Abstract
Good quality deoxyribonucleic acid (DNA) is the pre-requisite for its downstream applications. The presence of high concentrations of polysaccharides, polyphenols, proteins, and other secondary metabolites in mango leaves poses problem in getting good quality DNA fit for polymerase chain reaction (PCR) applications. The problem is exacerbated when DNA is extracted from mature mango leaves. A reliable and modified protocol based on the cetyltrimethylammonium bromide (CTAB) method for DNA extraction from mature mango leaves is described here. High concentrations of inert salt were used to remove polysaccharides; Polyvinylpyrrolidone (PVP) and β-mercaptoethanol were employed to manage phenolic compounds. Extended chloroform-isoamyl alcohol treatment followed by RNase treatment yielded 950‒1050 µg of good quality DNA, free of protein and RNA. The problems of DNA degradation, contamination, and low yield due to irreversible binding of phenolic compounds and coprecipitation of polysaccharides with DNA were avoided by this method. The DNA isolated by the modified method showed good PCR amplification using simple sequence repeat (SSR) primers. This modified protocol can also be used to extract DNA from other woody plants having similar problems.
Keywords: Extraction buffer, Mango, Polyphenols, DNA isolation, Simple sequence repeat (SSR), Secondary metabolites
1. Introduction
Mango (Mangifera indica L.) is a favorite fruit in the world especially in the Indo-Pakistan sub-continent. It is a rich source of vitamins, β-carotene, minerals, and antioxidants. Mango is known as the “king of fruits” for its unmatchable taste and flavor (Singh, 1996). Pakistan is situated at the western edge of the natural range of monoembryonic mango and is thus a centre of diversity for the crop (Bompard, 1993). The native Mangifera germplasm needs to be evaluated and conserved, not only for its intrinsic worth, but also because of the potential presence of valuable resistance against different diseases.
Characterization of mango germplasm based on morphological features is inefficient; this problem is further compounded by the perennial and monoembryonic nature of mango. Significant genetic improvement in mango can be accelerated by using genomic-based approaches. The isolation of good quality deoxyribonucleic acid (DNA) is the pre-requisite for molecular research. Like in other woody plants, DNA extraction from mango is problematic. The existing protocols (Dellaporta et al., 1983; Doyle and Doyle, 1990; Davis et al., 1995) are inefficient for extracting DNA from mature mango leaves. Therefore, there was a need to optimize the protocols for DNA extraction from mature leaf samples to yield high concentrations of good quality DNA fit for polymerase chain reaction (PCR) applications. The presence of high amounts of contaminants, mainly phenolic compounds, polysaccharides, and secondary metabolites impedes the DNA isolation procedure and inhibits analytical studies on the isolated DNA (Pirttilä et al., 2001). Polyphenolic compounds interact irreversibly with nucleic acid, resulting in the inability of different modifying enzymes to manipulate the DNA (Manoj et al., 2007). Polysaccharides are also problematic as they make the DNA unruly during pipetting and hinder the activity of polymerases, ligases, and restriction endonucleases (Fang et al., 1992; Sharma et al., 2002).
The majority of mango germplasm in Pakistan is monoembryonic; therefore seedlings are not representative of the parent trees (Schnell and Knight, 1992). Moreover, young leaves are not always available, and so the ability to use mature leaves would provide a good alternative for DNA extraction. Mature leaves of mango contain comparatively high quantities of polysaccharides and different secondary metabolites, posing problems in downstream applications of DNA.
The cetyltrimethylammonium bromide (CTAB) method of DNA isolation developed for young plant tissues (Doyle and Doyle, 1987) was modified. The aim of this study was to develop a protocol for DNA extraction using fully expanded mature leaves. Modifications were focused on minimizing phenolic compounds and polysaccharide co-isolation in DNA, maintaining the good quality of the DNA. The procedure described here is easy, efficient, and cost-effective and more importantly, yields good quality DNA from mature mango leaves.
2. Materials and methods
2.1. Reagents
The reagents used in our experiment are as follows: extraction buffer [including CTAB 0.04 g/ml, NaCl 3 mol/L, β-mercaptoethanol 3% (added just before use), ethylenediaminetetraacetic acid (EDTA) 20 mmol/L, Tris-HCl 100 mmol/L (pH 8.0), and PVP-40 (polyvinylpyrrolidone, molar weight 40 000) 0.025 g/ml]; chloroform-isoamyl alcohol 24:1 (v/v); ethanol (70%, 100%); sodium acetate 3 mol/L; RNase A 10 mg/ml; proteinase K 1 mg/ml; TE buffer [including Tris-HCl 10 mmol/L (pH 8.4), EDTA 1 mmol/L (pH 8.5)]; PCR buffer 10× (Fermentas, USA); deoxynucleotide triphosphates (dNTPs) 10 mmol/L (Fermentas, USA); MgCl2 25 mmol/L; simple sequence repeat (SSR) primers (MiSHRS series).
2.2. Plant materials
Mature and young leaf samples from ten commercial cultivars of mango were collected from the Germplasm Unit, Khanewal and Mango Research Station, Shujahabad while mature leaf samples of Eugenia jambolana, Psidium guajava, and Dalbergia sissoo were collected from the Forestry Area of Agriculture University, Faisalabad (Table 1). The leaf samples were washed, dried, sealed in zipper bags, and stored at −80 °C until used.
Table 1.
DNA yield and purity from mature and young leaves of ten commercial mango cultivars and three other woody plant species
| No. | Cultivar name | Leaf type | cDNA (µg/µl) | A260/A280 |
| 1 | Malda | Mature | 9.67 | 1.8 |
| 2 | Langra | Mature | 9.86 | 1.8 |
| 3 | Dusehri | Mature | 10.13 | 1.9 |
| 4 | Anwar Ratole | Mature | 9.50 | 1.9 |
| 5 | Samar Bahisht Chaunsa | Mature | 9.67 | 1.9 |
| 6 | Fajri | Mature | 9.55 | 1.8 |
| 7 | Sensation | Mature | 10.33 | 1.8 |
| 8 | Sufaid Chaunsa | Mature | 10.50 | 1.9 |
| 9 | Kala Chaunsa | Mature | 9.83 | 1.8 |
| 10 | Late Ratole No. 12 | Mature | 9.97 | 1.9 |
| 11 | Late Ratole No. 12 | Young | 10.10 | 1.8 |
| 12 | Eugenia jambolana | Mature | 9.84 | 1.9 |
| 13 | Psidium guajava | Mature | 9.21 | 1.8 |
| 14 | Dalbergia sissoo | Mature | 9.21 | 1.8 |
c DNA: DNA concentration
2.3. Extraction protocol
The following protocols were adopted for extracting good quality DNA from mature leaves. (1) Preheat the extraction buffer to 65 °C. (2) Crush the leaf samples (2 g) stored in zipper bags at −80 °C between the palms. (3) Place the crushed leaf samples in a pre-cooled pestle and mortar and grind to very fine powder using liquid nitrogen. (4) Transfer the powdered leaf sample to a 50-ml Falcon tube and add 10 ml of hot extraction buffer to the tube before the frozen powder starts thawing. (5) Invert the tubes several times to mix the ingredients thoroughly before incubation at 65 °C for 60‒80 min. (6) Add an equal volume of chloroform-isoamyl alcohol (24:1, v/v) and mix gently by inverting the tubes to form an emulsion. (7) Spin for 15 min at 5 200×g at room temperature. (8) Carefully transfer the supernatant solution (the top aqueous phase) to a new Falcon tube and discard the remaining organic phase. (9) Perform Steps 6 to 8 two or three times (for young leaves, 1‒2 treatments yield equivalent results). (10) Add 6 ml of isopropanol (prechilled) and mix gently by inverting the tube; place the tubes at −20 °C for 10‒15 min (the precipitated DNA will be visible at this step). (11) Spin at 8 500×g for 10 min and discard the supernatant solution. (12) Wash the pellets two or three times with 70% ethanol. (13) Invert the tube on a paper towel and let the pellets air-dry. (14) Resuspend the DNA in 200 µl 0.1× TE buffer, which was then treated with RNase (add 1 µl of stock RNase per 20 µl of DNA solution) and incubated at 37 °C for 2 h. (15) Treat with 200 µl of chloroform-isoamyl alcohol and mix gently. (16) Spin for 10 min at 5 200×g and transfer the supernatant into a new Eppendorf tube. (17) Add 1/10 volume of 3 mol/L NaCl or 1/10 volume of 3 mol/L Na acetate (equivalent to NaCl in performance), mix and precipitate the DNA with two volumes of chilled 100% ethanol (an equal volume of chilled isopropanol can also be used). (18) Spin for 10 min at 8 500×g, discard the supernatant and wash the pellets with 70% ethanol. (19) Air-dry the pellet and dissolve in 100 µl 0.1× TE. (20) The DNA concentration can be measured by taking absorbance at 260 nm, according to Sambrook et al. (1989) or by running aliquots on a 1% agarose gel (0.01 g/ml). (21) The purity of DNA can be determined by estimating the ratio of absorbance at 260 nm to that at 280 nm (A 260/A 280).
2.4. PCR amplification
A set of 35 SSR primers (Duval et al., 2005; Schnell et al., 2005) was used for amplification. PCR reaction mixture (20 µl) contained 20 ng/µl DNA, 10× PCR buffer, 10 mmol/L dNTPs, 50 mmol/L MgCl2, and 10 µmol/L each of forward and reverse primers. The amplification was carried out in a thermal cycler (Bio-Rad C-1000) using a program configured with a denaturation step of 5 min at 94 °C followed by 30 cycles of 30 s at 94 °C, 30 s at an appropriate annealing temperature, and 1 min at 72 °C. The program ended with one final extension at 72 °C for 8 min. The amplified products were separated by electrophoresis on a 3% agarose gel (0.03 g/ml) containing ethidium bromide.
3. Results
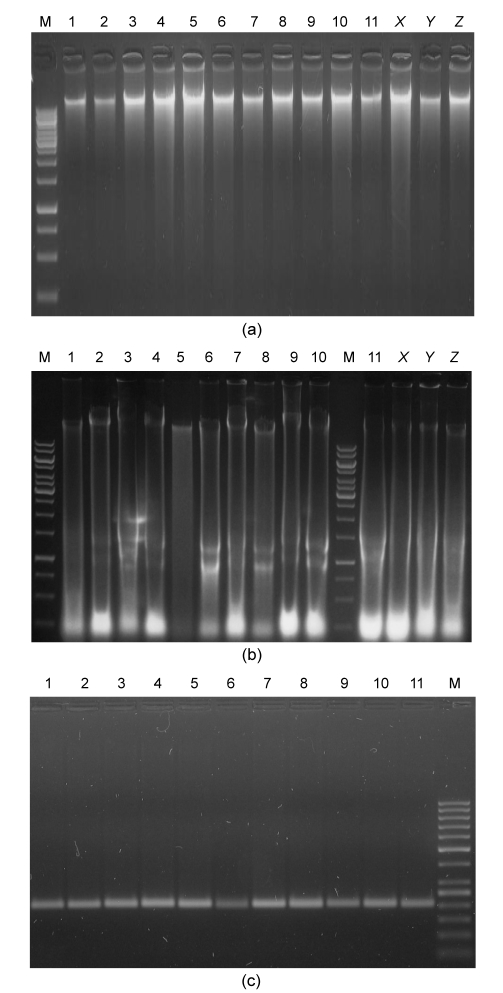
The modified protocol yielded high concentrations (950‒1 050 µg) of good quality DNA (A 260/A 280 1.80‒1.95) fit for PCR applications (Table 1 and Fig. 1). A total of 35 SSR primers were used in PCR amplification, of which 33 yielded good amplifications (Fig. 1).
Fig. 1.
Efficiency of modified DNA extraction protocol developed mainly for mature leaves of mango and other phenol- and polysaccharide-rich woody plant species
(a) DNA extraction from mature and young leaves of mango and other woody plants using the modified protocol; (b) DNA extraction from mature and young leaves of mango and other woody plants using a method as described by Doyle and Doyle (1987); (c) SSR-PCR of extracted DNA using SSR primers. M: (a, b) 1 kb DNA ladder, (c) 50 bp DNA ladder; 1: Malda; 2: Langra; 3: Dusehri; 4: Anwar Ratole; 5: Samar Bahisht Chaunsa; 6: Fajri; 7: Sensation; 8: Sufaid Chaunsa; 9: Kala Chaunsa; 10: Late Ratole No. 12 (mature); 11: Late Ratole No. 12 (young); X: Eugenia jambolana; Y: Psidium guajava; Z: Dalbergia sissoo
4. Discussion
Generally, mature plant tissues are not preferred for DNA extraction due mainly to the presence of high concentrations of polysaccharides, polyphenols, and other secondary metabolites (Dabo et al., 1993; Zhang and McD. Steward, 2000). This problem is exacerbated in the case of fully expanded mature mango leaves.
The CTAB extraction method originally developed by Doyle and Doyle (1987) was modified to remove polysaccharide, polyphenols, and other secondary metabolites. Polysaccharides inhibit PCR amplifications and can lead to erroneous interpretations (Kotchoni et al., 2003). Co-precipitation of polysaccharide was avoided by adding higher concentrations of selective precipitants of nucleic acid, CTAB (0.04 g/ml), and NaCl (3 mol/L) (Dellaporta et al., 1983). Long-tail surfactants (such as CTAB) produce a conformational change in the DNA from “random coil” to “compact globule”, making DNA precipitation more effective. Phenolic compounds are powerful oxidizing agents and bind covalently to the extracted DNA, making it useless for most of molecular manipulations (Porebski et al., 1997; Padmalatha and Prasad, 2006). A high concentration (0.025 g/ml) of PVP mixed in the extraction buffer (Fang et al., 1992; John, 1992; Moller et al., 1992; Lodhi et al., 1995) binds to phenolic compounds and helps in their removal. The superfluous quantities of cellular proteins were managed by triple extended treatment with chloroform-isoamyl alcohol. In addition to the removal of proteins, this treatment also helped to remove different coloring substances such as chlorophyll, pigments, and dyes. An extended 2-h RNase treatment followed by chloroform-isoamyl alcohol treatment ensured RNA- and protein-free DNA product fit for PCR amplification (Saghai-Maroof et al., 1984).
DNA isolated by this method yielded reproducible and consistent amplification products proving its compatibility for PCR applications using SSR primers (Fig. 1).
The modifications described above provide the opportunity to successfully collect good quality DNA from mature mango leaves for PCR applications. This protocol has the potential to extract DNA from the mature leaves of other species high in polysaccharides and polyphenols, when sufficient young leaf tissue is not available.
Acknowledgments
Positive inputs from Mr. Usman ASLAM (Centre of Agricultural Biochemistry and Biotechnology, University of Agriculture, Faisalabad, Pakistan), Ms. Sana IKRAM (Institute of Horticultural Sciences, University of Agriculture, Faisalabad, Pakistan), and Ms. Roomana ANJUM (Department of Plant Pathology, University of Agriculture, Faisalabad, Pakistan) are acknowledged.
Footnotes
Project supported by Punjab Agricultural Research Board (PARB), and the project No. 150 awarded to Dr. Iqrar Ahmad KHAN, Pakistan
References
- 1.Bompard JM. The genus Mangifera rediscovered: the potential contribution of wild species to mango cultivation. Acta Hort. 1993;341:69–77. [Google Scholar]
- 2.Dabo SM, Mitchell ED, Melcher U. A method for the isolation of nuclear DNA from cotton (Gossypium) leaves. Anal Biochem. 1993;210(1):34–38. doi: 10.1006/abio.1993.1146. [DOI] [PubMed] [Google Scholar]
- 3.Davis TM, Yu H, Haigis KM, McGowan PJ. Template mixing: a method of enhancing detection and interpretation of codominant RAPD markers. Theor Appl Genet. 1995;91(4):582–588. doi: 10.1007/BF00223283. [DOI] [PubMed] [Google Scholar]
- 4.Dellaporta SL, Wood J, Hicks JB. A plant DNA mini-preparation: version 2. Plant Mol Biol Rep. 1983;1(4):19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 5.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 6.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 7.Duval MF, Bunel J, Sitbon C, Risterucci AM. Development of microsatellite markers for mango (Mangifera indica L.) Mol Ecol Notes. 2005;5(4):824–826. doi: 10.1111/j.1471-8286.2005.01076.x. [DOI] [Google Scholar]
- 8.Fang G, Hamrnar S, Grumet R. A quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biofeedback. 1992;13(1):52–54. [PubMed] [Google Scholar]
- 9.John ME. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucleic Acid Res. 1992;20(9):2381. doi: 10.1093/nar/20.9.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotchoni SO, Gachomo EW, Betiku E, Shonukan OO. A home made kit for plasmid DNA mini preparation. Afr J Biotechnol. 2003;2(4):88–90. [Google Scholar]
- 11.Lodhi MA, Daly MA, Weeden NF, Reisch BI. A molecular marker based linkage map of Vitis . Genome. 1995;38(4):786–794. doi: 10.1139/g95-100. [DOI] [PubMed] [Google Scholar]
- 12.Manoj K, Tushar B, Sushama C. Isolation and purification of genomic DNA from Black Plum (Eugenia jambolana Lam.) for analytical applications. Int J Biotechnol Biochem. 2007;3(1):49–55. [Google Scholar]
- 13.Moller EM, Bahnweg G, Sandermann H, Geiger HH. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies and infected plant tissues. Nucleic Acids Res. 1992;20(22):6115–6116. doi: 10.1093/nar/20.22.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmalatha K, Prasad MNV. Optimization of DNA isolation and PCR protocol for RAPD analysis of selected medicinal and aromatic plants of conservation concern from Peninsular India. Afr J Biotechnol. 2006;5(3):230–234. [Google Scholar]
- 15.Pirttilä MA, Hirsikorpi M, Kämäräinen T, Jaakola L, Hohtola A. DNA isolation methods for medicinal and aromatic plants. Plant Mol Biol Rep. 2001;19(3):273. doi: 10.1007/BF02772901. [DOI] [Google Scholar]
- 16.Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15(1):8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- 17.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. PNAS. 1984;81(24):8014–8019. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring, Harbour, New York, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Schnell RJ, Knight RJ., Jr Frequency of zygotic seedlings from polyembryonic mango rootstocks. HortScience. 1992;27(2):174–176. [Google Scholar]
- 20.Schnell RJ, Olano CT, Quintanilla WE, Meerow AW. Isolation and characterization of 15 microsatellite loci from mango (Mangifera indica L.) and cross-species amplification in closely related taxa. Mol Ecol Notes. 2005;5(3):625–627. doi: 10.1111/j.1471-8286.2005.01018.x. [DOI] [Google Scholar]
- 21.Sharma AD, Gill PK, Singh P. DNA isolation from dry and fresh samples of polysaccharide-rich plants. Plant Mol Biol Rep. 2002;20(4):415. doi: 10.1007/BF02772129. [DOI] [Google Scholar]
- 22.Singh RN. Mango. New Delhi: ICAR; 1996. [Google Scholar]
- 23.Zhang J, McD. Steward JD. Economical and rapid method for extracting cotton genomic DNA. J Cotton Sci. 2000;4(3):193–201. [Google Scholar]
Recommended reading
- 24.Chen XD, Sun DF, Rong DF, Peng JH, Li CD. A recessive gene controlling male sterility sensitive to short daylength/low temperature in wheat (Triticum aestivum L.) J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2011;12(11):943–950. doi: 10.1631/jzus.B1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li YD. Developing new SSR markers from ESTs of pea (Pisum sativum L.) J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2010;11(9):702–707. doi: 10.1631/jzus.B1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou YF, Zhang XY, Xue QZ. Fine mapping and candidate gene prediction of the photoperiod and thermo-sensitive genic male sterile gene pms1(t) in rice. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2011;12(6):436–447. doi: 10.1631/jzus.B1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]