Abstract
Colorectal cancer is one of the most common cancers in the world. Histoclinical staging is efficient, but combination with molecular markers may improve the classification of stage II cancers. Several tumor-suppressor genes have been associated with colorectal cancer, and the most frequent allelic losses have been extensively studied for their prognosis effect, but the results remain controversial. In a previous study, we found a possible influence of the chromosome 5 status in the development of liver metastases in stage II colon cancers. We have here investigated the role of the APC gene, located in chromosome arm 5q, in a series of 183 colon adenocarcinomas through a combined analysis of gene expression, mutation, allelic loss and promoter methylation, and metastasis occurrence. Point mutations were found in 73% of cases and allelic losses were found in 39%; 59% of tumors presented with a biallelic inactivation, with a very strong interdependence of the two APC hits (P = 2.1 x 10-9). No association was found between expression, number and type of APC alterations, and metastatic evolution. Our results show that the determination of APC status cannot help in the prediction of metastasis and cannot be used to subclassify stage II colon cancers.
Introduction
APC is often cited as a prime example of a tumor-suppressor gene. Germline mutations occur in familial adenomatous polyposis (FAP) and somatic mutations in tumors of the digestive tract [1]. Most of the mutations occur within the first half of the coding sequence and result in the truncation of the APC gene product. Somatic mutations in colorectal tumors are clustered in a particular region called the mutation cluster region (MCR) [2]. Inactivation of both alleles of the APC gene is required as an early event to develop most of adenomas and carcinomas in the colon and rectum [3]. In sporadic microsatellite stable tumors (MSSs), the second hit at the APC locus consists in either loss of heterozygosity (LOH) or a second mutation, each in roughly 40% of the cases [4]. Promoter methylation has been proposed as an alternative to mutation or LOH also leading to gene inactivation [5]. In FAP, the second APC hit is determined by the site of the first germline event [6,7].
Studies of somatic APC inactivation in sporadic colorectal carcinomas confirmed and reinforced the two-hit interdependence theory [8–10]. The preferred explanation is that APC somatic mutations are selected according to a specific level of β-catenin signaling that induces tumor formation [11]. Because allelic losses have been identified in tumors exhibiting two APC somatic mutations [12,13], a new theory progressively emerged, that is, tumor severity unusually requires three APC hits instead of two and that the most frequent third hit was copy number gain or deletion [14,15]. If tumors develop and progress through stepwise accumulation of mutations in different functional pathways, with APC, it also seems that repeated targeting of the same pathway and/or gene is selected in some cancers.
Colorectal cancer is associated with therapeutic challenges. Half of the patients die within 5 years after diagnosis despite regular progress in adjuvant chemotherapy protocols and the use of targeted drugs. To date, it is not recommended to enter systematically stage II colorectal cancer patients (30% of cases) in adjuvant chemotherapy protocols, although more than 20% will develop distant metastases. The monitoring of genetic alterations could provide independent prognostic information, but frequent discrepancies have limited their use in medical practice.
In a previous study, we showed that metastatic risk in stage II colon adenocarcinomas increased 3.4-fold when both chromosome 5q arms were retained [16]. To determine what type of 5q and APC gene alteration could influence clinical evolution, we studied an independent series of 183 stage II colon adenocarcinomas combining clinical, genomic, and expression profiles.
Materials and Methods
Ethic Statements
The institutional review board (COS, Comité d'Orientation Stratégique) approved the project in September 27, 2007. A total of 183 colon cancer patients were enrolled in the study. Inclusions ended November 5, 2008. All patients signed a written informed consent to authorize further research studies on their tumor material before storage in the respective biological resources centers.
Patients and Biological Samples
Clinical data and biologic samples were provided by the members of the CIT3 program (colon cancer identity card) driven by a national consortium in France. Patients were selected through the different projects developed by the five consortium teams for having been diagnosed with sporadic colon adenocarcinomas classified as stage II and of MSS type at primary surgery. The MSS status was assessed by multiplex polymerase chain reaction (PCR) genotyping using the MSI Analysis System v1.2 (Promega, Charbonnières, France). Disease-free patients were followed up for a minimum of 36 months after surgery, and the occurrence of liver metastasis within the interval was recorded.
Tissue samples were collected on fresh surgical specimens by pathologists within 1 hour after surgery from patients who underwent curative colonic resection, then flash frozen in liquid nitrogen, and stored at -80°C until nucleic acids extraction. Nucleic acids were extracted using QIAamp and RNeasy kits (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. Quantification of samples was done with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Illkirch, France). RNA integrity was controlled on an Agilent Bioanalyzer (Agilent Technologies, Massy, France).
APC, KRAS, and BRAF Alterations Detection
The APC (exons 3 to 15 part J) [17], KRAS (exon 2), and BRAF (exon 15) genes were analyzed by direct sequencing of after PCR amplification. The presence of mutations was scored using the PhrepPhrapConsed v11 package then encoded according to the HGVS recommendations.
The allelic status of the APC locus was derived from comparative genomic hybridization analyses performed on 44K BAC arrays and scored after normalization with comparative genomic hybridization Analytics v2.4 software (Agilent technologies, Massy France). It was systematically checked by genotyping of the microsatellite locus D5S346 using fluorescent primers.
The methylation status of the APC promoter was determined using a pyrosequencing technique. The analysis was restricted to the 1A region known as the major APC promoter [18]. Zymo EZ Methylation Direct kit was used for bisulfite conversion according to the manufacturer's recommendations (Zymo Research Corp, Orange, CA). Modified DNA was amplified by PCR with primers 5′TTGTTTGTTGGGGATTGG and 5′biotin TCCAACACCTACCCCATTTC for 45 cycles at an annealing temperature of 56°C. The biotinylated strand was isolated using a Vacuum Prep Tool (Qiagen, Inc, Valencia, CA) according to the manufacturer's protocol. Pyrosequencing reactions were performed using a PyroMark Q96 MD Pyrosequencing instrument (Qiagen, Inc) with 500 nM of sequencing primer 5′GAGAGAAGTAGTTGTGTAAT that allowed the quantification of the methylation proportion of seven CpG. The global methylation level was calculated as the mean of each individual value.
Gene Expression Profiling
Expression profiles were established with Affymetrix U133 Plus 2.0 human oligonucleotide microarrays [19]. Scanning was done with an Affymetrix GeneArray scanner and quantification with the Affymetrix GeneChip Operating Software (Affymetrix, Paris, France). CEL files were processed in the R/Bioconductor environment. Preprocessing steps (background adjustment, normalization, and summarization) were performed with the GCRMA package [20]. Expression measures were handled on log2 scale for statistical computations as well as for display purposes. Genes with overall low intensity were removed from further analysis. All remaining genes had an expression value greater than log2 (100) in at least 10% of the samples. As tumor samples and/or RNA samples were provided by four different centers (Bordeaux, Marseille, Nice, and Paris AP-HP), genes were then median centered within each center individually. Data from the single sample from Nantes were pooled with that of the smallest group, that is, Paris AP-HP. Hierarchical clustering analyses were done with the R's hclust function. Genes and arrays dissimilarity matrices were computed using Pearson correlation and Euclidian distance, respectively. Both trees were built with Ward's linkage. Supervised analyses were done in the R/Bioconductor environment using the Limma moderated t test [21]. Nominal P values were adjusted with the Benjamini-Hochberg method controlling the false discovery rate.
Results
Biallelic APC Inactivation
A total of 184 DNA were screened for APC mutations and allelic loss. Analysis was not complete because of insufficient quantity or bad quality of DNA in 31 cases, which were no longer considered. A total of 59 losses of the long arm of chromosome 5 (5q LOH), 154 truncating mutations, and 1 APC microdeletion were detected. No alteration was evidenced in 29 samples. Single allelic loss or mutation was found in 12 and 22 samples, respectively. Two mutations were recorded in 43 tumors and 47 cases exhibited one mutation and one allelic loss; thus, 90 tumors showed two hits at the APC locus (Table W1). According to the position of the seven β-catenin degradation sites starting at codon 1265, the nature of the two hits was analyzed and revealed a highly specific spectrum (Table 1; P = 2.1 x 10-9).
Relationship between APC Status and Expression Profile
Expression profiles of 86 samples from the COL2 project (Table W2) were available to examine correlations with the APC status. In this series, 46 samples exhibited two hits at the APC locus. To explore the contribution of the APC promoter methylation to global APC inactivation, the methylation level was determined in the remaining 40 samples. Methylation values ranged from 1% to 69%. Because negative controls did not exceed 2%, tumors were scored unmethylated when the value was in this range (16 samples); 14 samples exhibited a methylation level greater than 10% and were thus considered as methylated; 10 samples had a value between 2% and 3.3% and were considered as unmethylated. Considering methylation as an APC inactivation mode, 13 tumors showed complete APC inactivation: 11 with a mutation and 2 with a 5q allelic loss solely.
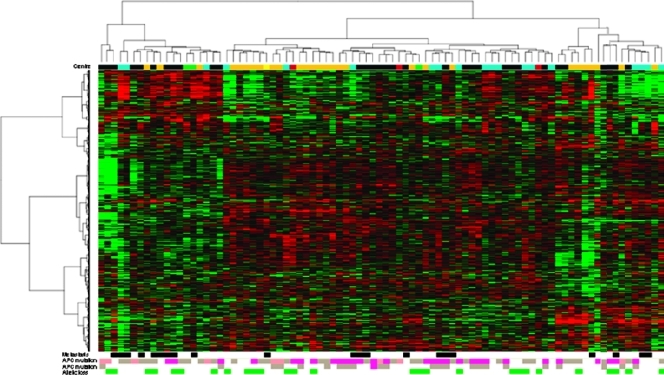
Unsupervised hierarchical clustering done after normalization (GEO access number GSE26905) showed no clear evidence that APC status could play a key role in shaping the expression profiles (Figure 1). To address more specific issues, several supervised analyses were done. They included the presence or absence of 5q LOH (25 vs 61 cases), of at least one APC mutation (68 vs 18 cases), of one APC alteration, that is, 5q LOH or APC mutation (73 vs 13 cases), the position of at least one APC mutation before or after codon 1265 (38 vs 30 cases), and, finally, the APC inactivation mode in case of two hits (25 samples with two APC mutations vs 21 cases with one APC mutation + one 5q LOH). None of them did generate a discriminating genes list significant enough to enable further investigation. When considering the methylation status, complete APC inactivation was found in 59 tumors, but their expression profile did not differ from that of tumors without APC alteration (13 cases).
Figure 1.
Hierarchical clustering of 86 stage II colon adenocarcinomas of MSS type. Unsupervised analysis was done on the 86 RNA selected according to the inclusion criteria (stage II, colon, MSS type) within tumor samples of the COL2 project (Table W2). Tumors from different origins were equally distributed within the different branches (top of the clustering): 32 from Bordeaux (BER, black), 28 from Nice (CAL, orange), 19 from Marseille (IPC, cyan), 6 from Paris AP-HP (3 from Lariboisière hospital [LAR, green] and 3 from Saint-Antoine hospital [STA, brown]), and 1 from Nantes (NAN, yellow). The occurrence (black) or not (white) of distant metastases is given at the bottom followed by the APC status. Mutations are presented in pink or light brown if occurring before or after codon 1265, respectively; dark pink shows mutations leading before the β-catenin-binding domain starting at codon 1020.
Expression Profile and Metastases Occurrence
The 86 patients whose tumor expression profile was available were diagnosed at a mean age of 66.5 years (range = 31–94 years). They were 42 men and 44 women, with a tumor localized in the ascending or descending colon in 22 and 64 cases, respectively. None of them received adjuvant chemotherapy. Twenty-one developed liver metastasis within 36 months after diagnosis. The others were disease free at this end point. Expression profiles depending on the presence or absence of distant metastasis were not associated with selective patterns. KRAS and BRAF mutations did not discriminate any subgroup according to the disease evolution.
Discussion
Allelic loss on chromosome arm 5q was found in 39% of 153 stage II colon adenocarcinomas of MSS type. This proportion is comparable to that found in other studies including the initial full description [22]. At least one mutation of the APC gene was observed in 73% of tumors, being concentrated in the MCR for 62% as expected [2]. Biallelic inactivation was found in 59% of cases, equally distributed in a combination of one APC mutation plus one 5q allelic loss or two APC mutations. This frequency is slightly lower than that previously reported, as well as the proportion of tumors harboring one APC mutation and 5q LOH [8,23]. The preferential association of mutations occurring in the MCR with 5q allelic loss reinforces the theory of the two-hit interdependence observed in sporadic colorectal cancers as well as in FAP-derived cancers.
No difference in expression profiles was observed when comparing biallelic inactivation modes. The most probable explanation is that RNA predicted to encode a much-shortened protein is unstable. This decreased stability has been shown to preclude protein truncation tests for the detection of mutations leading the first 14 exons of the APC gene [24,25]. Non-sense-mediated messenger RNA (mRNA) decay might hide here the possible differences between cells with biallelic inactivation due to one APC mutation before the MCR (with subsequent non-sense-mediated mRNA decay) plus one within the MCR or one mutation within the MCR plus one 5q allelic loss (with no mRNA at all). Proteins resulting from APC mutations should thus lead in both situations to half protein dosage in tumor cells compared to normal cells and to the conservation of APC ability to bind β-catenin. This observation suggests that a residual activity of β-catenin binding is probably necessary for tumor growth [11].
A more surprising observation was that no difference exists between tumors exhibiting at least one APC alteration (APC mutation and/or 5q allelic loss and/or APC promoter methylation) or none. Among 86 expression profiles available to analyze this parameter, only 13 were derived from tumors with no APC alteration, a proportion that probably is too low to document such effects; furthermore, this group of “no-mutation” cases is a fortiori not big enough to look at differences if we hypothesize that part of them have missed APC mutations and a part developed through a β-catenin-independent mechanism. This issue could be partially resolved by analyzing the APC protein expression, but formalin-fixed tissues were generally not available for this series of tumors.
Twenty-one patients developed distant metastases, that is, 24% of cases, an expected proportion in stage II colon adenocarcinomas. We were not able to detect differences in expression profiles between the groups with and without metastases. Tumor cells studied were part of primary lesions that did not metastasize in lymph nodes at the initial resection. It is possible that this type of sample is poor in cells prone to expand in a distant host organ through blood invasion several years later. Global analysis of the primary tumor is thus unable to detect their specific characteristics. An efficient option would be to compare primary tumors from 3-year disease-free patients and metastatic samples from patients with metastases at diagnosis.
In summary, our study shows that, although the APC gene seems to play a central role in primary tumor formation through a fine regulation of the β-catenin pathway, it is probably not directly or mainly involved in the metastatic process.
Supplementary Material
Table 1.
APC Inactivation Mode in Tumors with Two Hits.
| Hits | Mutation | ||
| Hits | |||
| <Codon 1265 | >Codon 1265 | ||
| LOH | 14 | 33 | |
| Mutation | <Codon 1265 | 0 | 39 |
| >Codon 1265 | -(39) | 4 | |
Acknowledgments
The authors thank MP Buisine who kindly provided us the opportunity to rapidly set up the specific methylation analysis.
Footnotes
This work was supported by LNCC (Ligue Nationale Contre le Cancer). All contributors to the consortium on colon cancer identity card CIT3-colon funded by LNCC are gratefully acknowledged: J.F. Mosnier (Nantes), M. Longy (Bordeaux), J.F. Fléjou (Paris), N. Sevenet (Paris), and F. Viret (Marseille) for COL2; G. Milano and M.C. Etienne (Nice) for COL2 and CRAL; J. Selves and S. Kirzin (Toulouse) for CCOR; V. Boige (Villejuif) and P. Laurent-Puig (Paris) for COLI; A. Duval (Paris) and M.P. Gaub (Strasbourg) for CORE. D.B. was granted by Association pour la Recherche sur le Cancer.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and are available online at www.transonc.com.
References
- 1.Ilyas M, Straub J, Tomlinson IPM, Bodmer WF. Genetic pathways in colorectal and other cancers. Eur J Cancer. 1999;35:335–351. doi: 10.1016/s0959-8049(98)00431-6. [DOI] [PubMed] [Google Scholar]
- 2.Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 3.Powell SM, Petersen GM, Krush AJ, Booker S, Jen J, Giardiello FM, Hamilton SR, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature. 1192;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 4.Olschwang S, Hamelin R, Laurent-Puig P, Thuille B, De Rycke Y, Li YJ, Muzeau F, Girodet J, Salmon RJ, Thomas G. Alternative genetic pathways in colorectal carcinogenesis. Proc Natl Acad Sci USA. 1997;94:12122–12127. doi: 10.1073/pnas.94.22.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segditsas S, Sieber OM, Rowan A, Setien F, Neale K, Phillips RK, Ward R, Esteller M, Tomlinson IP. Promoter hypermethylation leads to decreased APC mRNA expression in familial polyposis and sporadic colorectal tumours, but does not substitute for truncating mutations. Exp Mol Pathol. 2008;85:201–206. doi: 10.1016/j.yexmp.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Spirio LN, Samowitz W, Robertson J, Robertson M, Burt RW, Leppert M, White R. Alleles of APC modulate the frequency and classes of mutations that lead to colon polyps. Nat Genet. 1998;20:385–388. doi: 10.1038/3865. [DOI] [PubMed] [Google Scholar]
- 7.Lamlum H, Ilyas M, Rowan A, Clark S, Johnson V, Bell J, Frayling I, Efstathiou J, Pack K, Payne S, et al. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germ-line mutation: a new facet to Knudson's “two-hit” hypothesis. Nat Med. 1999;5:1071–1075. doi: 10.1038/12511. [DOI] [PubMed] [Google Scholar]
- 8.Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer WF, Tomlinson IP. APC mutations in sporadic colorectal tumors: a mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci USA. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheadle JP, Krawczak M, Thomas MW, Hodges AK, Al-Tassan N, Fleming N, Sampson JR. Different combinations of biallelic APC mutation confer different growth advantages in colorectal tumours. Cancer Res. 2002;62:363–366. [PubMed] [Google Scholar]
- 10.Crabtree M, Sieber OM, Lipton L, Hodgson SV, Lamlum H, Thomas HJ, Neale K, Phillips RK, Heinimann K, Tomlinson IP. Refining the relation between “first hits” and “second hits” at the APC locus: the “loose fit” model and evidence for differences in somatic mutation spectra among patients. Oncogene. 2003;22:4257–4265. doi: 10.1038/sj.onc.1206471. [DOI] [PubMed] [Google Scholar]
- 11.Albuquerque C, Breukel C, van der Luijt R, Fidalgo P, Lage P, Slors FJ, Leitão CN, Fodde R, Smits R. The “just-right” signaling model: APC somaticmutations areselected based on aspecificlevel of activationofthe β-catenin signaling cascade. Hum Mol Genet. 2002;11:1549–1560. doi: 10.1093/hmg/11.13.1549. [DOI] [PubMed] [Google Scholar]
- 12.Tomlinson IP, Roylance R, Houlston RS. Two hits revisited again. J Med Genet. 2001;38:81–85. doi: 10.1136/jmg.38.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sieber OM, Heinimann K, Gorman P, Lamlum H, Crabtree M, Simpson CA, Davies D, Neale K, Hodgson SV, Roylance RR, et al. Analysis of chromosomal instability in human colorectal adenomas with two mutational hits at APC. Proc Natl Acad Sci USA. 2002;99:16910–16915. doi: 10.1073/pnas.012679099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sieber OM, Segditsas S, Knudsen AL, Zhang J, Luz J, Rowan AJ, Spain SL, Thirlwell C, Howarth KM, Jaeger EE, et al. Disease severity and genetic pathways in attenuated familial adenomatous polyposis vary greatly but depend on the site of the germline mutation. Gut. 2006;55:1440–1448. doi: 10.1136/gut.2005.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segditsas S, Rowan AJ, Howarth K, Jones A, Leedham S, Wright NA, Gorman P, Chambers W, Domingo E, Roylance RR, et al. APC and the three-hit hypothesis. Oncogene. 2009;28:146–155. doi: 10.1038/onc.2008.361. [DOI] [PubMed] [Google Scholar]
- 16.Zeitoun G, Buecher B, Bayer J, Tanguy ML, Thomas G, Olschwang S. Retention of chromosome arm 5q in stage II colon cancers identifies 83% of liver metastasis occurrences. Genes Chromosomes Cancer. 2006;45:94–102. doi: 10.1002/gcc.20269. [DOI] [PubMed] [Google Scholar]
- 17.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 18.Horii A, Nakatsuru S, Ichii S, Nagase H, Nakamura Y. Multiple forms of the APC gene transcripts and their tissue-specific expression. Hum Mol Genet. 1993;2:283–287. doi: 10.1093/hmg/2.3.283. [DOI] [PubMed] [Google Scholar]
- 19.Bertucci F, Finetti P, Cervera N, Charafe-Jauffret E, Mamessier E, Adélaïde J, Debono S, Houvenaeghel G, Maraninchi D, Viens P, et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 2006;66:4636–4644. doi: 10.1158/0008-5472.CAN-06-0031. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 21.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 22.Ashton-Rickardt PG, Dunlop MG, Morris RG, Purdie CA, Steel CM, Evans HJ, Bird CC, Wyllie AH. High frequency of APC loss in sporadic colorectal carcinoma due to breaks clustered in 5q21-22. Oncogene. 1989;4:1169–1174. [PubMed] [Google Scholar]
- 23.Miyaki M, Konishi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, Muraoka M, Takahashi H, Amada Y, Fukayama M, et al. Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer Res. 1994;54:3011–3020. [PubMed] [Google Scholar]
- 24.Powell SM, Petersen GM, Krush AJ, Booker S, Jen J, Giardiello FM, Hamilton SR, Vogelstein B, Kinzler KW. Molecular diagnosis of familial adenomatous polyposis. N Engl J Med. 1993;329:1982–1987. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- 25.van der Luijt R, Khan PM, Vasen H, van Leeuwen C, Tops C, Roest P, den Dunnen J, Fodde R. Rapid detection of translation-terminating mutations at the adenomatous polyposis coli (APC) gene by direct protein truncation test. Genomics. 1994;20:1–4. doi: 10.1006/geno.1994.1119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.