Abstract
This research was aimed at isolating and identifying the predominant lactic acid bacteria (LAB) in the traditional Chinese salt-fermented soybean food, douchi, from Yunnan, China. The predominant LAB present were isolated and identified by conventional culture-dependent methods combined with molecular biological methods. Two hundred and sixty isolates were obtained from thirty kinds of traditional fermented douchi from six cities and counties in Yunnan, and those strains were divided into twelve groups by their morphological and biochemical characteristics. Based on 16S ribosomal DNA (rDNA) sequencing and analysis, 56 representative strains were identified as belonging to 6 genera and 14 species: Lactobacillus (4 spp.), Weissella (3 spp.), Pediococcus (2 spp.), Staphylococcus (2 spp.), Enterococcus (1 sp.), and Bacillus (2 spp.). The results show that douchi contains a large natural population of LAB of diverse composition from which some strains could be selected as starters for functional fermented foods. This is the first study on the original douchi from Yunnan, and the results suggest that it may be a useful source for the isolation of LAB. This study has also laid a foundation for further research on developing functional douchi products.
Keywords: Fermented food, Douchi, Lactic acid bacteria, Culture-dependent methods, 16S rDNA
1. Introduction
Douchi is a traditional Chinese salt-fermented soybean food which has not only been very well known and popular in China since ancient times, but has also been used as a medicine and for food seasoning for millennia (Zhang et al., 2006). Yunnan lies in the plateau regions of southwest China, and people of over twenty ethnicities live there. Because of its diverse landforms, changeable climate, and abundant resources, Yunnan is rich in douchi resources. Douchi is a regular part of the local diet and plays an important role in economic, social, and cultural development. Although it is very popular in Yunnan, douchi has not been studied in great detail to date. In Yunnan, it is made by a traditional processing method: soybean [Glycine max (L.)] seeds are washed and soaked in water for 24 h, then boiled for 1‒2 h, dried and packed tightly in a small bamboo basket layered with leaves of bamboo (Bambusoideae spp.) or banana (Musa spp.). The baskets are incubated and covered with the leaves of soybean plants to maintain an above-ambient temperature. After 3 to 5 d of fermenting, salt (NaCl) is added to about 12%‒15% (w/w), and spices (such as sugar, Chinese prickly ash, fresh hot pepper paste, or dry hot pepper powder) are added, and the mixture is packed in a tank for about one month.
Lactic acid bacteria (LAB) belong to a group of Gram-positive bacteria that excrete lactic acid as their main fermentation product into the culture medium, and are generally recognized as safe (Konings et al., 2000). Nowadays, LAB are important for the food and dairy industries because the lactic acid and other organic acids produced by these bacteria act as natural preservatives and flavour enhancers. LAB are also regarded as probiotics which are able to stimulate immune responses and prevent infections against enteropathogenic bacteria (Reid, 1999). Thus, LAB could contribute to food safety. Research has shown that LAB exist in fermented soybean food, such as tempeh and douchi in Taiwan (Moreno et al., 2002; Chen et al., 2006). It is important to define the exact composition of native douchi LAB and distinguish them down to the subspecies level, because this may raise the status of douchi, increase its marketability and profitability, and make it more commercializable.
Traditional bacterial classification methods based on morphological, physiological, and biochemical tests can be time-consuming, misleading, and laborious (Wattiau et al., 2001). To overcome these shortfalls, various methods that use DNA typing for molecular identification of microbial resources have been developed. More convenient and accurate identifications are achievable using nucleotide base sequencing of 16S ribosomal DNA (rDNA), which provides a basis for phylogenetic analysis and identification (Chin et al., 2006). Several studies have rapidly classified LAB based on 16S rDNA sequencing and this method can be used to identify and distinguish the LAB at the subspecies level (Kim B. et al., 2003; Chao et al., 2008).
This research was aimed at isolating and identifying the predominant LAB microflora from Yunnan traditional fermented douchi by conventional culture-dependent methods combined with molecular biological methods. Furthermore, the goal of this study was to determine the natural population of LAB in douchi and construct a phylogenetic tree of these microorganisms.
2. Materials and methods
2.1. Collection of samples
Thirty douchi samples were collected from six cities and counties within the main douchi-producing areas of Yunnan (Table 1). Each 100 g sample, prepared using traditional household methods, was collected and put into a sterilized polyethylene sampling bag, then transported to the laboratory where it was stored immediately at 4 °C. Analysis within two days of sample collection was considered optimal.
Table 1.
Average viable LAB count on MRS agar of different douchi samples from different producing areas
| Producing area | Sampling point | Douchi sample | Average viable cell count (CFU/ml) |
Number of isolates | |
| For each sample | For each sampling point | ||||
| Central Yunnan | Yimen | YM-1 | (7.9±0.4)×108 | (4.8±6.0)×108 | 13 |
| Yimen | YM-2 | (1.6±0.2)×109 | 9 | ||
| Yimen | YM-3 | (4.7±0.4)×108 | 7 | ||
| Yimen | YM-4 | (4.6±0.3)×107 | ND | ||
| Yimen | YM-5 | (7.5±0.5)×107 | 9 | ||
| Yimen | YM-6 | (1.0±0.1)×106 | 8 | ||
| Northeast Yunnan | Yongshan | ZT-1 | (3.6±1.1)×108 | (5.6±7.0)×108 | 8 |
| Yongshan | ZT-2 | (5.8±0.9)×108 | 18 | ||
| Yongshan | ZT-3 | (2.0±0.2)×109 | 10 | ||
| Yongshan | ZT-4 | (9.3±1.1)×107 | 10 | ||
| South Yunnan | Jianshui | JS-1 | (7.1±1.1)×107 | (5.2±5.9)×108 | 3 |
| Jianshui | JS-2 | (4.4±0.9)×107 | 2 | ||
| Jianshui | JS-3 | (1.2±0.3)×109 | 19 | ||
| Jianshui | JS-4 | (2.6±0.3)×108 | 5 | ||
| Jianshui | JS-5 | (9.3±1.3)×107 | 8 | ||
| Jianshui | JS-6 | (3.2±0.5)×108 | 1 | ||
| Jianshui | JS-7 | (1.8±0.1)×109 | 3 | ||
| Jianshui | JS-8 | (5.4±0.7)×108 | 5 | ||
| Jianshui | JS-9 | (3.3±0.3)×108 | 7 | ||
| Yuanyang | YH-10 | (1.5±0.4)×109 | (1.1±0.5)×109 | 5 | |
| Yuanyang | YH-11 | (8.5±0.5)×108 | 3 | ||
| Yuanyang | YH-12 | (1.2±0.2)×109 | 4 | ||
| Yuanyang | YH-13 | (1.4±0.3)×109 | 3 | ||
| Yuanyang | YH-14 | (4.3±0.4)×108 | 4 | ||
| Southeast Yunnan | Qiubei | QB-1 | (1.1±0.1)×109 | (1.1±0.3)×109 | 16 |
| Qiubei | QB-2 | (8.0±0.2)×108 | 13 | ||
| Qiubei | QB-3 | (1.3±0.4)×109 | 19 | ||
| West Yunnan | Shangri-la | XG-1 | (1.4±0.3)×109 | (1.4±0.6)×109 | 20 |
| Shangri-la | XG-2 | (7.4±0.4)×108 | 2 | ||
| Shangri-la | XG-3 | (2.0±0.2)×109 | 26 | ||
CFU: colony forming unit; ND: not determined, no strains were isolated from douchi samples due to lack of growth
2.2. Isolation of LAB
To isolate LAB from douchi, direct spreading and accumulation methods were used (Chen et al., 2005). For both methods, appropriate dilutions were spread onto acidic de Man-Rogosa-Sharpe (MRS) agar plates (pH 6.3, Oxoid Ltd., Basingtoke, Hampshire, England) supplemented with 0.04 g/L bromocresol purple and 0.01 g/ml CaCO3 (de Man et al., 1960; Lim and Im, 2009). All the plates were incubated under anaerobic conditions (AnaeroPack Rectangular Jar, Mitsubishi Gas Chemical Co. Inc., Japan) for 48‒72 h at 35 °C. After incubation, the colonies of LAB on the MRS agar plates were counted.
Typical colonies with different morphologies and which formed a clear zone were randomly isolated from the MRS agar plates using sterilized toothpicks. Five to ten LAB strains from MRS agar plates of each of the fermented douchi samples were randomly selected according to Harrigan and McMance (1976). All the isolates were cultured in MRS broth, and purified by replating on MRS agar plates (Chen et al., 2006). Prior to the molecular identification, the phenotypes of microbial isolates were determined based on colony and cell morphology. Then, the isolates were checked by Gram staining and catalase reaction (Mohd Adnan and Tan, 2007). Only isolates which had clear halos and were catalase-negative and Gram-positive, were chosen for further analysis. These isolates were stored at −80 °C in MRS broth containing 50% glycerol prior to molecular analysis (Benito et al., 2008).
2.3. Identification of LAB isolates
Total DNA was extracted from 1.5 ml of each isolate MRS broth, as described by Schmidt et al. (1991), with slight modification. Briefly, samples were centrifuged and the sediments were washed using water. The centrifuged cells were treated with lysozyme (1 mg/ml finally). After incubation for 2 h at 37 °C, samples were treated with sodium dodecyl sulfate [SDS, 1% (0.01 g/ml) finally] and proteinase K (0.4 mg/ml finally) and incubated for 1 h at 37 °C. Next, cetyltrimethylammonium bromide [CTAB, 1% (0.01 g/ml) finally] was added to the tubes which were then incubated at 65 °C in a water bath for 20 min. Cell lysates were extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, v/v/v), mixed thoroughly, and centrifuged at 15 000 r/min for 10 min. The upper phases of the centrifugation products were transferred to new tubes and an equal volume of chloroform/isoamyl alcohol (24:1, v/v) was added. Samples were then centrifuged at 15 000 r/min for 10 min and the upper phases were transferred to new tubes. A 0.6-volume of ice cold isopropanol was added to the samples. An additional centrifugation was performed at 12 000 r/min for 15 min at 4 °C. The pellet was then washed with ice-cold 70% ethanol, dried, and resuspended in TE buffer [10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L ethylenediaminetetraacetic acid (EDTA)].
Sequence analysis of the 16S rDNA was used to classify and identify the LAB isolates. The 16S rDNA polymerase chain reaction (PCR) amplifications of isolated strains were performed with two universal primers, 27F (5′-AGA GTT TGA TCA TGG CTC AG-3′) and 1492R (5′-TAC GGT TAC CTT GTT ACG ACT T-3′) (Kim et al., 2009). The PCR reaction was carried out using a TaKaRa Ex Taq gene amplification PCR kit (TaKaRa, Dalian, China) and performed on an ABI PCR System 2720 (Applied Biosystems, Singapore). The thermocycle program was as follows: 95 °C for 5 min; 35 cycles at 95 °C for 1 min, 60 °C for 1 min, and 72 °C for 2 min; and a final extension step at 72 °C for 10 min. After cycling, the PCR products were separated by electrophoresis on a 1% agarose gel (0.01 g/ml) containing ethidium bromide in 1× Tris-acetate-EDTA (TAE) buffer, and the results of the 1% agarose gel electrophoresis revealed a 1 466-bp PCR product. The bacterial 16S rDNA amplicons were purified using a PCR clean-up gel extraction kit (Macherey-Nagel GmbH, Germany) according to the manufacturer’s instructions, and were confirmed by 1% agarose gel electrophoresis. For phylogenic identification, the target regions of 16S rDNA were partially sequenced using a BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI 3730 PRISM genetic analyzer (Applied Biosystems, USA).
The 16S rDNA sequence homologies were examined by comparing the sequences obtained with those in the National Center for Biotechnology Information (NCBI) GenBank database and by using the online analysis tool EzTaxon (Chun et al., 2007). For phylogenetic analysis, sequences were aligned using the CLUSTAL X software (Thompson et al., 1997; Larkin et al., 2007). A phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei, 1987) with 500 bootstrap pseudoreplicates, using MEGA 4.0.1 (Tamura et al., 2007).
3. Results
3.1. LAB counts and strain isolation
LAB in thirty douchi samples were evaluated under anaerobic growth conditions on semi-selective MRS agar plates. All of the isolates which were Gram-positive produced white or light-yellowish colonies. Samples of YM-2, JS-3, JS-7, YH-10, YH-12, YH-13, ZT-3, QB-1, QB-3, XG-1, and XG-3 had the highest amounts of LAB on MRS agar, and among the samples ZT-3 and XG-3 have the most cells, at 2.0×109 colony forming unit per ml (CFU/ml). By contrast, sample YM-6 had the lowest cell count at 1.0×106 CFU/ml (Table 1). All of the samples from Shangri-la in west Yunnan had the highest average amount of LAB cells (Table 1). Two hundred and sixty acid-producing bacterial colonies were selected from all the plates randomly and were divided into twelve groups according to their morphological and biochemical characteristics, and then used for further studies.
3.2. Identification and phylogenetic analyses of LAB from douchi
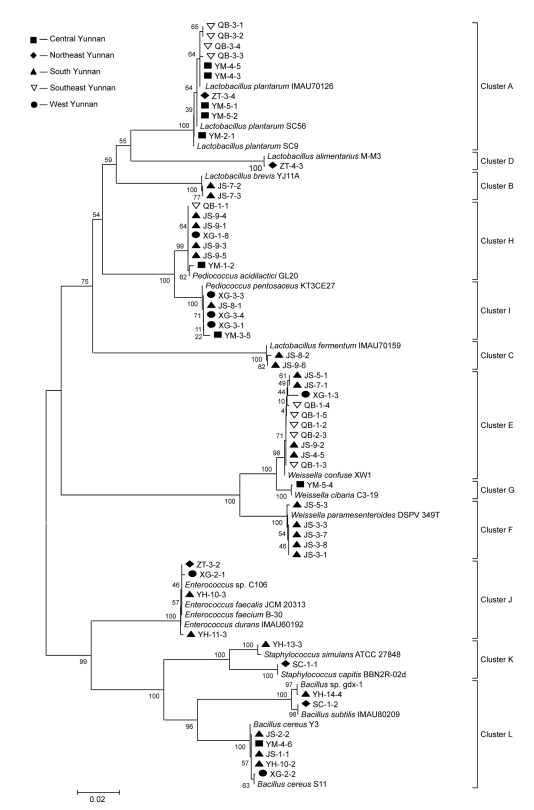
Fifty-six representative acid-producing bacterial strains which were Gram-positive and had clear halos on MRS agar were selected from the twelve bacterial groups, and were identified by their 16S rRNA gene sequences. The fifty-six representative strains isolated from the original fermented douchi were classified into six genera and fourteen species by partial sequencing of their 16S rDNA. Forty-nine of the strains were identified as LAB, including the following species: ten Lactobacillus plantarum, two L. brevis, two L. fermentum, one L. alimentarius, ten Weissella confuse, five W. paramesenteroides, one W. cibaria, seven Pediococcus acidilactici, five P. pentosaceus, four Enterococcus faecalis (identified by their biochemical characteristics, data not shown), one Staphylococcus simulans, and one S. capitis. The other acid-producing bacteria included seven strains of Bacillus sp. (Fig. 1).
Fig. 1.
A phylogenetic tree showing the relationship of 16S rDNA sequences in douchi samples from five producing areas in Yunnan
The tree was constructed using neighbor-joining analysis based on partial 16S rDNA sequences. Numbers at nodes indicate bootstrap values (n=500 replicates). Bar: 0.02 nucleotides substitution per site
The phylogenetic relationships among the partial 16S rRNA gene sequences of the 56 representative strains are shown in Fig. 1. The phylogenetic tree clearly shows that all the acid-producing bacterial strains were grouped into two categories at a similarity level of 76%, and all of the screened strains were assigned to twelve clusters (Fig. 1). The sequences affiliated to L. plantarum were isolated mostly from samples from central and southeast Yunnan. Sequences which were close to the genus Pediococcus were found in nearly all the samples from the different areas in Yunnan Province. The sequences which had more than 99% similarity to the genus Weissella were detected mostly in samples from south and southeast Yunnan. Some sequences from almost all the areas in Yunnan Province could be assigned to the genera Enterococcus, Staphylococcus, and Bacillus.
4. Discussion
In this study, the community structure of the predominant LAB in spontaneously fermented douchi was studied by conventional culture-dependent methods combined with molecular biological methods. LAB strains were identified on the basis of their genotypic characteristics, using direct DNA extraction, 16S rDNA amplification and sequencing. To our knowledge, this is the first study to investigate the natural populations of LAB in douchi using culture-dependent methods combined with molecular biological methods in China. This research will play an important role in the development of fermented soybean food. The subject of this study was the molecular identification of LAB populations obtained from 30 different douchi samples from Yunnan.
Based on these combined culture-dependent and molecular biological methods, a diverse range of LAB genera were detected in douchi. Six genera and over fourteen species were identified (Table 2). Based on the results (Fig. 1), Lactobacillus spp., Weissella spp., and Pediococcus spp. were the predominant LAB in douchi samples collected from Yunnan, China. The 16S rDNA sequences of the four strains from southeast Yunnan and the five strains from central Yunnan were affiliated to L. plantarum which has been found to play an important role in many types of fermented food (Briggiler Marcó et al., 2011; Liu et al., 2011; Won et al., 2011; Zago et al., 2011). Nearly all sequences which were associated with W. paramesenteroides were detected in samples from south Yunnan, and sequences affiliated to W. confuse were detected in the samples from south and southeast Yunnan. The genus Weissella has also been recognized as activated LAB in fermented food (Ndagano et al., 2011). The distribution of the strains did not show any significant trend among the sample areas. Some rare LAB strains were also found in the douchi samples, such as W. cibaria, S. simulans, and S. capitis (Table 2).
Table 2.
Numbers of LAB species isolated from different douchi producing areas of Yunnan
| Genus | Species | Number of LAB species |
||||
| Central Yunnan | Northeast Yunnan | South Yunnan | Southeast Yunnan | West Yunnan | ||
| Lactobacillus | L. plantarum | 5 | 1 | 0 | 4 | 0 |
| L. brevis | 0 | 0 | 2 | 0 | 0 | |
| L. fermentum | 0 | 0 | 2 | 0 | 0 | |
| L. alimentarius | 0 | 1 | 0 | 0 | 0 | |
| Weissella | W. confuse | 0 | 0 | 4 | 5 | 1 |
| W. paramesenteroides | 0 | 0 | 5 | 0 | 0 | |
| W. cibaria | 1 | 0 | 0 | 0 | 0 | |
| Pediococcus | P. acidilactici | 1 | 0 | 4 | 1 | 1 |
| P. pentosaceus | 1 | 0 | 1 | 0 | 3 | |
| Enterococcus | E. faecalis | 0 | 1 | 2 | 0 | 1 |
| Staphylococcus | S. simulans | 0 | 0 | 1 | 0 | 0 |
| S. capitis | 0 | 1 | 0 | 0 | 0 | |
| Others | Bacillus sp. | 1 | 1 | 4 | 0 | 1 |
The results also showed that different species were isolated from different samples (Table 2). This could be related to the different sampling points and processing technologies. However, it is necessary to confirm this trend by large-scale sampling and isolation. When phylogenetic analysis was carried out, a LAB population was found in all samples, and six genera were found in each douchi producing area (Fig. 1, Table 2). Nevertheless, some areas had specific LAB populations. Lactobacillus spp. were not found in samples from west Yunnan (Shangri-la); the southern producing areas (Jianshui and Yuanyang) had the largest populations of LAB, but no L. plantarum was found in samples from those areas; in northeast Yunnan (Yongshan), the isolated strains were significantly different from those found in other areas (Fig. 1). When identifying Enterococcus species from 16S rDNA sequences, high sequence homologies were observed among Enterococcus species (Table 2, Fig. 1 Cluster J). Other studies have shown the same results, and found it difficult to identify these strains to the species level based on the DNA sequences alone (Devriese et al., 1992; Yanagida et al., 2005). Finally, isolated Enterococcus sp. strains were identified as E. faecalis by their biochemical characteristics with an API50 CHL kit (data not shown). In most cases, strains of the same species were grouped into a unique cluster, and significant differences existed between these strains; these differences may relate to the douchi samples originating from different producing areas and processing technologies. These results demonstrate that differences in LAB species exist in spontaneously fermented douchi from Yunnan. The diversity of geographical factors and changing climatic conditions in the region may be the reasons for the diversity of LAB populations.
The other important outcome of this study relates to the LAB populations of commercially manufactured douchi and homemade douchi. Natto and tempeh are two kinds of fermented soybean food in Japan and Indonesia, respectively. Both are similar to douchi and are fermented by pure strains such as B. subtilis natto and Rhizopus oryzae, respectively. Although these two commercial soybean foods are becoming more and more popular among people around the world, there are few reports about their LAB flora. Compared with factory-produced douchi from Taiwan, Yunnan douchi has a diverse population of LAB (Chen et al., 2006).
Our results showed a different activated bacterial community structure from those found in previous studies related to fermented soybean. The predominant LAB of Taiwanese douchi are Enterococcus spp. (Tsai et al., 2007), whereas the predominant LAB in Yunnan douchi are Lactobacillus, Weissella, and Pediococcus. As to the famous fermented soybean food, natto, the activated bacteria are from the genus Bacillus (Kubo et al., 2011). These results suggest that the activated bacterial community structure varies greatly between different areas and fermentation methods. Furthermore, the samples collected from Jianshui in south Yunnan were poor in proteinase producing bacteria. It has been suggested that proteinase-producing bacteria are inhibited by the LAB from douchi. However, more studies are needed to confirm this phenomenon.
In conclusion, various LAB exist in douchi and identification of the LAB species that dominate the spontaneously fermented douchi is an important step in the development of new starter cultures for soybean fermentation. As for methods of isolation, accumulation under anaerobic conditions appears to be a suitable method for isolating LAB from douchi. Among the LAB strains, four E. faecalis, one S. capitis, and five B. cereus strains were detected in the douchi samples (Table 2). This indicates that consumption of douchi could be potentially hazardous, which might compromise the product in terms of food safety (Nout et al., 1998; Abriouel et al., 2008). To solve this problem, stricter hygienic measures during the manufacturing process and appropriate starter cultures should be adopted. Some douchi-isolated LAB strains showed antibacterial activities and other physiological functions. Future studies in our laboratory will characterize and identify the functional starter cultures and the physiologically active substances of these isolates. The data presented in this paper will provide a useful framework for further studies of the LAB population dynamics of douchi fermentation. To facilitate commercialization and increase consumer confidence in the fermented soybean food, douchi, in Yunnan, the quality control of starters and more accurate species labeling by molecular techniques are required.
Acknowledgments
The authors would like to thank Mr. Hong-wei LI, Qiubei No. 1 Middle School, Yunnan, China, and Ms. Qiong-ying LU, Jianshui No. 1 Middle School, Yunnan, China, for their help in the sample collection.
Footnotes
Project supported by the State Key Laboratory of Food Science and Technology of Jiangnan University (No. SKLF-KF-200805) and the Yunnan Provincial Science and Technology Department (No. KKSA200926038), China
References
- 1.Abriouel H, Martín-Platero A, Maqueda M, Valdivia E, Martínez-Bueno M. Biodiversity of the microbial community in a Spanish farmhouse cheese as revealed by culture-dependent and culture-independent methods. Int J Food Microbiol. 2008;127(3):200–208. doi: 10.1016/j.ijfoodmicro.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Benito MJ, Serradillaa MJ, Ruiz-Moyano S, Martín A, Pérez-Nevado F, Córdoba MG. Rapid differentiation of lactic acid bacteria from autochthonous fermentation of Iberian dry-fermented sausages. Meat Sci. 2008;80(3):656–661. doi: 10.1016/j.meatsci.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Briggiler Marcó M, Mercanti D, Reinheimer JA, Quiberoni A. Performance of spontaneous phage-resistant derivatives of Lactobacillus plantarum in fermented milk manufacture. Int Dairy J. 2011;21(11):857–862. doi: 10.1016/j.idairyj.2011.05.011. [DOI] [Google Scholar]
- 4.Chao SH, Tomii Y, Watanabe K, Tsai YC. Diversity of lactic acid bacteria in fermented brines used to make stinky tofu. Int J Food Microbiol. 2008;123(1-2):134–141. doi: 10.1016/j.ijfoodmicro.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Chen YS, Yanagida F, Shinohara T. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett Appl Microbiol. 2005;40(3):195–200. doi: 10.1111/j.1472-765X.2005.01653.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen YS, Yanagida F, Hsu JS. Isolation and characterization of lactic acid bacteria from dochi (fermented black beans), a traditional fermented food in Taiwan. Lett Appl Microbiol. 2006;43(2):229–235. doi: 10.1111/j.1472-765X.2006.01922.x. [DOI] [PubMed] [Google Scholar]
- 7.Chin HS, Breidt F, Fleming HP, Shin WC, Yoon SS. Identification of predominant bacterial isolates from the fermenting kimchi using ITS-PCR and partial 16S rDNA sequence analyses. J Microbiol Biotechnol. 2006;16(1):68–76. [Google Scholar]
- 8.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57(10):2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 9.de Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Microbiol. 1960;23(1):130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 10.Devriese LA, Laurier L, de Herdt P, Hasbrouck F. Enterococcal and Streptococcal species isolated from faeces of calves, young cattle and dairy cows. J Appl Microbiol. 1992;72(1):29–31. doi: 10.1111/j.1365-2672.1992.tb04877.x. [DOI] [PubMed] [Google Scholar]
- 11.Harrigan WF, McMance ME. Statistical Methods for the Selection and Examination of Microbial Colonies. In: Harrigan WF, McMance ME, editors. Laboratory Methods in Food and Dairy Microbiology. London: Academic Press; 1976. pp. 47–49. [Google Scholar]
- 12.Kim B, Lee J, Jang J, Kim J, Han H. Leuconostoc inhae sp. nov., a lactic acid bacterium isolated from kimchi. Int J Syst Evol Microbiol. 2003;53(4):1123–1126. doi: 10.1099/ijs.0.02463-0. [DOI] [PubMed] [Google Scholar]
- 13.Kim TW, Lee JH, Kim SE, Park MH, Chang HC, Kim HY. Analysis of microbial communities in doenjang, a Korean fermented soybean paste, using nested PCR-denaturing gradient gel electrophoresis. Int J Food Microbiol. 2009;131(2-3):265–271. doi: 10.1016/j.ijfoodmicro.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Konings WN, Kok J, Kuipers OP, Poolman B. Lactic acid bacteria: the bugs of the new millennium. Curr Opin Microbiol. 2000;3(3):276–282. doi: 10.1016/S1369-5274(00)00089-8. [DOI] [PubMed] [Google Scholar]
- 15.Kubo Y, Rooney AP, Tsukakoshi Y, Nakagawa R, Hasegawa H, Kimura K. Phylogenetic analysis of Bacillus subtilis strains applicable to natto (fermented soybean) production. Appl Environ Microbiol. 2011;77(18):6463–6469. doi: 10.1128/AEM.00448-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 17.Lim SM, Im DS. Screening and characterization of probiotic lactic acid bacteria isolated from Korean fermented foods. J Microbiol Biotechnol. 2009;19(2):178–186. doi: 10.4014/jmb.0804.269. [DOI] [PubMed] [Google Scholar]
- 18.Liu P, Shen SR, Ruan H, Zhou Q, Ma LL, He GQ. Production of conjugated linoleic acids by Lactobacillus plantarum strains isolated from naturally fermented Chinese pickles. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2011;12(11):923–930. doi: 10.1631/jzus.B1100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohd Adnan AF, Tan IK. Isolation of lactic acid bacteria from Malaysian foods and assessment of the isolates for industrial potential. Bioresour Technol. 2007;98(7):1380–1385. doi: 10.1016/j.biortech.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 20.Moreno MRF, Leisner JJ, Tee LK, Ley C, Radu S, Rusul G, Vancanneyt M, de Vuyst L. Microbial analysis of Malaysian tempeh, and characterization of two bacteriocins produced by isolates of Enterococcus faecium . J Appl Microbiol. 2002;92(1):147–157. doi: 10.1046/j.1365-2672.2002.01509.x. [DOI] [PubMed] [Google Scholar]
- 21.Ndagano D, Lamoureux T, Dortu C, Vandermoten S, Thonart P. Antifungal activity of 2 lactic acid bacteria of the Weissella genus isolated from food. J Food Sci. 2011;76(6):M305–M311. doi: 10.1111/j.1750-3841.2011.02257.x. [DOI] [PubMed] [Google Scholar]
- 22.Nout M, Jr, Bakshi D, Sarkar PK. Microbiological safety of kinema, a fermented soybean food. Food Control. 1998;9(6):357–362. doi: 10.1016/S0956-7135(98)00126-1. [DOI] [Google Scholar]
- 23.Reid G. The scientific basis for probiotic strains of Lactobacillus . Appl Environ Microbiol. 1999;65(9):3763–3766. doi: 10.1128/aem.65.9.3763-3766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):619–629. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173(14):4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Dudley J, Nei M, Kumar S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai YH, Kung HF, Chang SC, Lee TM, Wei CI. Histamine formation by histamine-forming bacteria in douche, a Chinese traditional fermented soybean product. Food Chem. 2007;103(4):1305–1311. doi: 10.1016/j.foodchem.2006.10.036. [DOI] [Google Scholar]
- 29.Wattiau P, Renard ME, Ledent P, Debois V, Blackkman G, Agathos SN. A PCR test to identify Bacillus subtilis and closely related species and its application to the monitoring of waste water treatment. Appl Microbiol Biotechnol. 2001;56(5-6):816–819. doi: 10.1007/s002530100691. [DOI] [PubMed] [Google Scholar]
- 30.Won TJ, Kim B, Oh ES, Bang JS, Lee YJ, Yoo JS, Yu H, Yoon J, Hyung KE, Park SY, et al. Immunomodulatory activity of Lactobacillus strains isolated from fermented vegetables and infant stool. Can J Physiol Pharmacol. 2011;89(6):429–434. doi: 10.1139/y11-047. [DOI] [PubMed] [Google Scholar]
- 31.Yanagida F, Chen YS, Shinohara T. Isolation and characterization of lactic acid bacteria from soils in vineyards. J Gen Appl Microbiol. 2005;51(5):313–318. doi: 10.2323/jgam.51.313. [DOI] [PubMed] [Google Scholar]
- 32.Zago M, Fornasari ME, Carminati D, Burns P, Suàrez V, Vinderola G, Reinheimer J, Giraffa G. Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011;28(5):1033–1040. doi: 10.1016/j.fm.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Zhang JH, Tatsumi E, Ding CH, Li LT. Angiotensin I-converting enzyme inhibitory peptides in douchi, a Chinese traditional fermented soybean product. Food Chem. 2006;98(3):551–557. doi: 10.1016/j.foodchem.2005.06.024. [DOI] [Google Scholar]
Recommended reading
- 34.Carraro L, Maifreni M, Bartolomeoli I, Martino ME, Novelli E, Frigo F, Marino M, Cardazzo B. Comparison of culture-dependent and -independent methods for bacterial community monitoring during Montasio cheese manufacturing. Res Microbiol. 2011;162:231–239. doi: 10.1016/j.resmic.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Liu P, Shen SR, Ruan H, Zhou Q, Ma LL, He GQ. Production of conjugated linoleic acids by Lactobacillus plantarum strains isolated from naturally fermented Chinese pickles. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2011;12(11):923–930. doi: 10.1631/jzus.B1100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YS, Kim MC, Kwon SW, Kim SJ, Park IC, Ka JO, Weon HY. Analyses of bacterial communities in meju, a Korean traditional fermented soybean bricks, by cultivation-based and pyrosequencing methods. J Microbiol. 2011;49(3):340–348. doi: 10.1007/s12275-011-0302-3. [DOI] [PubMed] [Google Scholar]
- 37.Martin-Platero AM, Valdivia E, Maqueda M, Martin-Sanchez I, Martinez-Bueno M. Polyphasic approach to bacterial dynamics during the ripening of Spanish farmhouse cheese, using culture-dependent and -independent methods. Appl Environ Microbiol. 2008;74(18):5662–5673. doi: 10.1128/aem.00418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi JH, Xiao YP, Li XR, Ma EB, Du XW, Quan ZX. Analyses of microbial consortia in the starter of Fen Liquor. Lett Appl Microbiol. 2009;48(4):478–485. doi: 10.1111/j.1472-765X.2009.02554.x. [DOI] [PubMed] [Google Scholar]