Abstract
The complexity of the process of metastasis is widely recognized. We report herein on a recurrent feature of high compared to low metastatic cells that is linked to their ability to survive early after their arrival at secondary sites. Using novel fluorescent-based imaging strategies that assess tumor cell interaction with the lung microenvironment, we have determined that most high and low metastatic cells can be distinguished within 6 hours of their arrival in the lung and further that this difference is defined by the ability of high metastatic cells to resist apoptosis at the secondary site. Despite the complexity of the metastatic cascade, the performance of cells during this critical window is highly defining of their metastatic proclivity. To explore mechanisms, we next evaluated biochemical pathways that may be linked to this survival phenotype in highly metastatic cells. Interestingly, we found no association between the Akt survival pathway and this metastatic phenotype. Of all pathways examined, only protein kinase C (PKC) activation was significantly linked to survival of highly metastatic cells. These data provide a conceptual understanding of a defining difference between high and low metastatic cells. The connection to PKC activation may provide a biologic rationale for the use of PKC inhibition in the prevention of metastatic progression.
Introduction
For many solid tumors, a common site of metastatic progression is the lung. Metastasis includes a complex set of steps that involves interaction between the tumor and the tumor microenvironment [1,2]. An area of considerable interest has been the identification of genes or proteins that contribute to or inhibit metastatic progression. High-throughput genomic and proteomic approaches have fueled the investigative pipeline with many candidate genes and proteins that may promote or inhibit metastasis [3–7]. It has been difficult to distinguish those candidates that are markers of metastatic progression from those that are causally linked to metastasis. One approach to understand these numerous candidates in a systemwide manner has been to identify pathways that may connect various candidates. This approach has the appeal of identifying points of convergence among various candidates that may be critical to a complex biologic process like metastasis. An alternative, non-mutually exclusive, and functionally driven approach has been to identify critical cellular processes or steps that most define the metastatic phenotype of cancers. In such an approach, it is understood that all biologic steps that are linked to metastatic progression are necessary; however, some of these steps may be readily achieved by both high and low metastatic cells. The corollary suggests that some steps are more defining of the metastatic phenotype and, as such, may be most valuable to target therapeutically. If a functional hierarchy of the steps of the metastatic cascade were available, an opportunity would exist to focus on those genes/proteins that are functionally related to higher-priority steps in metastasis. Such an approach would provide a basis to study of genes/proteins most critical to metastasis biology and therapy.
Progress toward such a goal of dissecting the steps in the metastatic cascade has been made possible using novel single-cell imaging strategies. Indeed, Chambers et al. [8] have evaluated discrete steps in the metastatic cascade using intravital imaging of metastatic cancer cells as they arrived in the liver. More recent studies in brain and in other tissues have allowed a similar understanding of critical steps in the metastatic cascade [9–11]. Such an understanding of the pulmonary metastatic phenotype is still lacking [12]. To help understand the biology of metastasis to the lung, we previously described a fluorescent imaging approach, single-cell videomicroscopy (SCVM) [13]. We first used SCVM to ask how the cytoskeleton linker protein, ezrin, contributed to metastasis of pediatric osteosarcoma and rhabdomyosarcoma. These studies indicated that ezrin played a critical role in metastasis within hours of the arrival of metastatic cells in the lung [13]. Since then, our experience with SCVM has expanded beyond the study of ezrin alone. In a series of unrelated publications, focused on sarcoma and breast cancer metastasis, we discovered that our findings with ezrin were not unique. Indeed, despite a diversity of species (i.e., mouse and human cells) and a variety of candidate proteins (both metastasis suppressors and metastasis associated), nearly all high compared to low metastatic cells could be distinguished within 6 hours of their entry to the lung. Collectively, these data suggested that the events occurring early after metastatic cells arrived in the lung are especially difficult for cancer cells to overcome and seemed to repeatedly distinguish high from low metastatic cells. It is reasonable that the study of these events may provide a context to prioritize future consideration of putative metastasis-associated genes related to both biology and therapy.
Using pairs of human and murine high and low metastatic breast cancer and sarcoma cells, two cancer types with a proclivity for metastasis to the lung, we now report on the specific cellular process that are most defining of metastasis and present data on the biochemical pathways linked to this phenotype. Using SCVM and a more recently reported ex vivo pulmonary metastasis assay (PuMA [14]) in which the metastatic progression of GFP-expressing cancer cells, from a single cell to the formation of multicellular colonies, in the mouse lung micro-environment can be assessed in real time for up to 30 days, we determined that a distinguishing feature was the ability of highly metastatic cells to avoid apoptosis within hours of their arrival in the lung. It is interesting to note that this resistance against apoptosis, disproportionately seen in highly metastatic cells, was not evident outside the context of metastasis because the high and low metastatic pairs were not distinguishable by growth or proliferation in vitro. The connection between apoptosis resistance and the cancer phenotype has been previously recognized [12,15–17]. Our data now provide a metastasis-specific context in which to consider the importance of apoptosis resistance as part of the metastatic phenotype. This priority on resistance to apoptosis early after arrival of cells in the lung now supports examination of molecular pathways causally linked to the drivers of the metastasis biology. We were surprised to find no association between the classic Akt survival pathway and this metastatic phenotype. Indeed, of all the pathways examined, only protein kinase C (PKC) activation was significantly linked to survival of highly metastatic breast cancer and sarcoma cells within hours of their arrival in the lung. The unique dependence on PKC activation was confirmed in preclinical in vivo studies of pulmonary metastasis using inhibitors of classic PKC isoforms.
Materials and Methods
Cell Lines and Media
The characterization and behavior of all the clonally related high and low metastatic cell lines has been previously reported as indicated below: murine osteosarcoma cells (high metastatic [met], K7M2; low met, K7M2 AS ezrin 1.46 [AS1.46] and 1.52 [AS1.52]) [13,18]; the human osteosarcoma cell lines (high met, HOS-MNNG [MNNG]; low met, HOS) [19]; the human soft tissue sarcoma cell lines (high met, HT1080 sh-control; low met, HT1080 sh-gp78), provided by Dr Allan M. Weissman (National Cancer Institute, Frederick, MD) [20]; human mammary cancer cell lines (high met, H1-177 EDG2; low met, H1-177), provided by Dr Patricia S. Steeg (National Cancer Institute, Frederick, MD)[21]; the murine osteosarcoma cell lines (high met, K7M3/FDN; low met, K7M3/neo), provided by Dr Eugenie S. Kleinerman (The University of Texas MD Anderson Cancer Center, Houston, TX) [22]; and the murine breast cancer cell lines (high met, 4T1; low met, 67NR), provided by Dr Lalage M. Wakefield (National Cancer Institute, Bethesda, MD) [23]. The human breast cancer cells were also provided by Dr Wakefield MDA-MB-231 All cell lines were maintained in vitro using Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA) culture medium containing 10% fetal bovine serum, l-glutamine (2 mmol), penicillin (100 U/ml), and streptomycin (100 mg/L; Invitrogen) at 37°C in a humidified 5% CO2 incubator.
Cell lines that expressed empty vector (EV AS1.46 and EV AS1.52) and activated Akt AS1.46 (act Akt AS1.46) were maintained in 6.7 µg/ml of puromycin (Sigma, St Louis, MO). Activated Akt AS1.52 (actAkt AS1.52) was maintained in 10 µg/ml of puromycin (Sigma). PT67 packaging cells (ATCC) were used for retroviral transfection according to the manufacturer's instruction. For all in vitro and in vivo assays, cells were harvested using Trypsin/Versene (Invitrogen) from 70% confluent cultures. Cell viability was assessed using trypan blue and all cell lines used were from less than 20 passages. All cells were verified mycoplasma free.
Retroviral Activated Akt Gene Infection
The murine stem cell virus (pMSCVpuro) (BD Biosciences, Palo Alto, CA) was used for the construction of the retrovirus-encoding activated Akt gene. Activated Akt DNA was amplified by standard polymerase chain reaction technique using activated Akt1 in pUSEamp complementary DNA (Upstate, Lake Placid, NY) as the template. The synthesized DNA was digested with XhoI and BglII enzymes, purified, and cloned to the pMSCVpuro vector. Packaging cells, PT67, were transfected with Lipofectamine 2000 (Invtrogen) transfection reagent following the manufacturer's protocol. Transfected cell lines were established by selection with 2.5 µg/ml of puromycin for 2 weeks, and single-cell clones were selected for determination of the virus titer. The virus titers were evaluated by culture with NIH3T3 cells. For infection, AS1.46 and AS1.52 cells were cultured with retrovirus containing PT67 cell culture supernatant in the presence of polybrane (4 mg/ml; Sigma).
In Vivo/Ex Vivo SCVM Imaging [13]
Cell lines were transduced with pSICO-eGFP or p960-X1-685eGFP or fluorescently labeled using 5 µM 5-chloromethylfluorescein diacetate (CMFDA; Invitrogen) according to the manufacturer's recommendation. At 1 and 6 hours, after tail vein injection of 2.0 x 105 cells, mice were euthanized by CO2 inhalation. Lungs were inflated using slow intratracheal injection of PBS and then imaged by inverted fluorescent videomicroscopy (Leica DM IRB, Buffalo Grove, IL) at a magnification of x100. Ten random frames were captured and analyzed using OpenLab software (Improvision, Waltham, MA) to define and count fluorescent (green) CMFDA tumor events larger than 10 pixels. The total numbers of events per lung per mouse were summarized as a mean of five mice per time point [13]. Animal care and use were in accordance with the guidelines of the National Institutes of Health's Animal Care and Use Committee.
Ex Vivo Terminal Transferase dUTP Nick End Labeling and In Vivo Apoptosis Detection Assay
Using 15-µm paraffin sections made from whole lung 1, 3, and 6 hours after CMFDA-or GFP-labeled tumor cell injection, terminal transferase dUTP nick end labeling (TUNEL) assay was performed using the in situ Cell Death Detection Kit, TMR red (Roche, Penzberg, Germany). Antigen retrieval solution (Dako, Carpinteria, CA) was preheated by microwave oven, and slides were incubated with the heating solution 10 minutes. The whole lung was examined using fluorescence microscopy to enumerate double-positive green/red apoptotic tumor cells and Zeiss LMS 510 NLO confocal microscopy system (Zeiss, Thornwood, NY). Apoptotic tumor cells were also imaged using fluorescent detection of activated caspase 3 (FLIVO; Immunohistochemistry Technologies, LLC, Bloomington, MN). One hour after tumor cell injection, whole lung fluorescence microscopy quantified double-positive green/red tumor cells experiencing caspase cleavage.
Western Blot Analysis and Kinase Assay
Cells were lysed in either SDS or RIPA buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 0.1% SDS, 0.5% deoxycholate, 1% NP-40) with proteinase inhibitor cocktail (Roche). Protein lysates (20–40 µg/lane), as determined by Bio-Rad protein assay, were separated in 4% to 20% or 6% SDS-PAGE followed by electrophoretic transfer to a nitrocellulose membrane (Invitrogen). The membranes were blocked with 5% nonfat dried milk in TBS-Tween 20 (20 mM Tris-HCl, pH 7.5, 8 g/L sodium chloride, 0.1% Tween 20) and incubated with primary antibody: 1:1000, anti-phospho-Akt (Ser473), anti-Akt, anti-p70 S6 kinase, anti-phospho-p70 S6 kinase, anti-ezrin/radixin/moesin (ERM), anti-phospho-ERM, anti-FAK, anti-phospho-FAK, antip44/42, anti-phospho-p44/42 (Cell Signaling, Beverly, MA); or 1:10,000, anti-β-actin (Sigma) overnight at 4°C. SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL) was used for detection of antibody. For Akt kinase, activity was assessed using phosphorylation of Akt substrate, glycogen synthase kinase-3, according to the manufacturer's instructions (Cell Signaling).
In Vivo/Ex Vivo PuMA [14]
GFP-positive tumor cells (K7M2, AS1.46, AS1.52, 4T1, and MDA-MB-231) (2 x 105) were delivered by tail vein injection to either female Balb/c (Taconic, Hudson, NY) or SCID/Beige (Charles Rivers, Wilmington, MA). Within 15 minutes of tumor cell injection, mice were euthanized with CO2 inhalation, and lungs were injected with a mixed agarose/medium solution. Lung sections for PuMA, generated as described [14], were incubated at 37°C in humidified conditions of 5% CO2. Pathway inhibitor doses that did not inhibit cell proliferation in vitro but did modulate intended targets were selected: 1 µM Akt inhibitor VIII (EMD Chemicals, Inc, Gibbstown, NJ), a PH domain-dependent inhibitor of Akt1/Akt2, 5 µMPI3 kinase activator (740 Y-P; Tocris Bioscience, Ellisville, MO), 25 nM mTOR inhibitor (Torin1; kind gift from Dr Nathanael S. Gray, Dana-Farber Cancer Institute and David M. Sabatini, Whitehead Institute/MIT/HHMI), PKC inhibitor (50 nM UCN-01 [EMD Chemicals, Inc] or 5 µM Ro31-8220 [Axxora Life Science, Inc, San Diego, CA]), 1 µM Src inhibitor (AZD0530/saracatinib; AstraZeneca Pharmaceuticals LP, Wilmington, DE), 10 µM MAPK inhibitor (EMD Chemicals, Inc), or DMSO was added to medium and changed every other day. PuMA images were captured and analyzed as previously described [14]. The inhibitor or activator index was calculated by dividing the tumor burden (area of fluorescence), in the inhibitor-or activator-treated lung section, by that of the control-treated lung section.
In Vivo Orthotopic Primary Tumor Growth and Experimental Metastasis Assays
Two million K7M2 murine osteosarcoma cells were injected into an orthotopic paraosseous location, adjacent to the left proximal tibia, in 4-week-old female Balb/c mice (Charles River, Wilmington, MA). After primary tumors reached 1 cm size, mice were treated with UCN-01 (7 mg/kg per day) or vehicle intraperitoneally, 5 days on and 2 days off. Longest tumor dimension was monitored by caliper every other day until humane end point of 1.5 cm tumor was achieved. Experimental metastases were achieved by tail vein injection of 1.0 x 106 cells. Eighteen days after cell injection, UCN-01 (7 or 14 mg/kg per day) or vehicle was administered for 4 days. Mice were euthanized, and lungs were extracted and fixed in formalin. The lung was stained hematoxylin and eosin and imaged with ImageScope (Aperio Technologies, Inc, Vista, CA). Pulmonary metastatic tumor area was quantified by measuring the tumor area in the whole lung.
Statistical Analysis
Differences in SCVM was analyzed by a nonparametric two-tailed t test. TUNEL and in vivo apoptosis detection were analyzed with a Fisher exact test. PuMA was analyzed by a one-way analysis of variance (ANOVA). Statistical analyses were performed using GraphPad Prism version 4.0C or InStat version 3.1a for Macintosh (GraphPad Software, La Jolla, CA). Statistical significance was defined as P ≤ .05.
Results
Defining the Phenotype of Highly Metastatic Cancer Cells
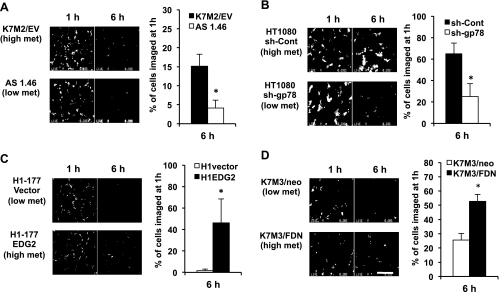
We previously reported the use of SCVM as a means to evaluate the influence of specific genes/proteins on the metastatic progression of both breast cancer and sarcoma cells to the lung (Figure 1). As expected, using this assay, we found that all cancer cell lines that we evaluated underwent significant attrition after their arrival in the lung (so called metastatic inefficiency); however, we were struck by the fact that, in five of the six studies of high and low isogenic metastatic cell lines, the high metastatic cells could be distinguished from the low metastatic cells within 6 hours of their arrival in the lung (Figures 1 and W1). We speculated that the cause of this early impact on metastatic efficiency may be the result of death of single metastatic cells in the lung or the dislodgement/migration of previously arrested metastatic cells back into the circulation. These data suggested that despite the complexity of the metastatic cascade, the events occurring early after the arrival of metastatic cells in the lung were unique and represented a stringent test for high versus low metastatic cells. The studies included two cancer types (breast cancer and sarcoma) recognized for progression to the lung. Exemplary of the behavior of the high and low metastatic cells, the K7M2 and AS1.46 pair is defined by differences in the expression of ezrin, a cytoskeleton linker protein associated with metastasis [13]. When using SCVM, the highly metastatic K7M2 cells yielded a predictable pattern of metastatic inefficiency with 15% of cells being retained in the lung within 6 hours compared with the less metastatic AS1.46 (Figure 1A) and AS1.52 cells (Figure W1A) where less than 4% retention occurred. Similar trends were seen in studies that included the inhibition of metastasis by the E3 ubiquitin ligase, gp78, and its association with the trans-membrane metastasis suppressor KAI1 (also known as CD82), in soft tissue sarcoma [20], the transcriptional regulation lysophosphatidic acid receptor by the metastasis suppressor Nm23-H1 in breast cancer cells [21], the role of Gemin5 as a regulator of the Nm23 phenotype in breast cancer cells [24], and the suppressive influence of the Fas signaling pathway in osteosarcoma [22]. The only biologic outlier in these studies of high and low metastatic cell line pairs was the comparison of the highly metastatic MNNG and less metastatic HOS osteosarcoma cells, which differ in the expression of an activated form of MET [25]. Both high (MNNG) and low (HOS) metastatic cells experienced similar levels of metastatic inefficiency (30% retention in the lung; Figure W1B), suggesting that events that occur later during metastatic progression in the lung distinguish this outlier pair of cells. As we pursued an understanding of the specific events linked to this early metastatic phenotype, we presumed that the MNNG and HOS cells should perform equally well in assays that modeled the early steps after the arrival in the lung. Indeed, if distinctions between MNNG and HOS were seen using these assays, we would question the relevance of these modeling approaches. As such, MNNG and HOS were used as internal controls of assay relevance.
Figure 1.
SCVM reliably distinguished high versus low metastatic (breast cancer and sarcoma) cells within 6 hours of their arrival in the lung. SCVM allows in vivo or ex vivo imaging of single fluorescently labeled metastatic cells in the mouse lung [13]. SCVM findings were seen in a variety of cancer types and biologic settings. (A) Ezrin in murine osteosarcoma (*P = .0043) [13]. (B) E-3 ligase in soft tissue sarcoma (*P = .0004) [20]. (C) EDG2 in human breast cancer (*P = .0056) [21]. (D) FAS-FAS-ligand in murine osteosarcoma (*P = .0007) [22]. Scale bars, 400 µm. Of the high and low metastatic cell line pairs examined, only the HOS-MNNG/HOS pair could not be distinguished at this early time point (Figure W1). All data represent the mean values ± SD from experiments performed at least three times; nonparametric two-tailed t test.
Metastatic Cells Preferentially Resist Apoptotic Death in the Lung
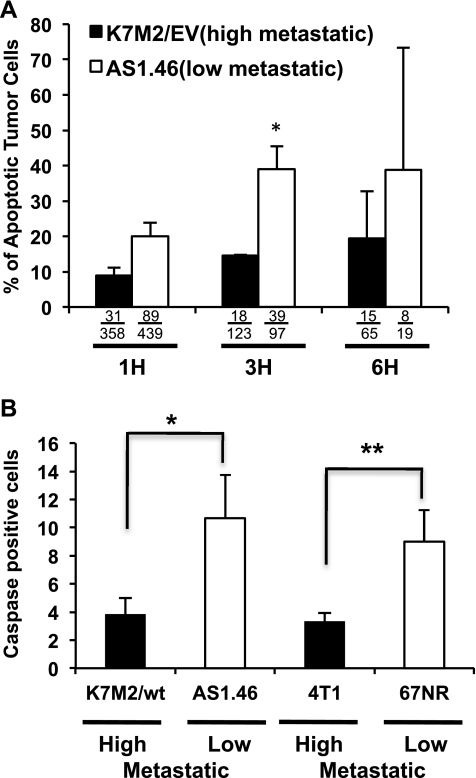
Continuing on our SCVM analysis of high and low metastatic cells, we next performed a TUNEL assay of high and low metastatic cells using two-color fluorescent analysis (green, tumor cell; TUNEL red, apoptotic cell) in fixed lung sections at discrete points after SCVM (Figures 2A and W2A). Highly metastatic osteosarcoma cells had significantly reduced double-positive fluorescent events compared with the less metastatic cells (Figures 2A and W2B). These differences in apoptotic rate were seen as early as 3 hours after the arrival of tumor cells in the lung (P < .001). Confirmatory studies using real time imaging of a fluorescent-labeled activated caspase 3, again demonstrated a reduction in apoptotic tumor cells in high compared with low metastatic osteosarcoma cells (Figures 2B and W2C). Studies in high and low metastatic murine breast cancer cells yielded similar results (Figure 2B). It is important to note that apoptotic rates of all high and low metastatic cells cultured in vitro were not distinguishable (data not shown). Using the same caspase activation assay and consistent with our prediction, we found equivalent rates of apoptosis in the lung (using SCVM) in the high (MNNG) and low (HOS) metastatic cells (data not shown). Collectively, these data suggest that early resistance against apoptotic death is a defining feature of highly metastatic cells.
Figure 2.
The highly metastatic phenotype is linked to an ability to resist apoptosis in the lung. (A) Red fluorescent TUNEL staining of lung, after SCVM, allowed acquisition and enumeration of double-positive (green/red) tumor cells (green) undergoing apoptosis (red) using confocal microscopy at 1, 3, and 6 hours after their arrival of cells in the lung (3 hours; *P < .0001). “% of Apoptotic Tumor Cells” was determined from the number of double-positive green and red apoptotic tumors over the total number of green tumor cells. Actual numbers of imaged green/red and green cells are provided below each bar. Representative images are presented in Figure W2. (B) The number of apoptotic (caspase 3 activated) tumor cells was significantly lower in high metastatic osteosarcoma cells (*P = .0015) and breast cancer cells (**P = .0223) 1 hour after tumor cell injection. As expected, no differences in caspase activation were observed between the MNNG and HOS osteosarcoma cell pairs, which were not distinguishable within 6 hours of their arrival in the lung. All data represent the mean values ± SD from experiments performed at least three times; Fisher exact test.
Akt Is Not, but PKC Is, Associated with the Early Metastatic Advantage of Highly Metastatic Cells
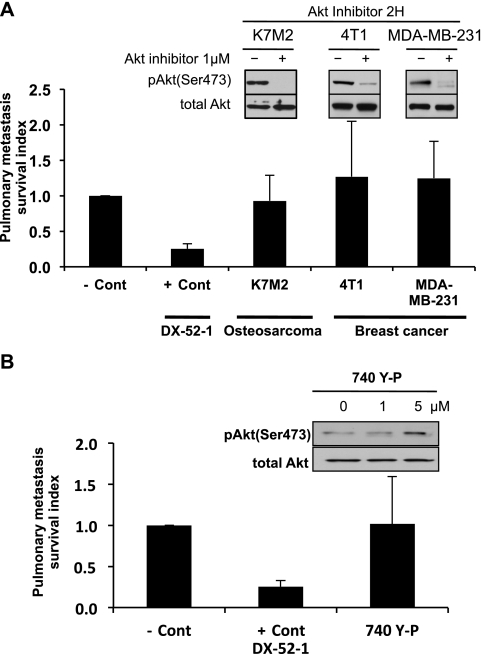
Using both genetic and pharmacologic approaches in sarcoma and breast carcinoma cells, we did not find evidence to link the Akt pathway with the observed survival phenotype of highly metastatic cells (Figures 3 and W3). We constitutively activated Akt1 in the less-metastatic AS1.46 cells by retrovirus infection of a myristoylated Akt construct (Figure W3, A and B). After Akt activation, we found no difference in the early metastatic phenotype in the lung using both SCVM and caspase activation assays (Figure W3, C and D). A pharmacological approach was then asked if an Akt1/2 inhibitor (PH domain dependent) would influence the early metastatic survival phenotype (Figure 3). Again, no difference in the metastatic behavior of highly metastatic (murine and human) osteosarcoma or breast cancer cells was seen after Akt pathway inhibitor exposure (Figure 3A). Concordantly, no enhancement of early metastatic phenotype was seen after exposure of the less-metastatic AS1.46 cells to the Akt pathway activator (Figure 3B).
Figure 3.
Modulation of the Akt survival pathway does not change the early metastatic phenotype. (A) PuMA was performed using various GFP-labeled highly metastatic cell lines treated with 1 µM Akt inhibitor or vehicle for 4 days. The antitumor antibiotic 7-cyanoquinocarcinol (DX-52-1; + Cont), an agent that markedly affected cell survival in vitro and reduced metastatic cell numbers in PuMA, was used as a positive control for maximal inhibition of cell survival compared to the untreated negative control (- Cont). The inhibition of Akt phosphorylation (AktSer473), after 1 µM Akt inhibitor exposure, was confirmed by Western analysis. (B) PuMA was performed by injection of GFP-labeled low metastatic AS1.46 cells for 4 days. PuMA cultures with GFP-labeled low metastatic AS1.46 cells were treated with vehicle, PI3 kinase activator (740-Y-P), or DX-52-1. The activation of Akt pathway, after 740-Y-P exposure for 2 hours, was confirmed by Western analysis. Specificity of this activation was confirmed by the lack of ERK1/2 phosphorylation at the same exposures. Pulmonary metastasis survival index was calculated by dividing the tumor burden (total fluorescence area) in the Akt inhibitor-treated lung section by that of control-treated lung section. All data represent the mean values ± SD from experiments performed at least three times; one-way ANOVA.
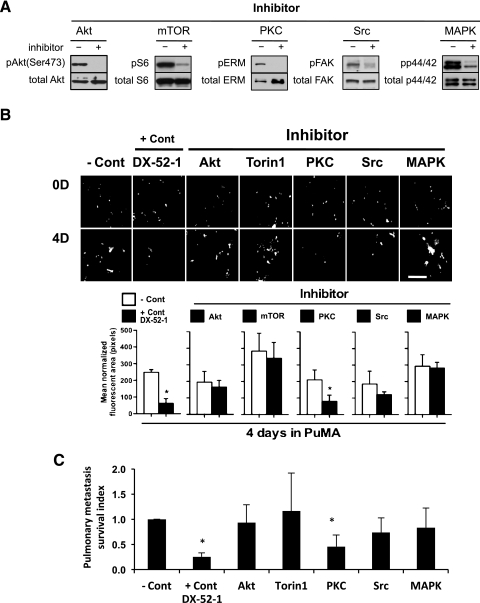
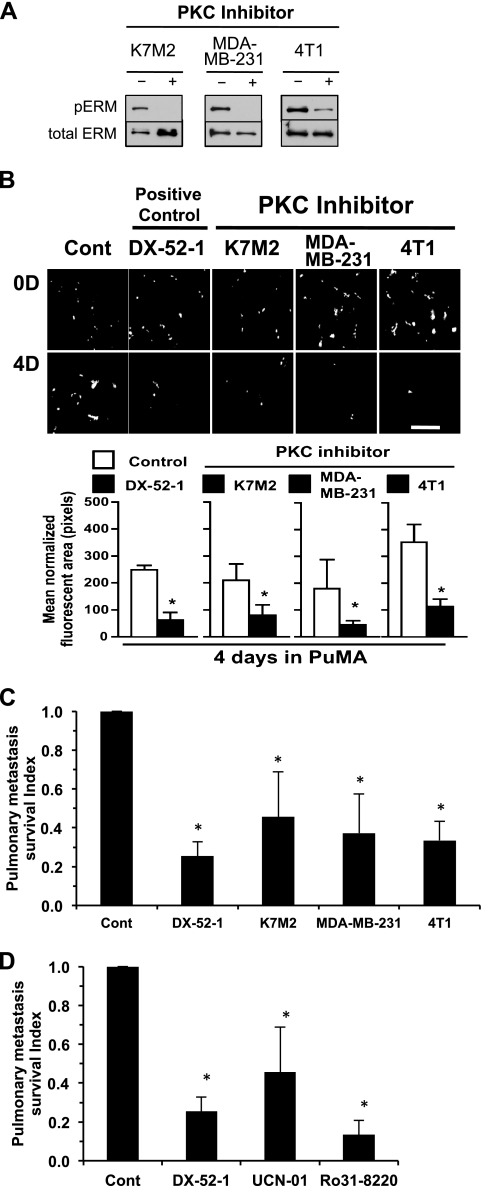
To begin to define alternative survival pathways responsible for the early metastatic resistance to apoptosis, we evaluated inhibitors of mTOR, PKC, Src, and MAPK within the PuMA. The activity of each inhibitor on the intended signaling pathway was confirmed by Western analysis of informative downstream targets at doses shown to have no effect on the cell line growth in vitro (Figure 4A). Of the agents screened, PKC inhibition with UCN-01 was found to most negatively influence the early metastatic phenotype of the breast cancer and sarcoma cell lines screened (P <.01; Figure 4, B and C). Consistent with these results, two distinct PKC inhibitors that both target classic PKC isoforms, UCN-01 and Ro31-8220, significantly inhibited early metastatic survival of murine and human osteosarcoma and breast cancer cells in the PuMA (P < .01; Figure 5).
Figure 4.
PKC pathway activation mediates survival of metastatic cancer cells. (A) The relevant target modulation of each pathway (Akt, mTOR, PKC, Src, and MAPK) inhibitor at the exposures that do not influence cell proliferation was confirmed by Western analysis. (B) PuMA cultures (K7M2 cells) were treated with pathway inhibitors or DX-52-1 as a positive control (+ Cont) compared to untreated cells (− Cont). PuMA metastatic burden was significantly lower in PKC inhibitor-treated group. Fluorescent cells (white arrows) have been confirmed as tumor cells by hematoxylin and eosin staining of lung sections in the PuMA (data not shown). Scale bar, 400 µm. (C) Pulmonary metastasis survival index was calculated by dividing the tumor burden (area of fluorescence) in the inhibitor-treated lung section by that of control-treated lung section. PKC-and DX-52-1-treated metastatic tumor burden was significantly lower than the control group (*P < .01). All data represent the mean values ± SD from experiments performed at least three times; one-way ANOVA.
Figure 5.
PKC activation is necessary for survival of single metastatic cells in the lung. (A) Western analysis for downstream target of PKC, phospho-ERM, was assessed in K7M2 osteosarcoma and MDA-MB-231 and 4T1 breast cancer cells using 50 nM exposures of UCN-01 (PKC inhibitor). (B) PuMA using GFP-labeled K7M2, MDA-MB-231, and 4T1 cells was followed for 4 days. PuMA cultures were treated with 50 nM PKC inhibitor or DX-52-1 as a positive control. The number of single metastatic cells (white arrows) in the lung is significantly decreased (quantification in lower panel) after exposure to PKC inhibitor. Scale bar, 400 µm. (C) Quantification of surviving single metastatic cells. Pulmonary metastasis survival index was calculated by dividing the tumor burden (area of fluorescence) in the PKC inhibitor-treated lung section by that of control-treated lung section. PKC-treated metastatic burden was significantly lower than control in K7M2 osteosarcoma and MDA-MB-231 and 4T1 breast cancer (*P < .01). (D) The use of two distinct PKC inhibitors (UCN-01 and Ro31-8220) significantly inhibited survival of single metastatic cells in the PuMA in K7M2 osteosarcoma cells (*P < .01). All data represent the mean values ± SD from experiments performed at least three times; one-way ANOVA.
The Process of Pulmonary Metastatic Progression Is Predominantly Sensitive to PKC Inhibition
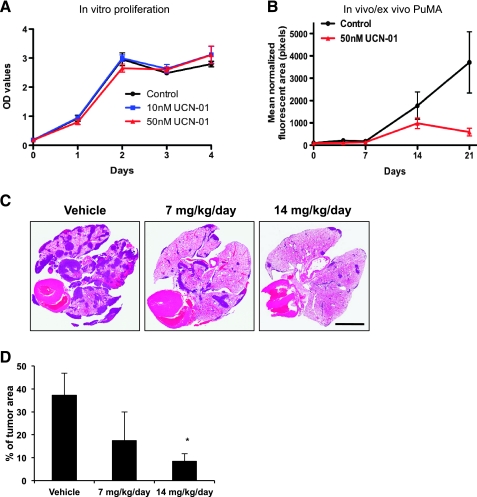
Given the previously recognized toxicity of PKC inhibitors, at their maximally tolerated dose, we first defined the minimum dose of PKC inhibitor that could target the PKC pathway in vivo (data not shown). At these effective targeting exposures, we found no effect of PKC inhibition on osteosarcoma cell growth in vitro (Figure 6A). However, in support of our hypothesis of the unique requirements of metastatic cells for PKC, we found a complete inhibition of metastatic progression in the PuMA (Figure 6B) and, more importantly, marked and effective reduction metastatic burden in vivo (Figure 6, C and D) after exposure to the PKC inhibitors.
Figure 6.
Pulmonary metastasis was uniquely inhibited after PKC inhibition. (A) The in vitro proliferation of tumor cells was not affected by PKC inhibitor (UCN-01) at 10 and 50 nM. A modest delay in orthotopic primary tumor growth to human end points was seen after PKC inhibitor treatment of mice (data not shown). (B) PKC inhibitor (UNC-01) used at exposures of 50 nM significantly inhibited pulmonary metastatic tumor burden within the PuMA. (C) In vivo experimental pulmonary metastasis was significantly inhibited after a 4-day treatment of mice with UCN-01 compared to vehicle control group. Scale bar, 5 mm. (D) Pulmonary metastatic tumor area was quantified by measuring the tumor area in the whole lung. PKC-treated metastatic tumor burden was significantly lower than vehicle-treated group in K7M2 osteosarcoma cells (*P < .05).
Discussion
An enhanced understanding of the biology of pulmonary metastasis is needed to improve outcomes for cancer patients. We previously studied the influence of several genes on the metastatic phenotype of cancer using a novel assay, SCVM, which allowed the evaluation of single metastatic cancer cells as they enter the lung [13]. A consistent post hoc observation from these imaging studies was that nearly all high versus low metastatic cells could be distinguished within the first 6 hours of the arrival of cells in the lung [13,20–22,26]. These observations, in both murine and human breast cancer and sarcoma, suggested that the metastatic phenotype was notably defined by the ability of metastatic cells to endure stresses early hours after their arrival in the lung.
On the basis of studies of metastatic inefficiency conducted in chick embryos, we then asked if the difference in early metastatic advantage for highly metastatic cells may be related to their ability to survive in the lung. We found that apoptotic rates, measured by caspase activation and TUNEL expression, are consistently lower in high compared to low metastatic cells. Previous reports have suggested the importance of apoptosis resistance as a feature of metastatic cells. Our data support and extend these findings by emphasizing the timing, context, and reproducibility of this important feature of metastatic cells (i.e., early after the arrival of cells at the secondary site) using relevant assays that model tumor interaction with the lung micro-environment in a number of isogenic cell line pairs. Collectively, these findings are consistent with a recurring hypothesis that the arrival of single cells in the lung is a stressful event. The stresses experienced by single cancer cells are likely related to differences in the micro-environment at the secondary site compared to the primary site, including differences in tissue oxygen tension [27], reactive species [28], inflammation [29–31], nutritional sources [32], pH, and other metabolic features [33]. We further hypothesize that cells that are able to resist these stresses, survive, progress, and yield metastasis [34]. It is important to note that the specific localization of single metastatic cells within the lung when they are forced to evade apoptosis is unknown. It is furthermore unclear how interaction of metastatic cells within the premetastatic and metastatic niches (endothelial cells, inflammatory cells, and other stromal cells) may prevent or trigger these apoptotic stresses.
Activation of Akt pathway is recognized as a critical survival signal of cancer cells [35]. Using both genetic and pharmacologic approaches in sarcoma and breast carcinoma cells, we could not link survival of highly metastatic cells, early after their arrival in the lung, with the Akt pathway. Our findings do not address the likely role that Akt plays later in progression of metastatic lesions [36–39]. In the absence of a connection with the Akt survival pathway, we then used the PuMA to screen other putative survival genes/pathways for an association with the survival phenotype of highly metastatic cells. Validated inhibitors of mTOR, PKC, Src, and MAPK were confirmed for their specific pathway inhibition in our cell systems. The only pathway inhibitor found to significantly influence metastatic progression was a PKC inhibitor. These results were found to be consistent across species (murine, human), cancer histologic types (osteosarcoma, breast carcinoma), and two distinct inhibitors of the classic PKC isoforms. It is important to note that the exposures of PKC inhibitor used in the PuMA had no influence on cell survival in conventional tissue culture conditions. PKC pathway activation has been associated with several steps of metastasis including resistance to apoptosis, migration, and invasion [40,41]. Recently, pharmacological inhibition of PKCα with αV5-3, a novel peptide inhibitor selective for PKCα, reduced mammary cancer metastasis by increasing their survival rate in mouse model of breast cancer [42]. The connection between PKC signaling and metastasis is also supported by studies from our group, suggesting that classic PKC isoforms regulate the phosphorylation of Ezrin, the prometastatic cytoskeleton linker protein. Because the PKC inhibitor UNC-01 is undergoing clinical development in patients with cancer, our data may be useful to conceive of optimized (tolerable and chronic) treatment schedules that may be active in limiting the survival of single metastatic cells in the lung. Such biologically based strategies may be quite distinct from the maximally tolerated dose-intensive schedules that are conventionally advanced into the clinical arena. It is understood that the metastatic seeding of secondary sites may occur before and after patients present with either localized or nonlocalized disease [43]. Our data describe the interaction of single cells within hours of their arrival in the lung; however, it is reasonable that metastatic cells are exposed to stresses and similar dependencies for PKC and other signals throughout metastatic progression. This may include times where single cells extend away from metastatic lesions during further metastatic progression into the surrounding lung micro-environment (when metastases metastasize) [44]. As such, there is not abasis to a priori determine that the targeting of “single” metastatic cells is “too late” to be helpful to patients. Furthermore, there is currently insufficient information to know when metastatic cells leave a primary tumor [45], whether they first seed a sanctuary site (i.e., the bone marrow) [46,47] or if subsequent seeding from a site of dormancy to a secondary organ (i.e., the lung) may occur after a patient presents with localized disease [48].
In summary, our results suggest the highly metastatic cells and not low metastatic cells survive and resist apoptosis as single cells early after their arrival in the lung. This survival advantage for highly metastatic cells is Akt independent but PKC and caspase 3 dependent. These data, emerging from studies of breast carcinoma and sarcoma cells, contribute to a detailed understanding of the steps that are most commonly defining of the metastatic phenotype and the pathways responsible for these steps. The translational value of these data suggests an opportunity to target the metastatic phenotype at critical and uniquely vulnerable states.
Supplementary Material
Acknowledgments
The authors thank Lee Helman, Su-Young Kim, Rosandra Kaplan, and Christina Mazcko for critical review of the article.
Abbreviations
- PKC
protein kinase C
- SCVM
single-cell videomicroscopy
- PuMA
pulmonary metastasis assay
- TUNEL
terminal transferase dUTP nick end labeling
Footnotes
This research was supported by the Intramural Research Program of the National Institutes of Health. The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Cawthorn TR, Amir E, Broom R, Freedman O, Gianfelice D, Barth D, Wang D, Holen I, Done SJ, Clemons M. Mechanisms and pathways of bone metastasis: challenges and pitfalls of performing molecular research on patient samples. Clin Exp Metastasis. 2009;26:935–943. doi: 10.1007/s10585-009-9284-5. [DOI] [PubMed] [Google Scholar]
- 4.Liu S, Sun MZ, Tang JW, Wang Z, Sun C, Greenaway FT. High-performance liquid chromatography/nano-electrospray ionization tandem mass spectrometry, two-dimensional difference in-gel electrophoresis and gene micro-array identification of lymphatic metastasis-associated biomarkers. Rapid Commun Mass Spectrom. 2008;22:3172–3178. doi: 10.1002/rcm.3725. [DOI] [PubMed] [Google Scholar]
- 5.Mira A, Isella C, Renzulli T, Cantarella D, Martelli ML, Medico E. The GAB2 signaling scaffold promotes anchorage independence and drives a transcriptional response associated with metastatic progression of breast cancer. Oncogene. 2009;28:4444–4455. doi: 10.1038/onc.2009.296. [DOI] [PubMed] [Google Scholar]
- 6.Morioka K, Tanikawa C, Ochi K, Daigo Y, Katagiri T, Kawano H, Kawaguchi H, Myoui A, Yoshikawa H, Naka N, et al. Orphan receptor tyrosine kinase ROR2 as a potential therapeutic target for osteosarcoma. Cancer Sci. 2009;100:1227–1233. doi: 10.1111/j.1349-7006.2009.01165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer KL, Eisenacher M, Braun Y, Brachwitz K, Wai DH, Dirksen U, Lanvers-Kaminsky C, Juergens H, Herrero D, Stegmaier S, et al. Microarray analysis of Ewing's sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. Eur J Cancer. 2008;44:699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Chambers AF, Naumov GN, Vantyghem SA, Tuck AB. Molecular biology of breast cancer metastasis. Clinical implications of experimental studies on metastatic inefficiency. Breast Cancer Res. 2000;2:400–407. doi: 10.1186/bcr86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gassmann P, Haier J, Schluter K, Domikowsky B, Wendel C, Wiesner U, Kubitza R, Engers R, Schneider SW, Homey B, et al. CXCR4 regulates the early extravasation of metastatic tumor cells in vivo. Neoplasia. 2009;11:651–661. doi: 10.1593/neo.09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedley BD, Vaidya KS, Phadke P, MacKenzie L, Dales DW, Postenka CO, MacDonald IC, Chambers AF. BRMS1 suppresses breast cancer metastasis in multiple experimental models of metastasis by reducing solitary cell survival and inhibiting growth initiation. Clin Exp Metastasis. 2008;25:727–740. doi: 10.1007/s10585-008-9184-0. [DOI] [PubMed] [Google Scholar]
- 11.Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 12.Wong CW, Lee A, Shientag L, Yu J, Dong Y, Kao G, Al-Mehdi AB, Bernhard EJ, Muschel RJ. Apoptosis: an early event in metastatic inefficiency. Cancer Res. 2001;61:333–338. [PubMed] [Google Scholar]
- 13.Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, Yeung C, Gorlick R, Hewitt SM, Helman LJ. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 14.Mendoza A, Hong SH, Osborne T, Khan MA, Campbell K, Briggs J, Eleswarapu A, Buquo L, Ren L, Hewitt SM, et al. Modeling metastasis biology and therapy in real time in the mouse lung. J Clin Invest. 2010;120:2979–2988. doi: 10.1172/JCI40252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glinsky GV, Glinsky VV. Apoptosis and metastasis: a superior resistance of metastatic cancer cells to programmed cell death. Cancer Lett. 1996;101:43–51. doi: 10.1016/0304-3835(96)04112-2. [DOI] [PubMed] [Google Scholar]
- 16.McConkey DJ, Greene G, Pettaway CA. Apoptosis resistance increases with metastatic potential in cells of the human LNCaP prostate carcinoma line. Cancer Res. 1996;56:5594–5599. [PubMed] [Google Scholar]
- 17.Glinsky GV, Glinsky VV, Ivanova AB, Hueser CJ. Apoptosis and metastasis: increased apoptosis resistance of metastatic cancer cells is associated with the profound deficiency of apoptosis execution mechanisms. Cancer Lett. 1997;115:185–193. doi: 10.1016/s0304-3835(97)04738-1. [DOI] [PubMed] [Google Scholar]
- 18.Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clin Exp Metastasis. 2000;18:261–271. doi: 10.1023/a:1006767007547. [DOI] [PubMed] [Google Scholar]
- 19.Rhim JS, Putman DL, Arnstein P, Huebner RJ, McAllister RM. Characterization of human cells transformed in vitro by N-methyl-N′-nitro-N-nitrosoguanidine. Int J Cancer. 1977;19:505–510. doi: 10.1002/ijc.2910190411. [DOI] [PubMed] [Google Scholar]
- 20.Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, Veenstra T, Hewitt SM, Helman LJ, Khanna C, et al. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- 21.Horak CE, Mendoza A, Vega-Valle E, Albaugh M, Graff-Cherry C, McDermott WG, Hua E, Merino MJ, Steinberg SM, Khanna C, et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:11751–11759. doi: 10.1158/0008-5472.CAN-07-3175. [DOI] [PubMed] [Google Scholar]
- 22.Koshkina NV, Khanna C, Mendoza A, Guan H, DeLauter L, Kleinerman ES. Fas-negative osteosarcoma tumor cells are selected during metastasis to the lungs: the role of the Fas pathway in the metastatic process of osteosarcoma. Mol Cancer Res. 2007;5:991–999. doi: 10.1158/1541-7786.MCR-07-0007. [DOI] [PubMed] [Google Scholar]
- 23.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 24.Lee JH, Horak CE, Khanna C, Meng Z, Yu LR, Veenstra TD, Steeg PS. Alterations in Gemin5 expression contribute to alternative mRNA splicing patterns and tumor cell motility. Cancer Res. 2008;68:639–644. doi: 10.1158/0008-5472.CAN-07-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper CS, Park M, Blair DG, ainsky MA, Huebner K, Croce CM, Vande Woude GF. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 26.Taylor JG, 6th, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, Yu Y, Chen QR, Shah K, Youngblood V, et al. Identification of FGFR4activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rofstad EK, Sundfor K, Lyng H, Trope CG. Hypoxia-induced treatment failure in advanced squamous cell carcinoma of the uterine cervix is primarily due to hypoxia-induced radiation resistance rather than hypoxia-induced metastasis. Br J Cancer. 2000;83:354–359. doi: 10.1054/bjoc.2000.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie K, Huang S. Regulation of cancer metastasis by stress pathways. Clin Exp Metastasis. 2003;20:31–43. doi: 10.1023/a:1022590402748. [DOI] [PubMed] [Google Scholar]
- 29.Brideau G, Makinen MJ, Elamaa H, Tu H, Nilsson G, Alitalo K, Pihlajaniemi T, Heljasvaara R. Endostatin overexpression inhibits lymphangiogenesis and lymph node metastasis in mice. Cancer Res. 2007;67:11528–11535. doi: 10.1158/0008-5472.CAN-07-1458. [DOI] [PubMed] [Google Scholar]
- 30.Caruso RA, Bellocco R, Pagano M, Bertoli G, Rigoli L, Inferrera C. Prognostic value of intratumoral neutrophils in advanced gastric carcinoma in a high-risk area in northern Italy. Mod Pathol. 2002;15:831–837. doi: 10.1097/01.MP.0000020391.98998.6B. [DOI] [PubMed] [Google Scholar]
- 31.Gorelik E, Wiltrout RH, Copeland D, Herberman RB. Modulation of formation of tumor metastases by peritoneal macrophages elicited by various agents. Cancer Immunol Immunother. 1985;19:35–42. doi: 10.1007/BF00199309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina RA, Owen GI. Glucose transporters: expression, regulation and cancer. Biol Res. 2002;35:9–26. doi: 10.4067/s0716-97602002000100004. [DOI] [PubMed] [Google Scholar]
- 33.Jentsch TJ, Stein V, Weinreich F, Zdebik AA. Molecular structure and physiological function of chloride channels. Physiol Rev. 2002;82:503–568. doi: 10.1152/physrev.00029.2001. [DOI] [PubMed] [Google Scholar]
- 34.Mendoza M, Khanna C. Revisiting the seed and soil in cancer metastasis. Int J Biochem Cell Biol. 2009;41:1452–1462. doi: 10.1016/j.biocel.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng S, Qiao M, Pardee AB. Metastasis and AKT activation. J Cell Physiol. 2009;218:451–454. doi: 10.1002/jcp.21616. [DOI] [PubMed] [Google Scholar]
- 36.Elliott BE, Meens JA, SenGupta SK, Louvard D, Arpin M. The membrane cytoskeletal crosslinker ezrin is required for metastasis of breast carcinoma cells. Breast Cancer Res. 2005;7:R365–R373. doi: 10.1186/bcr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukaya Y, Ishiguro N, Senga T, Ichigotani Y, Sohara Y, Tsutsui M, Shioura T, Iwamoto T, Hamaguchi M. A role for PI3K-Akt signaling in pulmonary metastatic nodule formation of the osteosarcoma cell line, LM8. Oncol Rep. 2005;14:847–852. [PubMed] [Google Scholar]
- 38.Kim CS, Vasko VV, Kato Y, Kruhlak M, Saji M, Cheng SY, Ringel MD. AKT activation promotes metastasis in a mouse model of follicular thyroid carcinoma. Endocrinology. 2005;146:4456–4463. doi: 10.1210/en.2005-0172. [DOI] [PubMed] [Google Scholar]
- 39.Krishnan K, Bruce B, Hewitt S, Thomas D, Khanna C, Helman LJ. Ezrin mediates growth and survival in Ewing's sarcoma through the AKT/mTOR, but not the MAPK, signaling pathway. Clin Exp Metastasis. 2006;23:227–236. doi: 10.1007/s10585-006-9033-y. [DOI] [PubMed] [Google Scholar]
- 40.Musashi M, Ota S, Shiroshita N. The role of protein kinase C isoforms in cell proliferation and apoptosis. Int J Hematol. 2000;72:12–19. [PubMed] [Google Scholar]
- 41.Sullivan RM, Stone M, Marshall JF, Uberall F, Rotenberg SA. Photo-induced inactivation of protein kinase Cα by dequalinium inhibits motility of murine melanoma cells. Mol Pharmacol. 2000;58:729–737. doi: 10.1124/mol.58.4.729. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Thorne SH, Sun L, Huang B, Mochly-Rosen D. Sustained inhibition of PKCα reduces intravasation and lung seeding during mammary tumor metastasis in an in vivo mouse model. Oncogene. 2011;30:323–333. doi: 10.1038/onc.2010.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khanna C. Novel targets with potential therapeutic applications in osteosarcoma. Curr Oncol Rep. 2008;10:350–358. doi: 10.1007/s11912-008-0054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren L, Hong SH, Cassavaugh J, Osborne T, Chou AJ, Kim SY, Gorlick R, Hewitt SM, Khanna C. The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene. 2009;28:792–802. doi: 10.1038/onc.2008.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai KL, Zhang ZY, Sahai E, Condeelis J, Segall JE. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66:192–197. doi: 10.1158/0008-5472.CAN-05-1242. [DOI] [PubMed] [Google Scholar]
- 46.Bruland OS, Hoifodt H, Saeter G, Smeland S, Fodstad O. Hematogenous micrometastases in osteosarcoma patients. Clin Cancer Res. 2005;11:4666–4673. doi: 10.1158/1078-0432.CCR-05-0165. [DOI] [PubMed] [Google Scholar]
- 47.Bruland OS, Hoifodt H, Hall KS, Smeland S, Fodstad O. Bone marrow micrometastases studied by an immunomagnetic isolation procedure in extremity localized non-metastatic osteosarcoma patients. Cancer Treat Res. 2009;152:509–515. doi: 10.1007/978-1-4419-0284-9_30. [DOI] [PubMed] [Google Scholar]
- 48.Barkan D, Kleinman H, Simmons JL, Asmussen H, Kamaraju AK, Hoenorhoff MJ, Liu ZY, Costes SV, Cho EH, Lockett S, et al. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res. 2008;68:6241–6250. doi: 10.1158/0008-5472.CAN-07-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.