Abstract
BACKGROUND: There is a need to identify new markers to assess recurrence risk in early-stage colorectal cancer (CRC) patients. We explored the prognostic impact of ether-a-gò-gò-related gene 1 channels and some hypoxia markers, in patients with nonmetastatic (stage I, II, and III) CRC. METHODS: The expression of hERG1, vascular endothelial growth factor A (VEGF-A), glucose transporter 1, carbonic anhydrase IX (CA-IX), epidermal growth factor receptor (EGF-R), and p53 was tested by immunohistochemistry in 135 patients. The median follow-up was 35 months. Clinicopathologic parameters and overall survival were evaluated. RESULTS: hERG1 displayed a statistically significant association with Glut-1, VEGF-A, CA-IX, and EGF-R; p53 with VEGF-A and CA-IX; Glut-1 with the age of the patients; and EGF-R with TNM and mucin content. TNM and CA-IX were prognostic factors at the univariate analysis; TNM, hERG1, and Glut-1, at the multivariate analysis. Risk scores calculated from the final multivariate model allowed to stratify patients into four different risk groups: A) stage I–II, Glut-1 positivity, any hERG1; B) stage I–II, Glut-1 and hERG1 negativity; C) stage I–II, Glut-1 negativity, hERG1 positivity; D) stage III, any Glut-1 and any hERG1. CONCLUSIONS: hERG1 positivity with Glut-1 negativity identifies a patient group with poor prognosis within stage I–II CRC. The possibility that these patients might benefit from adjuvant therapy, independently from the TNM stage, is discussed. IMPACT: More robust prognostic and predictive markers, supplementing standard clinical and pathologic staging, are needed for node-negative patients.
Introduction
Colorectal cancer (CRC) accounts for approximately 9.4% of total worldwide cancer causes and is the fourth most common cancer in men and the third in women [1]. Clinical and pathologic tumor staging, contributing to TNM classification, are nowadays the main predictor of prognosis and treatment in patients with CRC [2]. However, considerable stage-independent outcome variability is observed, which likely reflects molecular heterogeneity. In particular, stage II (node-negative) represents a wide spectrum of disease, where a 5-year survival can range from 85% to 50% [2]. Accordingly, the efficacy of adjuvant chemotherapy, although well established in node-positive CRC, needs to be fully validated for TNM stage II cases [3]. Hence, more robust prognostic and predictive markers, supplementing standard clinical and pathologic staging, are needed for node-negative patients.
Microsatellite instability and p53 alterations are among the most frequent tumor alterations associated with CRC oncogenesis [4,5]. A defect in mismatch repair accounts for approximately 15% colon cancers, whereas p53 alterations are found in half of all CRCs [6,7]. Indeed, microsatellite instability and p53 have been proposed to influence the clinical outcome of CRC, but their usefulness for clinical use is still under debate [8,9].
Molecular candidates implicated in CRC progression and potentially useful for prognostic purposes can be singled out from the hypoxia pathway. Tumor hypoxia is a key element of tumor progression, driving to a more aggressive phenotype and an increased propensity for metastases, as well as mediating radioresistance and chemoresistance [10,11]. Hypoxia can trigger such diverse effects because it switches on the expression of hypoxia-inducible factor 1 (HIF-1)-dependent genes, whose protein products allow tumor cells to survive the harsh tumor microenvironment [12]. HIF-1-dependent genes comprise those triggering the angiogenesis process [13]. Besides its relevance in tumor progression, the overlap between hypoxia and angiogenesis pathways can be exploited for prognostic and predicting purposes [14]. However, a thorough study evaluating the prognostic potential of hypoxia (and angiogenesis) markers in CRC is lacking so far.
For these reasons, we performed a pilot study aimed at exploring the prognostic impact of endogenous markers of tumor hypoxia in CRC. On the basis of what was indicated in the study of Goethals et al. [14], we determined the expression of 1) the vascular endothelial growth factor A (VEGF-A) [15], as an example of HIF-1-dependent gene; 2) carbonic anhydrase IX (CA-IX) [16], as a hypoxia marker involved in the pH homeostasis; 3) the glucose transporter 1 (Glut-1) [17], as a HIF-1-dependent gene and hypoxia marker involved in the control of the metabolic state of neoplastic cells; and 4) the epidermal growth factor receptor (EGF-R) [14,18], which triggers the intracellular signaling pathways regulating HIF-1 activity in solid tumors [19]. In this study, we also included the evaluation of the ether-a-gò-gò related gene (hERG1) potassium channel, which is expressed in CRC [20–22] and is functionally linked to hypoxia [23] and angiogenesis [24,25] in different types of cancer. The whole set of markers (VEGF-A, GLUT-1, CA-IX, EGF-R, and hERG1) were tested by immunohistochemistry (IHC) in surgical samples of nonmetastatic, TNM stages I to III, CRC patients. A classic biomolecular marker, p53, and clinicopathologic features were also included in the study. Overall survival (OS) was taken as the main prognostic indicator.
Materials and Methods
Patients
The study cohort included 135 untreated patients who underwent radical surgery with curative intent for colorectal adenocarcinomas at the Department of General Surgery and Surgical Oncology, Azienda Ospedaliero-Universitaria, Careggi, Florence. This cohort represents a subgroup of the patients operated on at the Department in the period September 2001 to July 2008 who were selected without any bias. Patients affected by hepatitis C viral infection or who had undergone preoperative radiotherapy or chemotherapy for rectal cancer were excluded. Samples of tumor were collected during surgery, after obtaining an informed written consent, and immediately processed for the sample storing (see next paragraphs). All the samples were classified as adenocarcinomas and staged according to the American Joint Committee on Cancer classification by experienced pathologists. Only samples that resulted to be TNM stages I to III were included in the study and further processed for IHC. Patients with stage III received adjuvant chemotherapy after surgery. All patients received the appropriate treatment after recurrence, according to the local guidelines.
Immunohistochemical Staining
IHC was carried out on 7-µm sections on positively charged slides. After dewaxing and dehydrating the sections, endogenous peroxidases were blocked with a 1% H2O2 solution in phosphate-buffered saline. Subsequently, antigen retrieval was performed 1) by treatment with proteinase K (5 µg/ml) (for hERG1, VEGF-A, CA-IX, and Glut-1 staining) or 2) by heating the samples in a microwave oven at 600 W in citrate buffer pH 6.0 for 10 (for EGF-R staining) or 20 minutes (for p53 staining). The following antibodies were used at the dilutions reported in parentheses: 1) anti-hERG1 monoclonal antibody (1:200; produced in our laboratory and distributed by Enzo Life Sciences, Lawsen, Switzerland); 2) anti-VEGF-A (1:100, polyclonal antibody anti-VEGF-A (A-20); Santa Cruz Biotechnology, Santa Cruz, CA); 3) anti-Glut-1 (1:100, polyclonal rabbit anti-human GLUT1; Dako Cytomation, Glostrup, Denmark); 4) anti-CA-IX monoclonal antibody (monoclonal murine antibody M75, 1:100; described in Pastorekova et al. [26]); 5) anti-EGF-R rabbit polyclonal IgG (1:100; Santa Cruz Biotechnology); and 6) anti-p53 monoclonal antibody (1:50; Dako Cytomation). Incubation with the primary antibody was carried out overnight at 4°C, except for the anti-p53 antibody, which was incubated for 30 minutes at room temperature. Immunostaining was performed with a commercially available kit (PicTure Plus kit and DAB; Zymed, Carlsbad, CA) according to the manufacturer's instructions.
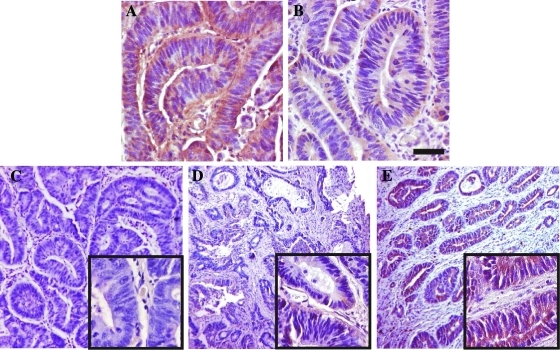
Specimens were evaluated using a quantitative assessment, using different scoring systems based on the determination of the percentage of positive cells. Among the scoring systems used, different cutoffs and a different number of groups were applied, the latter depending on the staining pattern and intensity of the marker under study. We adopted previously published scoring systems for VEGF-A, Glut-1, CA-IX, EGF-R, and p53. In particular, the assignment of a positive score was performed as follows: 1) For VEGF-A, when the sample showed more than 10% positive cells [27]. 2) For Glut-1, when areas displaying an unequivocal staining were detected, without using any scoring system. Red blood cells served as an internal positive control, whereas areas with normal epithelium, stroma, and edge effects were ignored as in Cooper et al. [28]. 3) For CA-IX, as reported in Korkeila et al. [29], without using a scoring system, samples were assessed as positive when more than 10% cells per microscopic field expressed the protein. 4) For EGF-R, when at least 1% positive cells were present in the sample [30]. 5) For p53, a 10% cutoff was adopted, as reported in Veloso et al. [31]. In any case, areas of necrosis, stroma, normal epithelium, and distinct edge effects were not scored. To assess hERG1 expression, we first performed IHC experiments following the same protocol published in Lastraioli et al. [20], using a polyclonal antibody raised in our laboratory. To further validate this procedure, we performed the same IHC using a monoclonal antibody recognizing a different epitope positioned in the S5 pore region of the channel [32]. Figure 1 shows representative sections stained with the anti-hERG1 polyclonal (A) and the anti-hERG1 monoclonal antibody (B). Both antibodies showed a clear and overlapping positivity in CRC cells, corroborated by the good concordance emerged by the statistical analysis carried out by χ2 test (P < .001). Because the monoclonal antibody gave a very low background signal, which allowed us to better evaluate the percentage of positive cells per microscopic field and therefore to assign an immunoreactivity score to each sample, we decided to use this antibody for further analyses. Stained sections were analyzed at a total magnification of 40x field by field, from top left to bottom right. Each field was assigned a percentage of positive tumor cells; hence, a semi-quantitative scoring method was used and a cutoff of 50% was applied. In other words, samples were classified as “score 0” when no staining was present, “score 1” when displaying a positivity in a percentage ranging from 1% to 49% of neoplastic cells, and “score 2” when the percentage of positive cells was more than 50%. Only samples displaying a “score 2,” for example, with a strong hERG1 expression, were considered “positive” in further analyses. Figure 1 (C–E) shows representative CRC sections displaying the different hERG1 scores.
Figure 1.
Scoring system assessment for hERG1 in CRC specimens. (A, B) Immunohistochemical detection of the hERG1 protein in the same, representative specimen of CRC using two different anti-hERG1 antibodies: the anti-hERG1 polyclonal antibody (A) and the anti-hERG1 monoclonal antibody (B). Bar, 100 µm. (C–E) Representative examples of hERG1 scoring (Materials and Methods) in CRC representative specimens using the anti-hERG1 monoclonal antibody; details at higher magnification are in the insets: (C) score 0 (0% of positive cells), (D) score 1 (1%–49% of positive cells per microscopic field), and (E) score 2 (>50% of positive cells per microscopic field). Note that only samples belonging to score 2 were considered positive. Bar, 200 µm (C, D, E); 50 µm (insets).
In any case, samples were evaluated by two independent investigators. Interobserver agreement was evaluated according to the simple Cohen κ of concordance and its 95% confidence interval (CI).
Statistical Analysis
The distributions of all studied patients were reported with respect to their demographic, clinical, and biologic characteristics and were summarized as frequencies and percentage. Continuous variables were reported as median and range of variation. The following demographic and clinical variables were investigated: age at the intervention, sex, site of tumor, TNM classification, and presence of colloid. The biologic assessment was evaluated through the expression of: hERG1 channels, Glut-1, VEGF-A, CA-IX, p53, and EGF-R markers. All markers were categorized as yes/no respect to their expression. Both in the association and survival analysis, age was categorized in two groups (<70 years vs ≥70 years). The presence of association between demographic, clinical, and biologic characteristics was evaluated by χ2 and Fisher exact tests when appropriate. A two-sided P ≤ .05 was considered significant. All the variables were investigated for their impact on OS. Because the study was retrospective, and the patients were subjected to different methods and timing of follow-up, we chose OS as the main end point. OS was defined as the time between intervention and death, whatever the cause. Observation time of patients alive at the last follow-up was censored. Median follow-up time was estimated according to the Kaplan-Meier inverse method [33]. In the univariate analysis, estimates of OS were calculated according to the Kaplan-Meier product-limit method [34]. Comparisons of estimated survival curves were performed by means of the log-rank test. Hazard ratios and appropriate 95% CIs were also calculated by means of the Cox proportional hazard model. A multivariate Cox regression model was fitted to evaluate the independent effect of each factor on OS. Starting from a full model, including sex, age, site of tumor, TNM, colloid, hERG1, Glut-1, VEGF-A, and CA-IX, nonsignificant variables were progressively removed according to a backward stepwise procedure based on the likelihood ratio test. A sensitivity analysis was also performed on 122 cases without missing variables and including p53 and EGF-R information. A probability of 0.10 was used both for removal and reentry criteria. Finally, we converted the Cox model predictor to a risk score that is directly related to an individual's probability of death from any cause, and starting from the distribution of risk scores and the strata identified by the combinations of the three predictors retained in the Cox model, we empirically classified all patients into four different risk groups.
Data were analyzed using the statistical software SAS 9.2 (SAS Corporation, Cary, NC).
Results
Immunohistologic Findings
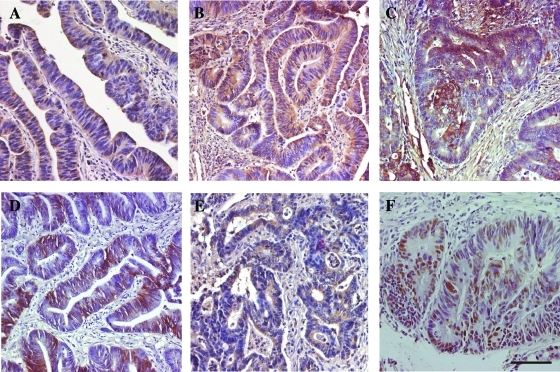
We studied 135 patients with stage I to III CRC, who underwent radical surgery for primary CRC at a single institution. In all samples, we determined the expression of hypoxia markers (VEGF-A, CA-IX, Glut-1, and EGF-R), of hERG1 potassium channels, as well as of a classic marker of CRC (p53) by IHC. IHC pictures relative to representative samples of the all markers are in Figure 2. hERG1 (Figure 2A) was expressed on the plasma membrane and in the cytoplasm of neoplastic cells, with a diffuse pattern of labeling and almost no staining in the tumor stroma. VEGF-A (Figure 2B) staining was intense and diffuse in the cytoplasm of tumor cells, with a weaker positivity in the stroma, as reported [27]. Glut-1 (Figure 2C) stained the plasma membrane and cytoplasm of tumor cells with a focal expression that might be confined either to small or to slightly bigger areas or to bigger ones, as reported [28]. CA-IX (Figure 2D) protein was expressed on the plasma membrane of tumor cells, with a focal expression pattern, as reported by Pastorekova et al. [26]. EGF-R (Figure 2E) staining was mainly membranous, diffused to all tumor cells, although with different intensities [30]. p53 (Figure 2F) stained tumor cell nuclei [31]. On the basis of the scoring system chosen for each marker, a score value was attributed to each sample. Note that only samples displaying a high hERG1 staining (score 2, see Figure 1E) were considered positive for further analyses. The κ value related to the interobserver measure of agreement was 0.91 (95% CI = 0.84–0.98).
Figure 2.
Immunohistochemical staining for hERG1 (A), VEGF-A (B), Glut-1 (C), CA-IX (D), EGF-R (E), and p53 (F) in CRC specimens. IHC experiments and assessment of score were performed according to what were reported in the Materials and Methods section. The description of the staining features is in the Results section. Bar, 100 µm.
Clinical Characteristics
Table 1 shows the clinicopathologic characteristics of the patients, as well as the distribution of the biologic markers under study. Of the 135 patients, 72 (53%) were female and 63 (47%) male. Median age was 68 years (range = 40–90 years). Fifty-seven tumors were located in the right colon, 14 in the transverse, 33 in the left, and 31 in the rectum.
Table 1.
Distribution of Clinicopathologic and Biomolecular Markers and Results of the Univariate Analysis.
| Parameter | No. Patients | 3-Year Survival | HR (95% CI) | P (LR Test) |
| Age | ||||
| <70 years | 73 (54.1%) | 0.59 | 1 (ref.) | .652 |
| ≥70 years | 62 (45.9%) | 0.55 | 1.14 (0.65–1.97) | |
| Sex | ||||
| Female | 72 (53.3%) | 0.55 | 1 (ref.) | .588 |
| Male | 63 (46.7%) | 0.59 | 1.16 (0.67–2.02) | |
| Tumor site | ||||
| Right colon | 57 (42.2%) | 0.53 | 1 (ref.) | .800 |
| Left colon | 33 (24.4%) | 0.77 | 0.71 (0.34–1.51) | |
| Transverse colon | 14 (10.4%) | 0.52 | 0.98 (0.40–2.44) | |
| Rectum | 31 (23.0%) | 0.50 | 0.80 (0.40–1.59) | |
| TNM stage | ||||
| I | 29 (21.5%) | 0.74 | 1 (ref.) | <.001 |
| II | 47 (34.8%) | 0.74 | 0.83 (0.33–2.06) | |
| III | 59 (43.7%) | 0.36 | 2.54 (1.17–5.52) | |
| Mucin | ||||
| No | 100 (74.1%) | 0.55 | 1 (ref.) | .504 |
| Yes | 35 (25.9%) | 0.60 | 0.81 (0.44–1.49) | |
| hERG1 | ||||
| Negative | 104 (77.0%) | 0.59 | 1 (ref.) | .203 |
| Positive | 31 (23.0%) | 0.50 | 1.49 (0.80–2.76) | |
| VEGF-A | ||||
| Negative | 25 (18.5%) | 0.52 | 1 (ref.) | .299 |
| Positive | 110 (81.5%) | 0.58 | 0.69 (0.34–1.39) | |
| Glut-1 | ||||
| Negative | 88 (65.2%) | 0.51 | 1 (ref.) | .111 |
| Positive | 47 (34.8%) | 0.68 | 0.60 (0.32–1.13) | |
| CA-IX | ||||
| Negative | 93 (68.9%) | 0.64 | 1 (ref.) | .022 |
| Positive | 42 (31.1%) | 0.31 | 2.02 (1.10–3.85) | |
| EGF-R | ||||
| Negative | 28 (22.2%) | 0.55 | 1 (ref.) | .845 |
| Positive | 98 (77.8%) | 0.58 | 0.94 (0.48–1.81) | |
| p53 | ||||
| Negative | 80 (61.1%) | 0.55 | 1 (ref.) | .876 |
| Positive | 51 (38.9%) | 0.63 | 1.05 (0.58–1.88) |
Relationship between Biologic Markers and Clinical Characteristics
An IHC signal positive for hERG1 was observed in 23% of patients (31/135). VEGF-A expression was detected in 82% (110/135), Glut-1 in 35% (47/135) of the samples, whereas CA-IX was expressed in 42 of 135 samples, accounting for 31%. EGF-R and p53 were positive in 78% (98/126) and 39% (51/131) of the samples, respectively (Table 1).
It emerged a statistically significant association between hERG1 and Glut-1 (P = .002), hERG1 and EGF-R (P = .050), hERG1 and CA-IX (P = .018), hERG1 and VEGF-A (P = .049), VEGF-A and p53 (P = .002), and CA-IX and p53 (P = .030). A marginally statistically significant association was observed between VEGF-A and CA-IX (P = .070) and between VEGF-A and Glut-1 (P = .080). The percentage of EGF receptor-expressing tumors significantly decreased according to TNM stage (96%, 83%, and 64% for stage I, II, and III, respectively; P for trend < .001) and inmucinous tumors (65% vs 83%, P = .032).A marginally statistically significant association was also found between Glut-1 and mucin, with Glut-1 less frequently expressed in mucinous tumors (23% vs 39%, P = .080).
Impact on Survival
Median follow-up was 35 months. Overall, 55 (41%) of 135 patients died during the period under investigation. At the univariate analysis, whose results are reported in Table 1, the following variables turned out to have a significant impact on OS: TNM stage and CA-IX expression.
Multivariate OS analysis, as reported in Table 2, identified the following independent prognostic factors: TNM, hERG1, and Glut-1. In particular, as evident from the HR values, whereas TNM stage III was confirmed to be an indicator of worse prognosis, hERG1 positivity emerged as a negative prognostic factor. Conversely, Glut-1 positivity had a positive impact of survival. The same results were confirmed by the sensitivity analysis including p53 and EGF-R information (Materials and Methods).
Table 2.
Multivariate Analysis.
| Variable | Coefficient (SE) | Hazard Ratio | 95% CI | P |
| TNM | ||||
| I | — | 1 (Ref.) | <.001 | |
| II | -0.28611 (0.46752) | 0.75 | 0.30–1.87 | |
| III | 1.26618 (0.40922) | 3.55 | 1.59–7.91 | |
| hERG1 | ||||
| Negative | — | 1 (Ref.) | ||
| Positive | 0.76697 (0.33345) | 2.15 | 1.12—4.13 | .021 |
| Glut-1 | ||||
| Negative | — | 1 (Ref.) | ||
| Positive | -1.15571 (0.35403) | 0.31 | 0.16–0.63 | .001 |
On the whole, from this analysis, it emerged that hERG1 positivity, in conjunction with the lack of Glut-1 expression, has a negative prognostic impact on survival that accompanies that of the TNM stage.
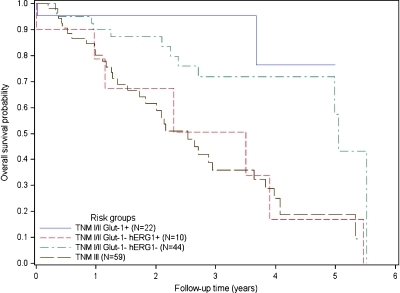
Starting from risk scores calculated from the final multivariate model, patients were stratified into four different risk groups, three of which comprised patients in TNM stages I and II, further subdivided on the basis of the staining for hERG1 and Glut-1, and the fourth group comprising stage III patients, as follows: A) TNM stage I–II, with Glut-1 positivity, any hERG1; B) TNM stage I–II with Glut-1 negativity and hERG1 negativity; C) TNM stage I–II, with Glut-1 negativity and hERG1 positivity; and D) TNM stage III, any Glut-1 and any hERG1. The relative percentage of stage I and stage II patients in the A to C groups was roughly the same (see legend to Figure 3). As evident from the Kaplan-Meier curves reported in Figure 3, the 3-year OS probabilities in the four groups were as follows: 95%, 72%, 51%, and 36%, respectively. It is worth noting that, in group C (i.e., TNM stages I and II, with Glut-1 negativity and hERG1 positivity) and group D (TNM stage III, independent of hERG1 and Glut-1 staining), survival curves almost overlapped.
Figure 3.
Kaplan-Meier curves of overall survival according to different combinations of tumor characteristics (TNM stage, Glut-1 status, and hERG1 status). Kaplan-Meier plots of overall survival probability for four different groups are reported. The number of patients in each group is reported in parenthesis. Blue curve indicates Glut-1-positive samples (10 TNM I, 12 TNM II); red curve, Glut-1-negative and hERG1-positive samples (5 TNM I, 5 TNM II); green curve, Glut-1-negative and hERG1-negative samples (14 TNM I, 30 TNM II); black curve, TNM 3 patients (59 TNM III).
Discussion
To our knowledge, this study is the first that evaluates the impact of different endogenous hypoxia markers, in conjunction with a K+ channel (hERG1), on OS of patients with nonmetastatic CRC. Our results, although still preliminary and with all the limits of a retrospective study, suggest for new clinical trials that the positivity for hERG1 and the negativity for Glut-1 allows to identify a patient's group, within stage I and II patients, which approaches the prognosis of stage III CRC.
The study evaluated the expression of Glut-1, CA-IX, VEGF-A, EGF-R, along with hERG1 K+ channels by IHC in surgical samples of nonmetastatic (TNM stages I to III) CRC patients, including a classic biomolecular marker, p53, and clinicopathologic features, and taking OS as the main prognostic indicator. The interobserver measure of agreement was almost perfect.
We focused on the analysis of the hypoxia pathway because of the relevance of this process and of the ensuing angiogenesis, in the malignant progression of cancer. Nevertheless, a thorough study assessing the prognostic and predictive impact of hypoxia/angiogenesis markers in CRC is lacking so far. Moreover, in our study, we also evaluated the expression of hERG1 K+ channels. The inclusion was dictated by the fact that K+ channels in general, and hERG1 in particular, are emerging as novel and believable biomarkers in several types of cancers, including CRC [19,22] (http://www.sophicalliance.com/). Moreover, our previous results indicated a functional link between hERG1 and the hypoxia and angiogenesis pathways in different cancer types [23–25] (Crociani et al., unpublished observations).
Indeed, a statistically significant association emerged between the expression of hERG1 and that of all the other hypoxia markers analyzed. This result further confirms, in the clinical setting, what emerging from experimental data, and candidates, for the first time, a potassium channel as a member of the endogenous hypoxia pathway, at least in CRC. VEGF-A expression, besides strongly associated to hERG1 expression, was also moderately associated with two other markers of tumor hypoxia (CA-IX and Glut-1), as well as with the presence of a mutated p53, witnessed by p53 IHC positivity. This confirms that hypoxia, maybe through an increased HIF-1-mediated transcriptional activity of the vegf-a gene, selects p53-mutated, hypoxia-resistant clones, thus contributing to CRC progression [8].
The univariate analysis of OS showed, in accordance with recent analyses, that p53 IHC positivity had no significant impact on survival of CRC patients, whereas it confirmed the prognostic role of the TNM stage [2]. In addition, at the univariate analysis, CA-IX emerged as an indicator of worse prognosis in CRC. The isozyme IX of carbonic anhydrases contributes to control the tumor extracellular pH, while being strongly inducible by hypoxia in tumors [35,36]. This is also relevant in CRC, where an increased CA-IX expression has been reported, especially in the tumor areas of high proliferation [37]. Consistent with our data, it was recently shown that CA-IX negativity is an independent predictor of longer disease-free survival and OS in rectal cancers [29].
The multivariate analysis, while confirming the prognostic relevance of the TNM stage, showed that only two of the markers we analyzed remained in the Cox model: hERG1 and Glut-1. Interestingly, whereas hERG1 positivity had a negative impact on OS, Glut-1, when positive, was a predictor of good prognosis. In other words, the positivity of hERG1 expression, in conjunction with the lack of Glut-1, reinforces the well-established negative prognostic impact of the clinicopathologic evaluation of the TNM stage.
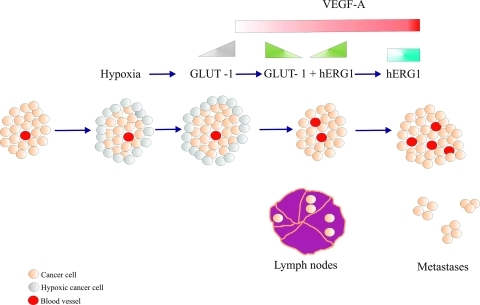
It is possible to conceive a hypothetical model, which also takes into account previously published results, where the interaction between Glut-1 and hERG1 is interpreted (Figure 4). Glut-1 expression is high as far as hypoxia is present within the tumor mass, whereas it is conceivable that its expression declines as soon as normoxic conditions are reestablished [38], and tumor cells restart growing and migrate outside the tumor mass, reaching regional lymph nodes. This can be consequence of the neoangiogenesis triggered by VEGF-A, whose secretion is regulated by hERG1 channels [24,25]. Hence, a condition is established, along tumor progression, which is characterized by the presence of hERG1 and the progressive disappearance of Glut-1. Such condition could allow the establishment of a more aggressive phenotype because hERG1 not only drives VEGF-A secretion and increases CRC cell invasiveness, a fact that explains its high expression in metastases [20].
Figure 4.
Model of the interplay between hERG1, Glut-1, and VEGF-A in CRC progression. When tumor mass (light pink circles) reaches a critical volume, hypoxic areas start to be present, mainly in peripheral areas of the tumor (grey circles). In such condition, Glut-1 expression is triggered (gray triangle at the top). Subsequently, normoxic conditions are restored due to VEGF-A-mediated angiogenesis (red circles inside the tumor mass and red rectangle at the top); VEGF-A secretion is regulated by hERG1 potassium channels, whose expression is switched on (green triangle at the top). In this phase of tumor progression, Glut-1 expression is reduced (green triangle at the top), tumor cells restart to grow and locally invade, reaching regional lymph nodes. In the latter stages of tumor progression, both VEGF-A and hERG1 are overexpressed (red and blue rectangles at the top, respectively), intratumoral angiogenesis increases (red circles in the tumor mass), and distant metastases occur.
Finally, the results of the multivariate analysis have some relevant clinical consequences. In fact, starting from risk scores calculated from the final multivariate model, patients were stratified into four different risk groups, three of which comprising stage I and II patients, further subgrouped on the basis of hERG1 and Glut-1 expression, and one group that comprises all the stage III patients (Figure 3, curves). The 3-year OS probabilities inside the groups allowed us to conclude that the patient group that displays positivity to hERG1 and a concomitant negativity to Glut-1, despite belonging to TNM stages I or II, has a risk probability that overlaps that of TNM stage III patients. This analysis was not biased by any evident disequilibrium between the groups, especially between stage I and stage II patients in the first three risk groups. Hence, this result gets statistical significance and acquires clinical relevance.
On the whole, the markers we propose (hERG1 and Glut-1), despite the small number of cases we analyzed and besides the use of a nonrandomized study group, might identify a subgroup of patients with regional, TNM stage I and II CRC, with a predictable worse prognosis. These results, to be further considered for clinical trials for stage II CRC patients, could conceivably provide an indication for a more aggressive adjuvant therapy in stage II, hERG1 positive and Glut-1 negative, patients group.
Acknowledgments
The anti-CA-IX antibody was a kind gift from Dr Pastorekova.
Footnotes
This work was supported by the Association for International Cancer Research (grant no. 06-0491), Associazione Italiana per la Ricerca sul Cancro (grant no. 1662), and Istituto Toscano Tumori (DD Regione Toscana no. 6888) to A.A., Ente Cassa di Risparmio di Firenze to F.D.C. No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer Sixth Edition Staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 3.Benson AB, III, Schrag D, Somerfield MR, Cohen AM, Figueredo AT, Flynn PJ, Krzyzanowska MK, Maroun J, McAllister P, Van Cutsem E, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 4.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 6.Kim GP, Colangelo LH, Wieand HS, Paik S, Kirsch IR, Wolmark N, Allegra CJ National Cancer Institute, author. Prognostic and predictive roles of high degree microsatellite instability in colon cancer: a National Cancer Institute - National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 7.Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, et al. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat. 2003;21:271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- 9.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris A. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL. Defining the role of hypoxia-inducible-factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26:281–290. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]
- 14.Goethals L, Debucquoy A, Perneel C, Geboes K, Ectors N, De Schutter H, Penninckx F, McBride WH, Begg AC, Haustermans KM. Hypoxia in human colorectal adenocarcinoma: comparison between extrinsic and potential intrinsic hypoxia markers. Int J Radiation Oncology Biol Phys. 2006;65:246–254. doi: 10.1016/j.ijrobp.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Ferrara N. Pathways mediating VEGF-independent tumor angiogenesis. Cytokine Growth Factor Rev. 2010;21:21–26. doi: 10.1016/j.cytogfr.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Kaluz S, Kaluzová M, Liao SY, Lerman M, Stanbridge EJ. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: a one transcription factor (HIF-1) show? Biochim Biophys Acta. 2009;1795:162–172. doi: 10.1016/j.bbcan.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuire BB, Fitzpatrick JM. Biomarkers in renal cell carcinoma. Curr Opin Urol. 2009;19:441–446. doi: 10.1097/MOU.0b013e32832f0c68. [DOI] [PubMed] [Google Scholar]
- 18.Steinbach JP, Klumpp A, Wolburg H, Weller M. Inhibition of epidermal growth factor receptor signaling protects human malignant glioma cells from hypoxia-induced cell death. Cancer Res. 2004;64:1575–1578. doi: 10.1158/0008-5472.can-03-3775. [DOI] [PubMed] [Google Scholar]
- 19.Franovic A, Gunaratnam L, Smith K, Robert I, Patten D, Lee S. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc Natl Acad Sci USA. 2007;104:13092–13097. doi: 10.1073/pnas.0702387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lastraioli E, Guasti L, Crociani O, Polvani S, Hofmann G, Witchel H, Bencini L, Calistri M, Messerini L, Scatizzi M, et al. herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells. Cancer Res. 2004;64:606–611. doi: 10.1158/0008-5472.can-03-2360. [DOI] [PubMed] [Google Scholar]
- 21.Ousingsawat J, Spitzner M, Puntheeranurak S, Terracciano L, Tornillo L, Bubendorf L, Kunzelmann K, Schreiber R. Expression of voltage-gated potassium channels in human and mouse colonic carcinoma. Clin Cancer Res. 2007;13:824–831. doi: 10.1158/1078-0432.CCR-06-1940. [DOI] [PubMed] [Google Scholar]
- 22.Dolderer JH, Schuldes CH, Bockhorn A, Altmannsberger M, Lambers C, von Zabern D, Jonas D, Schwegler H, Linke R, Schröder UH. HERG1 gene expression as a specific tumor marker in colorectal tissues. Eur J Surg Oncol. 2010;36:72–77. doi: 10.1016/j.ejso.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Fontana L, D'Amico M, Crociani O, Biagiotti T, Solazzo M, Rosati B, Arcangeli A, Wanke E, Olivotto M. Long-term modulation of HERG channel gating in hypoxia. Biochem Biophys Res Commun. 2001;286:857–862. doi: 10.1006/bbrc.2001.5464. [DOI] [PubMed] [Google Scholar]
- 24.Masi A, Becchetti A, Restano-Cassulini R, Polvani S, Hofmann G, Buccoliero AM, Paglierani M, Pollo B, Taddei GL, Gallina P, et al. hERG1 channels are overexpressed in glioblastoma multiforme and modulate VEGF secretion in glioblastoma cell lines. Br J Cancer. 2005;93:781–792. doi: 10.1038/sj.bjc.6602775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillozzi S, Brizzi MF, Bernabei PA, Bartolozzi B, Caporale R, Basile V, Boddi V, Pegoraro L, Becchetti A, Arcangeli A. VEGFR-1 (FLT-1), β1 integrin and hERG K+ channel form a macromolecular signaling complex in acute myeloid leukemia: role in cell migration and clinical outcome. Blood. 2007;110:1238–1250. doi: 10.1182/blood-2006-02-003772. [DOI] [PubMed] [Google Scholar]
- 26.Pastorekova S, Parkkila S, Parkkila AK, Opavský R, Zelník V, Saarnio J, Pastorek J. Carbonic anhydrase IX, MN/CA IX: analysis of stomach complementary DNA sequence and expression in human and rat alimentary tracts. Gastroenterology. 1997;112:398–408. doi: 10.1053/gast.1997.v112.pm9024293. [DOI] [PubMed] [Google Scholar]
- 27.Galizia G, Lieto E, Ferraraccio F, Orditura M, De Vita F, Castellano P, Imperatore V, Romano C, Ciardiello F, Agostini B, et al. Determination of molecular marker expression can predict clinical outcome in colon carcinomas. Clin Cancer Res. 2004;10:3490–3499. doi: 10.1158/1078-0432.CCR-0960-03. [DOI] [PubMed] [Google Scholar]
- 28.Cooper R, Sarioğlu S, Sökmen S, Füzün M, Küpelioğlu A, Valentine H, Görken IB, Airley R, West C. Glucose transporter-1 (GLUT-1): a potential marker of prognosis in rectal carcinoma? Br J Cancer. 2003;89:870–876. doi: 10.1038/sj.bjc.6601202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korkeila E, Talvinen K, Jaakkola PM, Minn H, Syrjänen K, Sundström J, Pyrhönen S. Expression of carbonic anhydrase IX suggests poor outcome in rectal cancer. Br J Cancer. 2009;100:874–880. doi: 10.1038/sj.bjc.6604949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galizia G, Lieto E, Ferraraccio F, De Vita F, Castellano P, Orditura M, Imperatore V, La Mura A, La Manna G, Pinto M, et al. Prognostic significance of epidermal growth factor receptor expression in colon cancer patients undergoing curative surgery. Ann Surg Oncol. 2006;13:823–835. doi: 10.1245/ASO.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 31.Veloso M, Wrba F, Kaserer K, Heinze G, Magalhães A, Herbst F, Teleky B. p53 gene status and expression of p53, mdm2, and p21Waf1/Cip1 proteins in colorectal cancer. Virchows Arch. 2000;437:241–247. doi: 10.1007/s004280000255. [DOI] [PubMed] [Google Scholar]
- 32.Guasti L, Crociani O, Redaelli E, Pillozzi S, Polvani S, Masselli M, Mello T, Galli A, Amedei A, Wymore RS, et al. Identification of a posttranslational mechanism for the regulation of hERG1 K+ channels expression and hERG1 current density in tumor cells. Mol Cell Biol. 2008;16:5043–5060. doi: 10.1128/MCB.00304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17:343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan EL, Meier P. Non parametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 35.Ivanov S, Liao S-Y, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905–919. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075–7083. [PubMed] [Google Scholar]
- 37.Saarnio J, Parkkila S, Parkkila AK, Haukipuro K, Pastoreková S, Pastorek J, Kairaluoma MI, Karttunen TJ. Immunohistochemical study of colorectal tumors for expression of a novel transmembrane carbonic anhydrase, MN/CA IX, with potential value as a marker of cell proliferation. Am J Pathol. 1998;153:279–285. doi: 10.1016/S0002-9440(10)65569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Airley R, Evans A, Mobasheri A, Hewitt SM. Glucose transporter Glut-1 is detectable in peri-necrotic regions in many human tumor types but not normal tissues: study using tissue microarrays. Ann Anat. 2010;192:133–138. doi: 10.1016/j.aanat.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]