by the Helmholtz Society (VH-NG-504), the DKFZ-MOST-Cooperation Program (Ca-135), the Marie Curie Program of the European Union (239308) and the German Research Foundation (TRR77 TP B03). The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 5.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 8.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 9.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 10.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 11.Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci USA. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 13.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koesters R, Adams V, Betts D, Moos R, Schmid M, Siermann A, Hassam S, Weitz S, Lichter P, Heitz PU, et al. Human eukaryotic initiation factor EIF2C1 gene: cDNA sequence, genomic organization, localization to chromosomal bands 1p34–p35, and expression. Genomics. 1999;61:210–218. doi: 10.1006/geno.1999.5951. [DOI] [PubMed] [Google Scholar]
- 15.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 18.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 20.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 21.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 22.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The DroshaDGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X, Zhang Y, Tucker L, Ramratnam B. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010;38:6610–6619. doi: 10.1093/nar/gkq547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 27.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westholm JO, Lai EC. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011;93:1897–1904. doi: 10.1016/j.biochi.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a R an GTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 35.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 36.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 37.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 41.Ugurel S, Thirumaran RK, Bloethner S, Gast A, Sucker A, Mueller-Berghaus J, Rittgen W, Hemminki K, Becker JC, Kumar R, et al. B-RAF and N-RAS mutations are preserved during short time in vitro propagation and differentially impact prognosis. PLoS One. 2007;2:e236. doi: 10.1371/journal.pone.0000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 43.Diederichs S, Haber DA. Sequence variations of microRNAs in human cancer: alterations in predicted secondary structure do not affect processing. Cancer Res. 2006;66:6097–6104. doi: 10.1158/0008-5472.CAN-06-0537. [DOI] [PubMed] [Google Scholar]
- 44.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39:356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF auto-regulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Allegra D, Mertens D. In-vivo quantification of primary microRNA processing by Drosha with a luciferase based system. Biochem Biophys Res Commun. 2011;406:501–505. doi: 10.1016/j.bbrc.2011.02.055. [DOI] [PubMed] [Google Scholar]
- 49.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 50.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Triboulet R, Chang HM, Lapierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT, II, Nelson S, Rosbash M. Genome-wide identification of targets of the Drosha-Pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 54.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 57.Potenza N, Papa U, Scaruffi P, Mosca N, Tonini GP, Russo A. A novel splice variant of the human dicer gene is expressed in neuroblastoma cells. FEBS Lett. 2010;584:3452–3457. doi: 10.1016/j.febslet.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 58.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choe J, Cho H, Lee HC, Kim YK. MicroRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO Rep. 2010;11:380–386. doi: 10.1038/embor.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 61.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, London WB, Chang CH, Yu AL. MicroRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.