Abstract
UHRF1 (Ubiquitin-like, with PHD and RING finger domains 1) plays an important role in DNA CpG methylation, heterochromatin function and gene expression. Overexpression of UHRF1 has been suggested to contribute to tumorigenesis. However, regulation of UHRF1 is largely unknown. Here we show that the deubiquitylase USP7 interacts with UHRF1. Using interaction-defective and catalytic mutants of USP7 for complementation experiments, we demonstrate that both physical interaction and catalytic activity of USP7 are necessary for UHRF1 ubiquitylation and stability regulation. Mass spectrometry analysis identified phosphorylation of serine (S) 652 within the USP7-interacting domain of UHRF1, which was further confirmed by a UHRF1 S652 phosphor (S652ph)-specific antibody. Importantly, the S652ph antibody identifies phosphorylated UHRF1 in mitotic cells and consistently S652 can be phosphorylated by the M phase-specific kinase CDK1-cyclin B in vitro. UHRF1 S652 phosphorylation significantly reduces UHRF1 interaction with USP7 in vitro and in vivo, which is correlated with a decreased UHRF1 stability in the M phase of the cell cycle. In contrast, UHRF1 carrying the S652A mutation, which renders UHRF1 resistant to phosphorylation at S652, is more stable. Importantly, cells carrying the S652A mutant grow more slowly suggesting that maintaining an appropriate level of UHRF1 is important for cell proliferation regulation. Taken together, our findings uncovered a cell cycle-specific signaling event that relieves UHRF1 from its interaction with USP7, thus exposing UHRF1 to proteasome-mediated degradation. These findings identify a molecular mechanism by which cellular UHRF1 level is regulated, which may impact cell proliferation.
Keywords: deubiquitination, phosphorylation UHRF1 S652
Epigenetic regulation has emerged as an important mechanism that regulates many chromatin template-based processes, including transcription, DNA replication, and repair. An important component of epigenetic regulation is DNA CpG methylation, which is mediated by DNA methyltransferases such as DMNT1 and DNMT3a/b and an accessory factor DNMT3L (1, 2). Recent studies demonstrate that maintenance of DNA methylation patterns requires UHRF1 (Ubiquitin-like, with PHD and RING finger domains 1) (also called Np95 and ICBP90). UHRF1 binds hemimethylated CpG and recruits DNMT1 to ensure faithful propagation of the DNA methylation patterns through DNA replication (3, 4). UHRF1 is also localized to euchromatic regions where it regulates transcription possibly by impacting DNA methylation and histone modifications (5, 6). UHRF1 has been shown to regulate cell proliferation, and its loss has been implicated in the mis-regulation of both G1 and G2/M phases of the cell cycle, respectively (7). However, very little is known how this important epigenetic regulator itself is regulated. To address this question, we have recently undertaken a proteomics approach and identified a cell cycle signaling-regulated physical interaction of UHRF1 with the deubiquitylase USP7 (HAUSP) (8, 9) and demonstrated that it is important for protecting UHRF1 from proteasomal degradation in a cell cycle-specific manner.
USP7 (HAUSP) functions as a deubiquitylase that regulates the stability of both p53 and MDM2 (9, 10) as well as a number of other proteins (11–13). We found that USP7 physically interacts with UHRF1. The complementation experiments using interaction-defective and catalytic mutants of USP7 firmly established the importance of physical interaction and catalytic activity of USP7 in regulating UHRF1 stability. Through physical interaction, USP7 mediates deubiquitylation of UHRF1, thus counteracting UHRF1 ubiquitylation and ubiquitin-mediated proteasomal degradation. Importantly, our data further suggest that UHRF1 is released from USP7 at the M phase of the cell cycle due to phosphorylation of UHRF1 at serine 652 located in the USP7-interacting domain. The release of UHRF1 from USP7 at the M phase is accompanied by UHRF1 degradation. Importantly, UHRF1 knockdown cells reconstituted with the phosphorylation-resistant therefore more stable UHRF1 mutant (S652A) grow more slowly than cells reconstituted with the wild-type UHRF1, suggesting the importance of maintaining an appropriate level of UHRF1 for cell proliferation.
In summary, our findings provide important insights into mechanisms that regulate UHRF1. Specifically, our results shed light on the underlying mechanism of the reduced UHRF1 stability in the M phase of the cell cycle by identifying a critical, M phase-specific phosphorylation event that releases UHRF1 from USP7, thus exposing UHRF1 to proteasomal degradation. Taken together, this study highlights UHRF1 turnover via a signaling pathway that controls protein–protein interaction as an important mode of regulation of UHRF1 function in cell proliferation.
Results
UHRF1 Affinity Purification Identifies Association with USP7 (HAUSP).
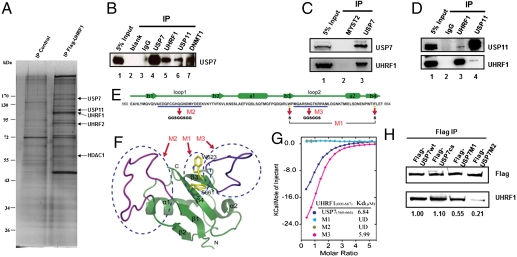
To understand UHRF1 regulation, we carried out affinity purification of FLAG epitope-tagged UHRF1 from 293T cells and identified known interacting proteins such as HDAC1 and PCNA as well as two deubiquitylases, UPS7 and USP11 (14, 15) (Fig. 1A and Table S1). Reciprocal coimmunoprecipitation (Co-IP) using antibodies directed against UHRF1, USP7, and USP11 confirmed the interaction between endogenous UHRF1 and USP7/11 (Fig. 1B, lanes 5–7, and Fig. 1 C and D), consistent with the recent reports (3, 16–19). Taken together, these results demonstrate physical interactions between UHRF1 and USP7/11 in vivo and suggest that UHRF1, DNMT1, and USP7/USP11 are in the same protein complex.
Fig. 1.
Discrete domains of UHRF1 and USP7 (HAUSP) mediate their direct interaction. (A) Tandem affinity purification of UHRF1. Human FLAG:HA tagged-UHRF1 was purified from whole cell extract of 293T cells and the associated proteins were identified by mass spectrometry using the ComPass program developed by Sowa and Harper (14). Shown here is a silver staining gel of the tagged UHRF1 from nuclear extracts. Associated polypeptides were detected by silver staining and the peptides indicated on the right were confirmed by Western blotting. (B–D) Reciprocal immunoprecipitation confirmed interaction of endogenous UHRF1 with USP7 and USP11. (B) HeLa cell lysates were immunoprecipitated with IgG, USP7, UHRF1, USP11, DNMT1 antibodies, followed by Western blot using USP7 antibody (B). (C and D) Endogenous USP7, but not USP11, interacts with UHRF1 in HeLa cells. Cell lysates were subjected to immunoprecipitation using antibodies indicated in the figures, followed by Western blotting using USP11 and UHRF1 antibody, respectively. (E) Primary sequence of UBLUSP7 is shown and secondary structural elements are indicated above the sequences. Three mutants, M1, M2, and M3, were generated and the sequence alternations were underlined. While M2 and M3 involve changes of multiple amino acids, M1 carries mutations of only two amino acids (W623S/F661S) of UBLUSP7. (F) Ribbon representation of NMR structure of UBLUSP7 with secondary structural elements indicated. NMR structure of UBLUSP7 (PDB ID code 2KVR) was used for modeling. The regions corresponding to M2 and M3 are colored in purple and blue, respectively. The two amino acid residue altered in the M1 mutant are colored in yellow. The mutations of UBLUSP7 were designed based on the previous knowledge of the reported Ubiquitin recognition (40) and the structural feature of UBLUSP7, which has the two extended loops regions that are predicated to be involved in protein–protein interaction. (G) Superimposed ITC enthalpy plots for the binding of SpacerURHF1 (syringe) with wild type and mutations of UBLUSP7 (Cell). The estimated binding affinity (Kd) numbers are listed in the insert. UD: undetectable. (H) Lysates from cells lines expressing wild-type and mutant USP7 proteins (cs, M1, and M2) (refer to Fig. 2C for details) were subjected to immunoprecipitation with Flag antibody beads, then blotted with either FLAG or UHRF1 antibodies, respectively.
Identification of Regions and Amino Acids Involved in Mediating UHRF1/USP7 Interaction.
We found recombinant UHRF1 and USP7 proteins purified from bacteria interacted with each other in vitro (Fig. S1A), suggesting that the physical interaction between these two proteins is likely to be direct. To identify the interaction domains, we first generated USP7 deletion mutants and found that the UBL (Ubiquitin-like) domain (Fig. S1B) is sufficient to mediate interactions with the full-length UHRF1 protein (Fig. S1C, lanes 7–11). Using the USP7 UBL domain (UBLUSP7), we next showed that the region of UHRF1 encompassing amino acids 600–687 (UHRF1600–687) is sufficient to interact with UBLUSP7 (Fig. S1D, lanes 7–12). Taken together, these results suggest that discrete regions of USP7 and UHRF1 mediate their physical interaction.
The solution structure of UBLUSP7 has been reported recently. The primary sequence of UBLUSP7 is shown in (Fig. 1E) and the structure is modeled in (Fig. 1F). The UBL structure is conserved in the UBL-containing proteins and the β3 and β4 turns have been shown to mediate protein–protein interactions via the conserved amino acids tryptophan (W) and phenylalanine (F). Furthermore, loops 1 and 2 represent flexible structures and may also be involved in protein–protein interactions. To address these possibilities, we generated three UBLUSP7 mutants. M1 contains the W and F to serine (W623S/F661S) substitutions in the third and fourth turns, while M2 and M3 contain multiple amino acid substitutions in loops 1 and 2, respectively. We next used isothermal titration calorimetry (ITC) to measure the interaction of wild-type and mutant UBLUSP7 with UHRF1600–687. As shown in (Fig. 1G and Table S2), the Kd of the interaction between the wild-type UBLUSP7 and UHRF1600–687 is approximately 7 μM, demonstrating robust protein–protein interactions. While mutations in loop 2 (M3) did not affect binding (Kd = ∼ 6 μM), mutations in loop 1 and the double point mutant abrogated binding, indicating that loop 1 and the third and fourth turns of UBLUSP7 are involved in physical interactions with UHRF1. Consistently, coimmunoprecipitation experiments using wild-type and USP7 mutants showed that both M1 and M2, but not the catalytic mutation, compromised USP7 interactions with UHRF1 in vivo (Fig. 1H).
Direct Physical Interaction Is Important for USP7 to Deubiquitlyate UHRF1 and to Regulate Its Stability.
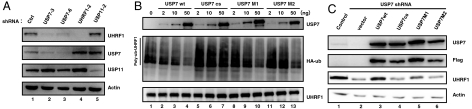
UHRF1 is ubiquitylated in vivo and is subject to regulation by ubiquitin-mediated proteolysis (20–22). As shown in Fig. S2A, Western blotting of the immunoprecipitated UHRF1 showed that UHRF1 is polyubiquitylated in vivo (compare lanes 2 and 1). Importantly, simultaneous transfection of wild-type USP7, but not the catalytically inactive mutant (C223S), significantly reduced UHRF1 ubiquitylation (Fig. S2A, compare lanes 3 and 4). Recombinant USP7 also mediated significantly more robust deubiquitylation of UHRF1 in vitro than the catalytically inactive mutant (Fig. S2B, compare lanes 2–4 with 5–7). These findings are consistent with a number of recent reports that USP7 regulates UHRF1 ubiquitylation in vitro and in vivo (19, 23, 24). Our data further demonstrated that down-regulation of USP7, but not USP11, is accompanied by a reduction of the UHRF1 protein (Fig. 2A, top panel, compare lanes 2 and 3 with lane 5), but not mRNA level (Fig. S2C), in HCT116p53+/+ as well as in a number of other cell lines, including HeLa, HT1080 and HCT116 p53-/- cells (Fig. S3 A–C). In contrast, inhibition of UHRF1 expression did not affect USP7 protein level (Fig. 2A, second panel, lane 4). Consistently, compromising endogenous USP7 function by overexpression of the catalytic inactive mutant (C223S) reduced UHRF1 steady level, while overexpression of wild-type USP7 elevated UHRF1, albeit moderately (Fig. S3D). Taken together, these findings suggest that USP7, but not USP11, regulates UHRF1 ubiquitylation and stability in vivo.
Fig. 2.
Physical interaction is important for USP7 to deubiquitylate UHRF1 and to regulate its stability. (A) Knockdown of USP7 is correlated with reduced UHRF1 steady state levels. UHRF1 USP7 and USP11 shRNAs were infected into HCT116 cells and cell lysates were subjected to Western blot analysis using UHRF1, USP7, USP11 antibodies, respectively. Actin is included as a loading control. (B) Impact of wild-type, catalytically inactive and interaction-defect mutant of USP7 on UHRF1 ubiquitylation in vitro. UHRF1-poly-Ub was purified from HEK293T cotransfected with His-UHRF1 and HA-Ub while His-USP7wt, USP7cs, USP7M1, USP7M2 were purified from insect cells. Purified UHRF1-poly-Ub was incubated with different amounts of either wild-type or mutant USP7 as indicated, and the reaction mixtures were subjected to Western blot with an HA antibody, which detects poly-ubiquitylated UHRF1. (C) Impact of wild-type, catalytically inactive and interaction-defective USP7 on UHRF1 steady state levels in vivo. HCT116 p53-/- cells stably expressing either control or USP7 shRNA-3 were cotransfected GFP empty vector with vectors expressing FLAG USP7 WT, USP7cs, USP7M1, or USP7 M2. Cells were FACS sorted 48 h posttransfection and the GFP positive cells were collected for Western blot and analysis using USP7, FLAG, UHRF1, and actin antibodies, respectively (indicated on the right).
We next investigated whether direct physical interaction of USP7 with UHRF1 is important for UHRF1 stability. As expected, wild-type but not the catalytically inactive USP7 (C223S) reduced UHRF1 ubiquitylation in vitro in a dose dependent manner (Fig. 2B, compare lanes 2–4 with 5–7). Importantly, USP7 mutants unable to interact with UHRF1 are significantly compromised in their ability to deubiquitylate UHRF1 (Fig. 2B, lanes 8–13), indicating that physical interaction between USP7 and UHRF1 is important for USP7 to mediate UHRF1 deubiquitylation. The same is true in vivo; while wild-type USP7 restored UHRF1 level in cells in which endogenous USP7 expression was inhibited by RNAi (compare lane 3 with lanes 1 and 2), the ability of the catalytically inactive (Fig. 2C, third panel, lane 4) and the interaction-defective USP7 mutants to restore UHRF1 level (Fig. 2C, third panel, lanes 5 and 6, M1 and M2) was compromised. Taken together, these results suggest that physical interaction between USP7 and UHRF1 is important for USP7 to regulate UHRF1 ubiquitylation and stability.
UHRF1 and USP7 Physical Interaction Is Regulated by Phosphorylation During Cell Cycle Progression.
What might be the physiological significance of the physical interaction between UHRF1 and USP7? While carrying out mass spectrometry analysis, we noticed phosphorylation of serine (S) 652 of UHRF1 (isoform 2) (Fig. 3A), consistent with recent proteomics studies (25, 26). Serine 652 falls within the USP7-interacting domain of UHRF1 (Fig. S1B), raising the possibility that phosphorylation of UHRF1 may regulate its association with USP7. To confirm, we raised UHRF1 S652-phosphor-specific antibodies, which only reacted with S652ph but not unmodified peptides in vitro (Fig. S4A). Furthermore, the pS652 antibodies only recognized the wild-type but not the S652A mutant of UHRF1 isolated from transfected cells by immunoprecipitation (Fig. S4B). In a third assay to determine antibody specificity, whole cell extracts were prepared from cells treated with either a UHRF1 or control shRNA and blotted by UHRF1 and pS652 antibodies, respectively. As shown in Fig. S4C, the intensity of the bands representing UHRF1 (marked by arrows) is significantly diminished by the UHRF1 but not control shRNA. It should be noted that a number of high molecular weight bands detected by the pS652 antibodies do not appear to be UHRF1, as they were not affected by the UHRF1 shRNA. Importantly, however, as shown in Fig. S4D, the pS652 antibodies mainly recognized S652 phosphorylated UHRF1 but not other phosphor proteins in vivo as the UHRF1 shRNA significantly reduced the immunostaining signal. Taken together, these findings suggest that the pS652 antibodies we developed mainly recognize phosphorylated UHRF1. We then prepared extracts from cells that were blocked with either double thymidine or thymidine-nocodazole (Fig. S4E), which arrest cells at the G1/S and M phases of the cell cycle, respectively (27). Western blot analysis showed that S652-phosphorylated UHRF1 is only found in the M but not G1/S enriched cell population (Fig. S4F). Time course analysis of cells released from G1/S by Western blotting showed that at the M phase, the pS652 level is significantly elevated while the UHRF1 protein level is diminished (about 60%) (Fig. 3B), and the latter observation is consistent with the recently published data (19). Collectively these results support the model that phosphorylation of S652 leads to a reduction of UHRF1 level. Importantly and consistently, the UHRF1 S652 phosphorylation signal is detectable by immunostaining in cells that are undergoing mitosis [Fig. 3C, Upper, DAPI-stained mitotic cells are indicated by arrows, 482 out of 529 S652ph-positive cells (91%) counted show clear chromosome condensation], coincident with a lower level of UHRF1 in mitosis cells (Fig. 3C, Lower). The amino acid sequence surrounding S652 resembles the consensus site for the cyclin-dependent kinase CDK1 (28), indeed, recombinant CDK1-cyclin B is capable of phosphorylating S652 of UHRF1 in vitro (Fig. 3D). Consistently, roscovitine, a known inhibitor of cyclin-dependent kinases including CDK1-cyclin B (29, 30), leads to an almost complete inhibition of UHRF1 S652 phosphorylation in vivo (Fig. 3E). These results suggest that UHRF1 S652 is phosphorylated by CDK1-cyclin B at the M phase of the cell cycle.
Fig. 3.
UHRF1 S652 is phophorylated in the M phase and is likely mediated by CDK1-cyclin B. (A) FLAG-UHRF1 was purified from transiently transfected 293T cells, digested with trypsin, and analyzed by LC. MS/MS spectrum of the peptides at m/z 836.83, identifying EEEEQQEGGFASPR phosphorylation on S652. (B) 293T cells were synchronized to G1/S by double thymidine block and then released. Cells collected at indicate time points were lysated for Western blot analysis using UHRF1, pS652, CCNB1, and actin antibodies. (C) HCT116 p53-/- cells were fixed and immunostained with the UHRF1 and S652 phospho-UHRF1 antibodies while DNA was stained with DAPI. In the immunostaining image some of the cells that are undergoing mitosis and show clear UHRF1 S652ph signals are circled. The same cells in the DAPI-stained image are indicated by arrows. Out of a total of 529 S652ph-positive cells counted, 428 showed clear chromosome condensation (91%) indicating that they are in the M phase of the cell cycle. Scale bars, 10 μm. (D) Wild-type UHRF1 (aa 600–687) and S652A mutants were purified from bacteria and subjected to phosphorylation by CDK1-cyclin B (purchased from NEB) in vitro and were analyzed by Western blotting using the S652 phospho-UHRF1 antibody. (E) HCT116 p53-/- cells were treated with 50 μM roscovitine (ROS) for the indicated time. Lysates were analyzed by Western blot using the S652 phospho-UHRF1, UHRF1, and actin antibodies. The S652 phospho-UHRF1 and UHRF1 band intensity was normalized against the internal actin controls.
What might be the function of this phosphorylation event? To address this issue, we first mutated S652 to aspartic acid (D), which mimics phosphorylation. As shown in Fig. 4A, ITC showed that while UHRF1600–687 interacts with UBLUSP7(aa 560–664) with a Kd of approximately 9 μM, S652D interaction with USP7 was reduced by approximately 16-fold (Kd = ∼ 150 μM), supporting the idea that S652 phosphorylation disrupts UHRF1 interaction with USP7. We also established stable cell lines expressing FLAG-tagged, wild-type, S652A and S652D, respectively. Importantly, the S652A mutation, which renders UHRF1 resistant to phosphorylation at S652, appears to be more stable than wildtype UHRF1 as evidenced by a reduced turnover rate of the mutant, while UHRF1 carrying the S652D mutation, which mimics S652 phosphorylation, is indeed intrinsically less unstable than wild-type UHRF1 (Fig. 4B). Consistently with our model, the S652D mutation, which mimics phosphorylation at S652, significantly reduced the interaction of with USP7 as determined by Co-IP (Fig. S4G). Importantly, the endogenous UHRF1, which is phosphorylated at S652, also showed reduced physical interaction with USP7 (Fig. 4C) and enhanced turnover at the M phase of the cell cycle (Fig. 4D). Inhibition of cyclin-dependent kinase activities by roscovitine led to an increase, albeit modest, of the steady state level of UHRF1 (Fig. 3E). To investigate the functional significance of this phosphorylation event further, we carried out complementation experiments by introducing either wild-type or the S652A mutant into cells where the endogenous UHRF1 is knocked down by RNAi. As shown in Fig. 4E, knocking down UHRF1 expression reduced cell proliferation over a course of 4 d, consistent with a proproliferation role of UHRF1 reported by previous studies (31). Reintroduction of wild-type UHRF1, but not the S652A mutant, partially restored proliferation. As controls, both wild type and S652A are expressed at comparable levels in these cells (Fig. S4H). Taken together, these findings suggest that UHRF1 interaction with USP7 is regulated by phosphorylation during cell cycle and is important for proteasomal degradation of UHRF1 in the M phase of the cell cycle. Importantly, regulation of the UHRF1 level at the M phase through S652 phosphorylation may be important for cell proliferation control.
Fig. 4.
UHRF1 S652 phophorylation disrupts interaction with USP7 and decreases UHRF1 stability. (A) Superimposed ITC enthalpy plots for the binding of SpacerUSP7 560–664 (syringe) with UHRF1 600–687 and mutations of UHRF1 S652D (Cell). The estimated binding affinity (Kd) numbers are listed in the inset. (B) Stable cell lines were established that express either FLAG-tagged, wild-type or S652 mutants (S652A and S652D). Cells were treated with 50 ug/mL cycloheximide (CHX) as indicated. Extracts were prepared at the indicated time points (top) and used for Western blotting. The amount of UHRF1 was normalized against the corresponding actin signal and the quantitations were shown at the bottom of each panel. (C) HCT116 p53-/- cells were treated as in Fig. S4D. The extracts were immunoprecipitated with a UHRF1 antibody, followed by Western blotting using USP7 and UHRF1 antibody, respectively. (D) HCT116 p53-/- cells were treated as in Fig. S4D and released into 50 μg/mL cycloheximide (CHX) containing culture as indicated. Lysates were analyzed by Western blot with UHRF1 and actin antibodies, respectively. UHRF1 band intensity was normalized against the internal actin controls. (E) HCT116 p53-/- stable cell lines were established that coexpress control or UHRF1 shRNA with indicated Flag-tagged UHRF1 (wild type or mutant). They were seeded at 1 × 104 cells in triplicate 60 mm plates. Cells were trypsinized and counted at indicated time points. Standard deviation bars were obtained from the triplicate counts. (F) A working model. At the G1 and S phases of the cell cycle, UHRF1 level is regulated by the balance of its ubiquitylation and deubiquituylation (mediated by USP7), with deubiquitylation inhibiting UHRF1 proteasomal degradation. CDK1-cyclin B mediates phosphorylation of UHRF1 at M phase, which disrupts the interaction with USP7 leading to an increased turnover and thus reduces steady state levels of UHRF1.
Discussion
In this report, we have identified an important mechanism that regulates the epigenetic regulator UHRF1. Specifically, we have provided biochemical and kinetic data that establish the physical interaction between UHRF1 and the deubiquitylase USP7 and demonstrated by complementation experiments that such an interaction is critical for protecting UHRF1 from proteasome-mediated degradation. Importantly, we have shown that this interaction is regulated by a cell cycle-specific mechanism; UHRF1 is specifically phosphorylated at S652 in the M phase of the cell cycle resulting in a reduced physical interaction of UHRF1 with USP7, which leads to a reduced steady state level of UHRF1. In summary, our findings highlight an intricate mechanism that regulates UHRF1 level in the cell; USP7 protects UHRF1 via direct protein–protein interaction and deubiquitylation, but M phase-specific phosphorylation causes the release of UHRF1 from USP7, leading to its M phase-specific destruction. Importantly, the S652A phosphorylation defective mutant renders slower cell proliferation than wild-type UHRF1, suggesting that maintaining appropriate levels of UHRF1 is important for cell proliferation regulation.
USP7 was initially shown to be as an enzyme (known as HAUSP) that mediates deubiquitylation of both p53 and MDM2, thus playing a critical role in regulating p53 steady state level in the cell (9, 32). More recently studies also provided additional examples of USP7 regulation of protein stability, including DNMT1, a partner of UHRF1 in the regulation of CpG methylation (16, 18) as well as PTEN (33), H2B (34), Claspin (11), and REST (35). Together with this study, these findings establish a paradigm where the stability of a plethora of important regulators such as p53 and UHRF1 are regulated by USP7 via physical interactions and deubiquitylation.
An important and perhaps distinguishing feature of the UHRF1 and USP7 interaction is its cell cycle regulation. We found that UHRF1 level is regulated during cell cycle where it is degraded in the M phase. Our mass spectrometry effort and the development of the serine 652 phosphor-specific antibodies allowed us to unequivocally demonstrate UHRF1 S652 phosphorylation in the M phase of the cell cycle, consistent with a recent report (26). Mutational and protein–protein interaction assays provided further evidence that this phosphorylation event compromises the physical interaction between UHRF1 and USP7, both in vitro and in vivo. Importantly, the reduced interaction with USP7 is coincident with a reduced stability of UHRF1 in the M phase of the cell cycle, reinforcing the notion that phosphorylation of this site regulates UHRF1 stability. Changing UHRF1 stability leads to an altered cell proliferation (Fig. 4E), indicating the importance of maintaining appropriate level of UHRF1 through S652 phosphorylation. It should be noted that a recent study also identified S652 phosphorylation as a potential mTOR target (25), suggesting that S652 phosphorylation may play additional biological roles. Interestingly, the adjacent S674 of UHRF1, which also falls within the USP7-interacting domain, is a potential CDK1-cyclin B phosphorylation site. Indeed, a recent study shows that the S674 equivalent of the zebrafish UHRF1 is phosphorylated by ccna2/cdk2 (cyclinA/cdk2), but this enzyme is only active in S but not M phase of the cell cycle (36). Thus, it remains to be determined whether S674 is phosphorylated by CDK1-cyclin B in vivo and whether S674 phosphorylation also contributes to the regulation of UHRF1-USP7 physical interaction at the M phase of the cell cycle.
Taken together, our findings support the model (Fig. 4F) whereby UHRF1 is protected by USP7 during G1 and S phases of the cell cycle. Upon entering M phase, phosphorylation of S652 of UHRF1 by CDK1-cyclin B disrupts the interaction with USP7. The unbound UHRF1 subsequently undergoes proteasomal degradation, which may be important for the cell to enter the next round of the cell cycle where UHRF1 level is restored as a result of the loss of S652 phosphorylation and the regaining of interaction with USP7. The fact that altering UHRF1 stability (Fig. 4 B and E) impacts cell proliferation is consistent with this model.
Materials and Methods
Cell Culture and Transfection.
HCT116 p53+/+ and HCT116 p53-/- cells were obtained from Bert Vogelstein’ s lab and maintained in McCoy’s 5A medium containing 10% fetal bovine serum (FBS) (Hyclone) and 0.1% Pen-Strep. HeLa cells and human embryonic kidney 293T cell were grown in DMEM supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 1% Pen-Strep.
Plasmids and Antibodies.
UHRF1 was cloned into pET-28a (Novagen), pMSCV (Clontech), pcDNA4 (Invitrogen), pLenti 6.2 (Invitrogen). USP7wt and mutants were cloned into pPB-CAG vector. Rabbit anti-UHRF1 antibodies were raised by immunizing rabbits with full-length His-UHRF1 protein and mouse anti-UHRF1 (BD 612264) was used for immunostainning. Anti-FLAG (m2) antibody was purchased from Sigma. Anti-HA antibody and beads were purchased from Santa Cruz (sc-7392, sc-7392ac) while anti-USP7 and anti-USP11 antibodies were obtained from Santa Cruz (H-200) and Bethyl Laboratories A031-613A), respectively. S-652ph antibodies were raised in rabbits using the prephosphorylated peptide (CQEGGFAS(p)PRTGKG-NH2) as an antigen.
RNA Interference.
USP7, UHRF1, USP11 shRNA were purchased from Open Biosystems USP7 shRNA-3: tgtatctattgactgcccttt. USP7 shRNA-6: cgtggtgtcaaggtgtactaa. UHRF1 shRNA-2: gcctttgattcgttccttctt. UHRF1 shRNA-4: tgtgaaatactggcccgagaa. USP11 shRNA-2: ccgtgatgatatcttcgtcta. Lentivirus of USP7, UHRF1, USP11 shRNAs were made according to the protocol on Open Biosystems Web site.
Primers for RT-PCR, UHRF1(forward): 5′gcagaggctgttctacaggg; UHRF1 (reverse): 5′ gtgtcggagagctcggagt; USP7 (forward): 5′gagtgatggacacaacaccg; USP7 (reverse): aaacacggagggctaaggac; GAPDH (forward): 5′tgatgacatcaagaaggtggtgaag; GAPDH (reverse): 5′tccttggaggccatgtgggccat.
Protein Complex Purification and Data Analysis.
FLAG-tagged UHRF1 was purified from 293T cell using whole cell as well as nuclear extracts (37). The immunoprecipitated material was analyzed by mass spectrometry and the ComPass program (14).
GST Pull-Down Assays.
GST- USP7 fusion proteins (50 µg) were immobilized to 25 µL of glutathione beads (GE Healthcare). Purified UHRF1 proteins (at least 500 μg each) were incubated with GST-USP7 truncations in 300 µL binding buffer [50 mM Tris-HCl pH 8.0 (pH 8.8 for UHRF1 full length), 150 mM NaCl, 0.1% TritonX-100] for 1 h at 4 °C. Glutathione beads were then washed five times with 500 µL binding buffer. The bound proteins were analyzed by SDS/PAGE and stained using Commassie Blue.
Isothermal Titration Calorimetry (ITC).
To obtain a direct binding affinity between SpacerUHRF1 and UBLUSP7, UBLUSP7 were titrated with SpacerUHRF1 using ITC-200 microcalorimeter (GE Healthcare) at 10 °C. UBLUSP7 mutants were also titrated with SpacerUHRF1 to verify whether specific mutations affected the binding affinity. All proteins and peptides were exchanged to a buffer containing 10 mM HEPES, pH 8.0, and 0.1 M NaCl by gel-filtration chromatography. ITC data were analyzed and fit using software Origin 7.0 (OriginLab Corporation). The results are summarized in Table S2.
Coimmunoprecipitation.
HeLa nuclear proteins were extracted as described (38), diluted with buffer A to 150 mM KCl , and NP40 was added to the final concentration of 0.1%. The extract was spun at 14,000 rpm for 15 min at 4 °C, 5% was kept for input while the rest was incubated with anti-USP7 (Santa Cruz, H-200), anti-UHRF1 (Bethyl Laboratories, A301-470A), anti-USP11 (Bethyl Laboratories, A301-613A), anti-DNMT1 (from Guoliang Xu’s lab) or rabbit IgG antibody for 1 h at 4 °C. Protein A/G (Santa Cruz) beads were then added for overnight incubation at 4 °C. The beads were washed stringently, and the bound proteins were boiled in SDS sample buffer and Western blot using anti-USP7, anti-UHRF1, and anti-USP11 antibodies.
In Vivo Ubiquitination Assay.
293T cells were transfected with His-UHRF1, HA-ubiquitin, Myc-USP7wt, or Myc-USP7cs. At 48 h posttransfection, cells were washed with PBS twice and solubilized in 1% SDS concentration to inactivate deubiquitinateing enzymes and diluted with low salt buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% NP40) to make final SDS concentration 0.1%. Equal amounts of proteins from the extract were immunoprecipitated with anti-HA(SC-7392AC) resin at 4 °C for 3 h, washed three times with RAIP (50 mM Tris-HCl, 150 mM NaCl, 0.1% SDS) buffer and boiled in SDS sample buffer for Western blot analysis with UHRF1 antibody.
In Vitro Deubiquitination Assay.
His-USP7 WT, cs, M1, M2 were purified from Tn5 insect cells. Poly-ub-UHRF1 was purified from 293T cells cotransfected with His-UHRF1 and HA-Ub, using Ni-NTA Agarose beads, and eluted with imidazole. Purified UHRF1 was subjected to dialysis overnight and then used as substrate for deubiquitinate reactions. The purified Poly-ub-UHRF1 protein was incubated with purified USP7 WT, cs, M1 or M2 protein in the deubiquitination buffer (50 mM Tris-HCl, pH 7.4, 0.5 mM EDTA, 10 mM β-mercaptoethanol) at 32 °C for 45 min (39). The reactions were terminated by the addition of SDS sample loading buffer and proteins were resolved on 8% SDS/PAGE and blotted with the anti-HA antibody.
FACS Analysis.
HCT116 p53-/- cells were treated with double thymidine or thymidine and nocodazle as described previously (27). Cells were fixed in 70% ethanol, cellular DNA was stained with propidium iodide (Sigma 81845), and analyzed by FACSCalibur flowcytometer (Becton Dickinson).
Immunostaining.
HCT116 p53-/- cells were washed with TBS buffer twice, fixed with 4% paraformaldehyde in TBS for 10 min at room temperature, and permeabilizated with 0.5% NP-40 in TBS for 15 min. Cells were washed with TBST and incubated in 5% BSA in TBST overnight, the S-652ph antibodies were added for 1.5 h, then washed with TBST and incubated with Fluor-conjugated secondary antibodies and 4′,6′-diamidino-2-phenylindole (DAPI) for 1 h. Following TBST washing, the images were captured with Olympus BX51 microscope.
Supplementary Material
Acknowledgments.
We thank Dr. Bert Vogelstein for HCT116 p53+/+ and HCT116 p53-/- cell lines, Dr. Madelon M. Maurice for the Myc-USP7 wild type and Myc-USP7 C223S mutant plasmids, and Dr. Guoliang Xu for DNMT1 antibodies. This work was supported by the 985 Program from the Chinese Ministry of Education and 973 State Key Development Program of Basic Research of China (2009CB825602, 2009CB825603) to the Epigenetics Laboratory at the Institute of Biomedical Sciences, Fudan University, Shanghai, China. This work was also supported in part by the National Basic Research Program of China (2009CB918600, 2011CB965300) and the National Natural Science Foundation of China (30870493, 31030019, 11079016).
Footnotes
Conflict of interest statement: Fei Lan is an employee of Constellation Pharmaceuticals Inc., and Yang Shi is a cofounder of Constellation Pharmaceuticals Inc. and a member of its scientific advisory board. The remaining authors have declared no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116349109/-/DCSupplemental.
References
- 1.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 2.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 3.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 4.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–U925. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 5.Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2008;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajakumara E, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins Y, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett RD, et al. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 11.Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCF beta TrCP-but not APC(Cdh1)-mediated proteolysis of Claspin. J Cell Biol. 2009;184:13–19. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Horst A, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–U1040. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 13.Kessler BM, et al. Proteome changes induced by knock-down of the deubiquitylating enzyme HAUSP/USP7. J Proteome Res. 2007;6:4163–4172. doi: 10.1021/pr0702161. [DOI] [PubMed] [Google Scholar]
- 14.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 17.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112:439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 18.Felle M, et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Citterio E, et al. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins Y, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2007;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt. J Cell Biochem. 2011;112:439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 24.Mittler G, Butter F, Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2008;19:284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 27.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 29.Planchais S, et al. Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. Plant J. 1997;12:191–202. doi: 10.1046/j.1365-313x.1997.12010191.x. [DOI] [PubMed] [Google Scholar]
- 30.Wu SC, Zhang Y. CDK1-mediated phosphorylation of Ezh2 regulates its stability. J Biol Chem. 2011;286:28511–28519. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 32.Meulmeester E, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Knaap JA, et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Huang Z, et al. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zindy F, et al. Cyclin A is required in S phase in normal epithelial cells. Biochem Biophys Res Commun. 1992;182:1144–1154. doi: 10.1016/0006-291x(92)91851-g. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 38.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joo HY, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 40.Winget JM, Mayor T. The diversity of ubiquitin recognition: Hot spots and varied specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.