Abstract
SgrS RNA is a model for the large class of Hfq-associated small RNAs that act to posttranscriptionally regulate bacterial mRNAs. The function of SgrS is well-characterized in nonpathogenic Escherichia coli, where it was originally shown to counteract glucose-phosphate stress by acting as a repressor of the ptsG mRNA, which encodes the major glucose transporter. We have discovered additional SgrS targets in Salmonella Typhimurium, a pathogen related to E. coli that recently acquired one-quarter of all genes by horizontal gene transfer. We show that the conserved short seed region of SgrS that recognizes ptsG was recruited to target the Salmonella-specific sopD mRNA of a secreted virulence protein. The SgrS–sopD interaction is exceptionally selective; we find that sopD2 mRNA, whose gene arose from sopD duplication during Salmonella evolution, is deaf to SgrS because of a nonproductive G-U pair in the potential SgrS-sopD2 RNA duplex vs. G-C in SgrS-sopD. In other words, SgrS discriminates the two virulence factor mRNAs at the level of a single hydrogen bond. Our study suggests that bacterial pathogens use their large suites of conserved Hfq-associated regulators to integrate horizontally acquired genes into existing posttranscriptional networks, just as conserved transcription factors are recruited to tame foreign genes at the DNA level. The results graphically illustrate the importance of the seed regions of bacterial small RNAs to select new targets with high fidelity and suggest that target predictions must consider all or none decisions by individual seed nucleotides.
Keywords: seed pairing, small noncoding RNA, target discrimination
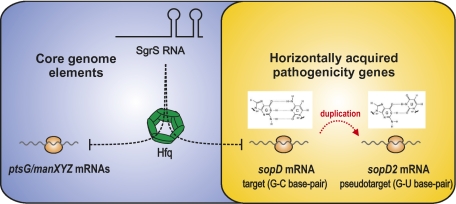
Horizontal gene transfer (HGT) is a driving force in general microbial evolution that allows bacterial pathogens to acquire new virulence factors from exogenous sources (1). The Salmonella enterica species is a group of enterobacterial pathogens that cause a range of diseases from gastroenteritis to typhoid fever, and it has horizontally acquired >25% of the total genetic material since the time Salmonella and Escherichia coli shared a common ancestor (2). The new genes include all virulence factors that Salmonella secrete into mammalian host cells through the two type 3 secretion systems (T3SSs) encoded on the Salmonella pathogenicity islands, SPI-1 and SPI-2 (3–5). The products of HGT genes generally have a fitness cost for recipient bacteria, and therefore, it is crucial that these genes are integrated into existing regulatory networks to prevent inappropriate expression (6, 7). Studies of bacterial regulators recruited to regulate HGT genes have identified signaling events that promote or suppress virulence, and they contributed to our understanding of the DNA recognition preferences of transcriptional regulators that mediate the repression or activation of newly acquired genes (8–10).
Small noncoding RNAs (sRNAs) are an emerging and abundant class of gene expression regulators that control many branches of cellular physiology. Most of the ∼100 sRNAs known in Salmonella act on trans-encoded target mRNAs through short base-pairing interactions that are aided by the RNA chaperone, Hfq (11–16). It is not known whether and how these Hfq-dependent sRNAs exert control of HGT genes. Evidence has been circumstantial and based on observations that disruption of hfq alters expression of many Salmonella HGT loci (13, 17, 18, 19) and that Hfq binds many virulence factor mRNAs, suggesting that they might be targets of sRNAs (13).
The study of HGT targets could help better understand the building plan of sRNAs and how bona fide targets are discriminated from thousands of other cellular transcripts. For example, despite the great diversity in length (50–250 nt) and structure, increasing numbers of Hfq-dependent sRNAs are found to rely on a few highly conserved nucleotides—the seed—for binding to conserved targets. If new HGT targets were also recognized by the seed, this recognition would define the seed as the sRNA region that is generally responsible for mRNA binding.
In this paper, we show that the Hfq-associated SgrS RNA, present in both pathogenic and nonpathogenic enterobacteria (20, 21), was recruited to posttranscriptionally repress the synthesis of SopD, a recently acquired Salmonella-specific virulence protein that is secreted by both T3SSs (22). Pioneering work in E. coli had established SgrS as the centerpiece of a stress response to the accumulation of phosphorylated sugars, especially glucose (21, 23). This prior work also showed that the ∼240-nt SgrS RNA is bifunctional (Fig. 1A): the 5′-located sgrT ORF encodes a 40-aa peptide that blocks glucose import by an unknown mechanism (24), whereas a 3′-located conserved region inhibits de novo synthesis of major sugar uptake proteins by direct base pairing with the ptsG and manXYZ mRNAs (21, 25). The SgrS–ptsG mRNA interaction has been exceptionally well-characterized and was shown to rely on only six nonredundant base pairs (26).
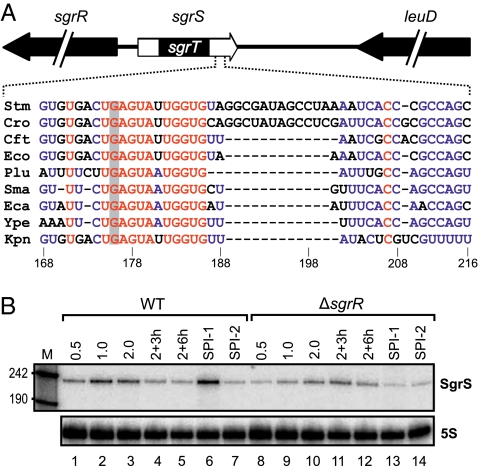
Fig. 1.
Expression of SgrS in Salmonella. (A) Genomic location of Salmonella sgrS and conservation of the antisense domain in enterobacteria. (Stm: Salmonella typhimurium LT2; Cro: Citrobacter rodentium; Cft: E. coli CFT 073; Eco: Escherichia coli K12; Plu: Photorhabdus luminescens; Sma: Serratia marcescens; Eca: Erwinia carotovora; Ype: Yersinia pestis; Kpn: Klebsiella pneumoniae). The SgrS 5′ end also encodes the SgrT ORF (24) and is flanked by the sgrR gene encoding its transcription factor. The antisense domain is located in the 3′ end of the molecule and characterized by a stretch of 6 + 5 conserved nucleotides. The gray bar indicates the G to C single-nucleotide exchange in SgrS* (G176 → C). (B) Northern blot analysis of WT and sgrR mutant Salmonella collected from various stages of growth (OD600 of 0.5, 1.0, 2.0, 3, and 6 h after cells had reached OD600 = 2.0 as well as SPI-1 and SPI-2 induction conditions). 5S rRNA served as loading control.
We show that, in Salmonella, similar regions of SgrS interact with both the ptsG and sopD mRNAs, suggesting that sRNAs preferably use their preestablished seed regions to sample incoming HGT mRNAs for potential regulation. Intriguingly, the closely related sopD2 mRNA that contains an almost identical target site is refractory to SgrS regulation. Our analyses reveal that a single C to T transition in sopD2, resulting in a nonproductive G-U wobble base pair, enables SgrS to discriminate these similar mRNAs at the level of a single hydrogen bond. The results have ramifications for the prediction of sRNA target interactions.
Results
SgrS Targets in Salmonella.
The sgrS gene (also known as ryaA) was originally identified in a global screen for Hfq-binding sRNAs in E. coli and resides between setA and sgrR (27); the latter gene encodes a transcription factor that activates SgrS synthesis when high levels of phosphorylated sugars threaten to poison the cell (21, 28). We confirmed that SgrS is expressed in Salmonella Typhimurium strain SL1344 during exponential and early stationary phase (Fig. 1B, lanes 1–3), and as expected, it was greatly diminished in the absence of SgrR (Fig. 1B, lanes 8–10). In addition, we discovered that SPI-1–inducing conditions (high salt and low oxygen), known to activate Salmonella invasion genes, strongly induced SgrS expression, an effect that was abolished in the sgrR mutant strain (Fig. 1B, lane 6 vs. 13).
To identify the target suite of SgrS in Salmonella, we used a pulse expression approach (29, 30) and scored global changes in mRNA abundance with microarrays after transient (10 min) overexpression of the sRNA from a PBAD promoter. Of the 4,716 ORFs represented on the arrays, just six mRNAs were altered more than or equal to threefold by SgrS (Table 1). In accordance with previous findings in E. coli (21, 31), overexpressed SgrS repressed the ptsG mRNA by 10-fold as well as the manXYZ operon, which encodes a mannose-specific uptake system (25). The yigL mRNA is an additional candidate target that was up-regulated by SgrS; it is present in both Salmonella and E. coli, and it encodes a potential haloacid dehalogenase (HAD)-like hydrolase with a predicted role in sugar metabolism (32). Most importantly, however, we discovered a significant down-regulation of the Salmonella-specific sopD gene. We followed up these global observations by repeating the SgrS pulse expression in a ΔsgrS strain and quantified transcript changes by quantitative RT-PCR, which showed a sevenfold reduction of both the ptsG and sopD mRNAs (Table 1). This finding identified the horizontally acquired sopD mRNA as a candidate target of SgrS.
Table 1.
Genes differentially regulated on SgrS pulse expression
| Gene | ID | Fold regulation* | Fold regulation† | Description‡ |
| manX | STM1830 | −10.3 | −34.9 | Mannose-specific enzyme IIAB |
| manY | STM1831 | −12.4 | −20.2 | Mannose-specific enzyme IIC |
| manZ | STM1832 | −5.5 | −13.1 | Mannose-specific phosphotransferase system protein IID |
| ptsG | STM1203 | −10.2 | −7.1 | Glucose-specific phosphotransferase system IIBC components |
| sopD | STM2945 | −5.3 | −7.0 | Secreted effector protein |
| yigL | STM3962 | +3.7 | +4.0 | Putative sugar phosphatase |
*Fold regulation obtained by transcriptomic analysis of pBAD-driven SgrS expression on Salmonella-specific microarrays. Genes that were at least threefold differentially regulated and had a P value ≤ 0.01 are listed.
†Fold regulation obtained by quantitative RT-PCR.
‡Description is based on the annotation at Colibase (http://xbase.bham.ac.uk/colibase/).
SgrS Controls the Synthesis of Virulence Factor SopD.
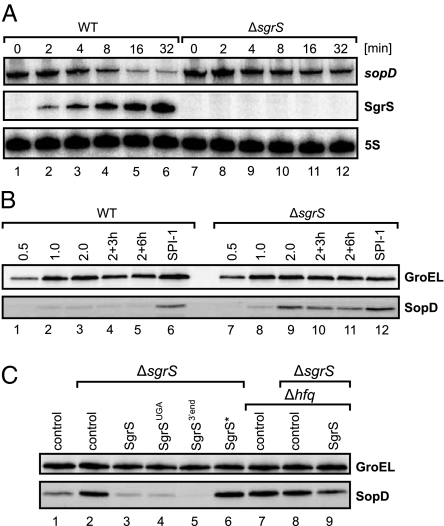
To address whether sopD is also regulated under physiologically relevant conditions, we treated Salmonella with the nonmetabolizable glucose analog α-methyl glucoside (αMG), a strong inducer of the chromosomal sgrS gene (21). Northern blots showed that addition of αMG to exponentially growing cells strongly up-regulated SgrS within 16 min and also reduced the levels of the monocistronic sopD mRNA more than fivefold (Fig. 2A, lanes 1–6). We identified SgrS as the silencer responsible for the reduction in sopD expression, because in the presence of αMG, sopD levels were only slightly reduced in a ΔsgrS strain (∼1.3-fold) (Fig. 2A, lane 7 vs. 12). Carbon source availability can modulate SPI-1 gene expression (33) and might account for this basal reduction at the level of sopD transcription.
Fig. 2.
The SgrS antisense domain is required for SopD repression. (A) Northern blot analysis of WT and sgrS mutant Salmonella challenged with the glucose analog αMG. Samples were withdrawn before and at the indicated time points posttreatment. Total RNA isolates were inspected for sopD and SgrS expression using gene-specific probes; 5S rRNA served as a loading control. (B) Western blot analysis of SopD::3×FLAG protein in WT and ΔsgrS strains at various conditions of Salmonella growth (OD600 of 0.5, 1.0, 2.0, 3, and 6 h after cells had reached OD600 = 2.0 as well as SPI-1–inducing conditions). GroEL served as an internal loading control. (C) Western blot analysis of SopD::3×FLAG protein in WT, ΔsgrS, Δhfq, or ΔsgrS/Δhfq bacteria. Strains were transformed with the indicated plasmids and grown under SPI-1–inducing conditions. Total protein was extracted, and SopD::3×FLAG proteins levels were inspected by Western blot.
At the protein level, SgrS-dependent regulation was evident even under nonstress conditions, when higher levels of SopD protein were seen in ΔsgrS than WT bacteria throughout growth (Fig. 2B, lanes 1–5 vs. 7–11), as well as under the SPI-1–inducing conditions (Fig. 2B, lane 6 vs. 12). Collectively, these results identified SgrS as a repressor of SopD synthesis under both infection-relevant and standard in vitro conditions.
Regulation of SopD Requires the Seed Region of SgrS.
Given that SgrS is a bifunctional RNA (Fig. 1A) (21, 24, 26), we needed to define whether the sgrT ORF, seed pairing domain, or both regulate sopD expression. To address this question, we complemented the ΔsgrS strain with plasmids that constitutively expressed WT SgrS or several rationally designed mutants, and we assessed impact on SopD levels (Fig. 2C and RNA expression data in SI Appendix, Fig. S1). In agreement with our previous results (Fig. 2B), under the SPI-1–inducing conditions, sgrS mutants displayed ∼2.5-fold higher SopD levels compared with WT (Fig. 2C, lane 2 vs. 1). In contrast, a plasmid overexpressing WT SgrS caused an approximately sevenfold reduction in SopD levels (Fig. 2C, lane 3 vs. 2), and similar repressions were mediated by SgrS with a severely truncated SgrT peptide (stop mutation at fifth codon) (Fig. 2C, lane 4) or just the 3′ region of the sRNA (Fig. 2C, lane 5). Thus, the 3′ end of the sRNA mediated sopD regulation. Furthermore, an SgrS* mutant RNA with a G176 → C point mutation in the 3′-located seed (Fig. 1A) fully abrogated regulation (Fig. 2C, lane 6). Importantly, G176 is essential for recognition of ptsG mRNA in E. coli (26), indicating that SgrS acted on sopD by direct base pairing.
To corroborate this finding, we tested the effect of SgrS in the absence of Hfq. A Salmonella Δhfq strain showed the same elevated SopD levels as ΔsgrS (compared with WT) (Fig. 2C, lanes 1, 2, and 7) with no additional increase in a ΔsgrS/Δhfq double mutant (Fig. 2C, lane 8), suggesting that both Hfq and SgrS are essential for sopD silencing. Overexpression of SgrS only caused a mild reduction in SopD expression (∼1.5-fold) in the absence of Hfq (Fig. 2C, lane 9). Collectively, these results suggested that SgrS uses its seed domain to regulate sopD posttranscriptionally by Hfq-dependent pairing.
ptsG and sopD mRNAs Are Regulated by Similar RNA Duplexes.
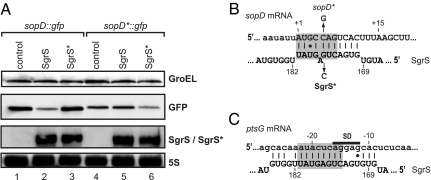
Most Hfq-associated sRNAs bind in the 5′ region of mRNAs, and similarly, SgrS down-regulated a translational sopD::gfp reporter containing only the 5′ UTR and first 20 codons of sopD (Fig. 3A, lanes 1 and 2). SgrS reduced the levels of SopD::GFP by approximately fivefold relative to sRNA control vector. We considered this regulation significant, particularly because it exceeded a previously observed 2.5-fold repression of a ptsG::gfp reporter by SgrS in E. coli (34).
Fig. 3.
SgrS binds sopD in the proximal coding sequence. (A) Western and Northern blot analyses of Salmonella harboring plasmid pPL-SgrS or mutant plasmid pPL-SgrS* in combination with either WT sopD::gfp or mutant sopD*::gfp fusion plasmids. Protein and RNA samples were collected at OD600 of 2.0, and SopD::GFP protein levels were determined by Western blot analysis. SgrS and SgrS* levels were detected by Northern blot using a gene-specific oligonucleotide. (B) Graphical presentation of the SgrS–sopD interaction. Numbering for sopD relative to the start-codon AUG (A is +1) and SgrS relative to +1 of the transcription start site. The sopD coding region and SgrS are shown in capital letters. Vertical arrows denote nucleotides introduced in sopD::gfp and SgrS, respectively. (C) Graphical presentation of the SgrS–ptsG interaction for comparison. The gray bars indicate SgrS nucleotides shared in the interactions sopD and ptsG.
Computer-aided analysis of antisense complementarity between the regions of the sRNA (3′ end) and target (5′ end) that were involved in regulation predicted an almost perfect 11-bp RNA duplex of SgrS with the early coding region of sopD (Fig. 3B). Importantly, this duplex involves all of the six SgrS nucleotides that form the critical core interaction with ptsG mRNA (Fig. 3 B and C), including G176, which is required for the repression of both ptsG (26) and sopD (Fig. 2C).
To validate this RNA interaction through compensatory nucleotide changes in vivo, we confirmed that the SgrS* mutant did not repress the sopD::gfp reporter and constructed an sopD*::gfp fusion with a compensatory C+5 → G point mutation (Fig. 3B). This mutant reporter was refractory to WT SgrS but fully regulated by the compensatory SgrS* RNA (Fig. 3A, lanes 5 and 6). These experiments show that the seed region of SgrS is used to target the laterally acquired sopD mRNA.
A single G-U Pair Prevents Regulation of the Nearly Identical sopD2 mRNA by SgrS.
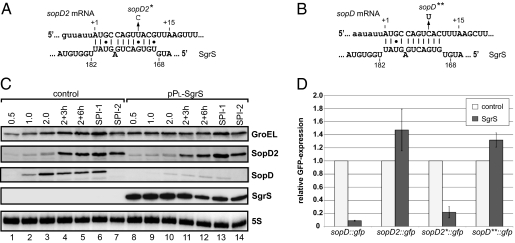
Salmonella encodes an additional effector protein, SopD2, that is 42% identical to SopD (22). Only S. enterica species that are very closely related to the ancestral S. bongori lack this effector, indicating that sopD2 arose from an early duplication of sopD and was followed by evolution to a divergent function (35). Intriguingly, the SgrS target site is almost identical within sopD and sopD2 (SI Appendix, Fig. S2). We predicted an 11-bp RNA duplex for SgrS-sopD2, which only differs from SgrS-sopD by U instead of C opposite to G172 of SgrS (Fig. 4 A and B). However, our microarray experiment (Table 1) revealed no regulation of sopD2 mRNA by SgrS. Similarly, the SgrS overexpression plasmid failed to down-regulate chromosomally expressed SopD2::FLAG protein under any growth condition tested (Fig. 4C), ruling out the possibility that regulation only occurred at the protein level. By contrast, SgrS depleted SopD::FLAG protein under all growth conditions (Fig. 4C), proving that the SgrS plasmid was functional.
Fig. 4.
A single nucleotide exchange protects sopD2 from targeting by SgrS. (A) Graphical presentation of predicted SgrS–sopD2 interaction. The vertical arrow denotes the U to C change in the sopD2* allele. (B) Graphical presentation of the SgrS–ptsG interaction. The vertical arrow denotes the C to U change in the sopD** allele. (C) Western and Northern blot analyses of strains harboring a chromosomal sopD2::3×FLAG allele transformed with a control plasmid (lanes 1–7) or the pPL-SgrS plasmid (lanes 8–14). Samples were collected at the indicated time points of growth in rich media or under SPI-1– and SPI-2–inducing conditions, and were subjected to Western and Northern blot analyses. Regulation of the SopD::3×FLAG is shown for comparison. (D) Quantitative Western blot analysis of sopD::gfp, sopD2::gfp, sopD2*::gfp, and sopD**::gfp reporters combined with a control vector or pPL-SgrS. Salmonella (ΔsgrS) double transformants were grown in rich media to OD600 of 2.0, and total protein samples were subjected to Western blot analysis of GFP protein. Relative GFP expression was calculated from the average of three independent experiments. The standard deviation is indicated by error bars.
Can a single noncanonical G-U instead of the canonical G-C pair suffice for discrimination and prevent sopD2 from being targeted by SgrS? To address this question, an sopD2::gfp reporter was constructed that (as with sopD::gfp) included the entire 5′ UTR and the first 20 codons of sopD2. As expected, this reporter was not regulated by SgrS (Fig. 4D). Next, we introduced a T+9 to C point mutation within the sopD2 region to generate essentially the same SgrS site as in the bona fide sopD target. Strikingly, this sopD2*::gfp reporter was fully regulated by SgrS, displaying approximately fivefold repression (Fig. 4D). In a reciprocal approach, we introduced a C+9 → T point mutation in sopD::gfp such that the duplex with SgrS would carry the sopD2-like G-U pair (Fig. 4B). The resulting sopD**::gfp fusion showed the same basal GFP activity as sopD::gfp but was not controlled by SgrS (Fig. 4D).
To prove that SgrS effectively discriminates between the native sopD and sopD2 mRNAs, we mutated the chromosomal sopD2 locus (sopD2*; T+9 → C). In contrast with the lack of regulation of the sopD2 WT gene (Fig. 4C), SopD2 protein synthesis from sopD2* was as strongly repressed by SgrS as the authentic SopD target (SI Appendix, Fig. S3). Thus, a single G-U pair in the seed duplex allows sopD2 to escape regulation by SgrS.
Target Discrimination by the SgrS Seed at the Level of Translation.
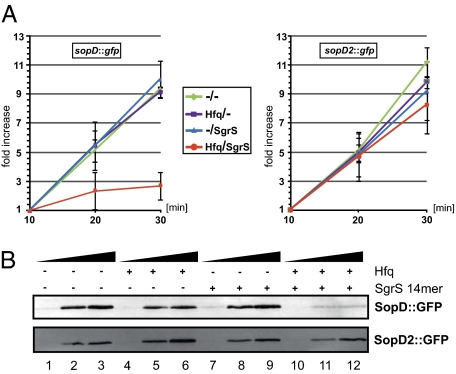
The target site location in the proximal coding sequence predicted that SgrS controlled the sopD mRNA at the level of translation (36, 37). We used a 70S ribosome translation assay (38–40) to determine the ability of SgrS to inhibit protein synthesis from in vitro synthesized sopD::gfp or sopD2::gfp mRNAs (Fig. 5A). In the absence of SgrS or Hfq, both mRNAs produced comparable amounts of protein with a linear increase over the course of a 30-min assay. Addition of Hfq protein or SgrS sRNA alone had a negligible effect on the translation, which was also reported for the ptsG mRNA in the work by Maki et al. (39). In contrast, the combined presence of SgrS and Hfq decreased SopD::GFP levels by ∼3.5-fold at the 30-min time point (Fig. 5A, Left) but failed to repress sopD2::gfp (Fig. 5A, Right). The in vitro translation system offered additional evidence for the discrimination between sopD and sopD2, and it suggested that, as with SgrS-ptsG (39), translational interference is the primary event in SgrS-mediated sopD repression.
Fig. 5.
In vitro translation assay of sopD::gfp and sopD2::gfp translation. (A) sopD::gfp (Left) and sopD2::gfp (Right) mRNAs were supplemented with mRNAs only, equimolar Hfq, 10-fold excess of SgrS, or a combination of Hfq and SgrS, and they were subjected to in vitro translation assays. Samples were collected at 10, 20, and 30 min postaddition of 70S ribosome, and GFP expression was monitored by Western blot. Experiments were carried out in triplicates, and translation efficiency was normalized to the levels of GFP observed after 10 min of translation. The error bars indicate the standard deviation. (B) Analogous to A; however, SgrS was exchanged by an RNA-oligo covering the SgrS antisense domain (SgrS residues 168–181) (Fig. 2A). Levels of SopD::GFP (Upper) and SopD2::GFP (Lower) were determined by Western blot, and triplicate results of these experiments are shown in SI Appendix, Fig. S4.
To narrow down a critical region of SgrS for sopD mRNA repression, we tested a seed-derived short SgrS RNA that the work by Maki et al. (38) had shown to suffice for ptsG inhibition in vitro. This 14-mer RNA (SgrS nucleotides 168–181) afforded the same fold regulation and selectivity as full-length SgrS with respect to sopD::gfp and sopD2::gfp translation, including the strict requirement for Hfq (Fig. 5B and SI Appendix, Fig. S4). The striking overlap with the previous ptsG data (38) lead us to conclude that the SgrS seed region, a critical RNA element that had already evolved in Salmonella, was acquired for the regulation of the horizontally acquired SopD virulence factor.
Seed Composition Affects Target Discrimination by the G-U Pair.
Successful seed pairing of small RNAs with targets has been proposed to largely depend on the thermodynamic stability of the RNA duplex that is formed (41, 42). Accordingly, most target search algorithms, including the popular RNAhybrid (43), calculate minimum free energy (MFE) to determine an energy optimum of intermolecular RNA hybridization. Using this algorithm, a typical MFE for bacterial seed pairing ranges from −24.2 to −18.8 kcal/mol−1, which was established with 10 targets of RybB sRNA (44).
The ability of the 14-mer seed to substitute for full-length SgrS in target regulation (see above) allows the 14-mer to be used as a proxy to calculate RNA duplex stability of SgrS and sopD/sopD2 (SI Appendix, Table S1). With an MFE of −17.4 kcal/mol, the nonproductive SgrS–sopD2 interaction was indeed slightly weaker than the productive SgrS–sopD pairing (−18.6 kcal/mol), and both were much weaker than SgrS–ptsG (−22.7 kcal/mol−1). However, the marginal difference (−1.2 kcal/mol) between SgrS-sopD and SgrS-sopD2 sheds doubt on the assertion that RNA duplex strength is solely responsible for the observed discrimination.
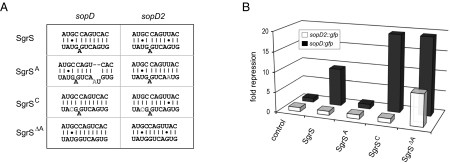
To address this issue, we looked for loss or gain of regulation of the sopD and sopD2 reporters after mutation of the SgrS seed at three selected positions (Fig. 6A). First, we changed G172 to A (SgrSA), which dramatically increases the MFE (to −13.4 kcal/mol) of the SgrS-sopD duplex and also changes its continuity; as expected, sopD was no longer regulated (Fig. 6B). The same mutation would endow SgrS-sopD2 with an A-U Watson–Crick pair at the critical G-U pair position. However, because no additional hydrogen bond forms (unlike the three-hydrogen bond G-C pair, both G-U and A-U make only two hydrogen bonds), this mutation does little to MFE and unsurprisingly, failed to bring about regulation of sopD2 (Fig. 6B). These data show that a mere discrimination of a wobble vs. Watson–Crick pair cannot be responsible for the target selectivity of SgrS.
Fig. 6.
Systematic analysis of the SgrS–sopD/sopD2 interaction. (A) Graphical presentation of the SgrS-sopD/sopD2 duplexes and SgrS mutants used in B. Nucleotides exchanged in the SgrS mutants have been marked in gray. (B) Quantitative Western blot analysis of sopD::gfp and sopD2::gfp reporters combined with a control vector, pPL-SgrS, pPL-SgrSA, pPL-SgrSC, or pPL-SgrSΔA. Salmonella (ΔsgrS) double transformants were grown in rich media to OD600 of 2.0, and total protein samples were subjected to Western blot analysis of GFP protein. Data are shown as fold repression relative to the control samples (average of three independent experiments). All SgrS variants are expressed equally (SI Appendix, Fig. S1).
Second, we sought to increase overall duplex strength with a U179 → C mutation in SgrS (SgrSC), which changes an unrelated G-U pair to G-C in both SgrS-sopD and SgrS-sopD2. This change lowered the predicted MFE of the SgrS-sopD duplex by −2 kcal/mol−1, and it doubled the regulation of the sopD reporter. This SgrS mutation also strengthened the interaction with sopD2 beyond the stability of the WT SgrS-sopD duplex (−19.4 vs. −18.6 kcal/mol) but again, it failed to bring about sopD2 regulation.
Third, we discovered how to regulate sopD2 with SgrS, by deleting A177 (SgrSΔA), a nucleotide that bulges out from the predicted SgrS-sopD and SgrS-sopD2 duplexes and weakens them by disrupting helix continuity. SgrSΔA has a predicted favorable change of MFE with sopD2 by −2.8 kcal/mol−1. This single-nucleotide deletion in SgrS brought sopD2 regulation from zero to eightfold (Fig. 6B) (in other words, similar to the level of repression achieved with WT SgrS on the sopD reporter). We note that, for sopD itself, despite a −1.8 kcal/mol difference in MFE, regulation by the SgrSA and SgrSC mutants of SgrS was the same. Both mutants afforded ∼18-fold repression, indicating potential saturation of regulation.
These data show that, in SgrS seed pairing, positional constraints rather than a continuum of MFE values are responsible for the all or none discrimination of individual seed nucleotides between cognate and near-cognate targets.
Discussion
The identification of the recently acquired sopD mRNA as an SgrS target in Salmonella combined with unexpected nonregulation of the almost identical sopD2 mRNA reveal that bacterial seed pairing can distinguish between mRNAs at the level of a single hydrogen bond. A critical wobble G-U pair, forming only two of three hydrogen bonds of Watson–Crick G-C, accounts for the deafness of sopD2 to SgrS. This unprecedented discrimination by a single hydrogen bond seems to be independent of base topology, which can allow noncanonical base pairs to impact the formation and strength of RNA duplexes (45). First, flipping the G-C pair in the SgrS-sopD duplex (SI Appendix, Fig. S5) maintains target regulation, despite a predicted spatial change of the seed helix. Second, an A-U pair at this position, which restores the glycosidic angle from 54° in G-U wobble to the general 65° of Watson–Crick pairs, does not rescue SgrS-sopD2 regulation (Fig. 6B). By contrast, amending the SgrS-sopD2 duplex with a single G-C pair allows SgrS to fully repress sopD2.
Target regulation by seed pairing is an emerging concept in bacteria. It was inspired by the ability of 22-nt microRNAs of eukaryotes to use their conserved seed region (nucleotides 2–7) to recognize mRNAs (46) and observations that Hfq-associated sRNAs carry conserved short regions that mediate target recognition (40, 44, 47, 48). The latter includes pioneering work on SgrS, in which the work of Vanderpool and Gottesman (21) predicted targeting of ptsG mRNA by highly conserved nucleotides of SgrS, which are now referred to as the seed. Subsequently, the work by Kawamoto et al. (26) showed that only six nucleotides of this region acted nonredundantly in ptsG repression. Our results with sopD/sopD2 shed light on the SgrS seed, showing that a single nucleotide can make an all or none decision through a critical Watson–Crick pair. This natural example of high-fidelity target selection illustrates how cells may reduce the potential regulatory noise from off-target interactions of sRNAs with near-cognate cellular transcripts. Moreover, our results suggest that bacterial sRNA target predictions, which have chiefly relied on thermodynamic stability of RNA duplexes, must pay attention to G-U pairs in seeds to reduce the number of false positives.
The SgrS–sopD interaction is a 7 + 4-bp RNA helix interrupted by a bulged adenosine (Fig. 3B); with respect to MFE, it is weaker than most Hfq-dependent sRNA-mRNA duplexes (44, 49) including the well-studied SgrS–ptsG interaction (SI Appendix, Table S1). In fact, weaker duplexes have been found to allow posttranscriptional control [e.g., FnrS-folE (50) or ArcZ-sdaC (37)]. Specifically, the MFE of ArcZ-sdaC (−17.4 kcal/mol) matches the predicted −17.5 kcal/mol of SgrS-sopD2, but SgrS cannot repress sopD2 (Fig. 4). A minor increase in MFE achieved by fortification of a distal base pair to G-C cannot compensate for the local weakness of the critical G-U pair, which was evident from failure to regulate sopD2 with the SgrS U179 → C variant (Fig. 6B). Increasing duplex stability closer to the critical G-U pair by removing the bulged A, however, did bring about regulation of sopD2 (Fig. 6B). Although a recent study of E. coli RyhB sRNA concluded that target selection reflected a thermodynamic continuum of general RNA duplex strength (42), our results with SgrS favor a model where the strength of critically positioned base pairs is pivotal to target selection. To test these results further, we monitored SgrS-mediated sopD vs. sopD2 discrimination at lower (20 °C) or elevated (44 °C) temperature. SgrS-mediated down-regulation of sopD2::gfp did not occur at either temperature, whereas repression of sopD::gfp was observed at both temperatures (SI Appendix, Fig. S6), supporting our conclusion that overall changes in RNA duplex stability play a lesser role in target selection. We caution, however, that changes in temperature might also affect RNA duplex formation at a different level, including sRNA-related activity of Hfq (51).
There is an intriguing commonality between bacterial and eukaryotic seed pairing. Animal microRNAs are most potent in gene silencing when fully composed of Watson–Crick base pairs (46, 52). Similarly, a recent survey of target interactions of Hfq-dependent sRNAs suggested that G-U pairs within the first three duplex positions (excluding terminal G-U pairs) are extremely rare (i.e., found in 2 of 49 validated interactions) (49). Nonetheless, judging from the failure to generate sopD2 regulation by simply changing the critical G-U pair to A-U (Fig. 6), we conclude that local duplex stability within the seed region is more relevant than a simple Watson–Crick vs. non-Watson–Crick discrimination. Additional experiments will be needed to fully understand the biochemical basis of target selection. The successful restoration of selective sopD::gfp vs. sopD2::gfp repression in an in vitro translation assay (Fig. 5) provides an important experimental framework to understand how intrinsic RNA elements, together with Hfq, determine productive regulation.
With a length of ∼240 nt, SgrS has much greater potential sequence space to bind mRNAs, but it uses only the short seed region to select the sopD target. Thus, one may liken the sRNA seed sequence to the DNA contact binding face of a transcription factor. Just as the binding of transcription factors is inherently constrained to a fixed pattern in DNA, Hfq-dependent sRNAs with a single seed region such as SgrS are inherently limited in their capability to incorporate new genes into their target suites. Importantly, this constrained capacity is crucially determined by a previously established lead target(s); in the case of SgrS, the ancestral regulation of the ptsG and/or manXYZ mRNAs in the context of phosphosugar stress (21, 25) has likely shaped the boundaries of flexibility for the evolution of new SgrS–mRNA interactions.
The biological reason for the regulation of sopD and not sopD2 by SgrS remains to be understood. Why is sopD regulated in this way? Intuitively, a repression of SopD would suggest a role of SgrS in Salmonella virulence, and a global screen in mice did show that sgrS played a minor role in virulence (53). We have yet to account for this mild attenuation by phenotypes caused by SgrS deficiency; the ΔsgrS strain did not show any attenuation during host cell invasion or intracellular replication assays. Interestingly, a comparative Northern blot analysis indicates that expression of SgrS is differentially controlled between Salmonella and nonpathogenic E. coli strain MG1655 (SI Appendix, Fig. S7). This finding would be consistent with a virulence-associated function of SgrS specific to enteric pathogens with lifestyles that involved colonization and invasion of the inflamed gut epithelium. SopD is known as a general virulence factor, with multiple roles in the development of gastroenteritis, replication in mouse macrophages, and systemic virulence of Salmonella (35, 54). SopD also acts as a dual effector delivered by both the SPI-1 and SPI-2 T3SSs that is expressed at later stages of infection when other SPI-1 effectors are no longer produced (22). Considering the broad expression and functions of SopD, it may be necessary to regulate this effector protein at multiple levels, including the posttranscriptional level at which SgrS and Hfq come into play. Intriguingly, although SgrS is primarily viewed as responding to sugar stress, its sugar-dependent expression may be exploited by Salmonella to recognize different host cell types and subcellular environments through carbon source availability and accordingly adjust the levels of SopD.
Why then is sopD2 not regulated by SgrS? The sopD2 gene is not present in the ancestral S. bongori strain (22), and the duplication event at which sopD gave rise to sopD2 likely occurred after or concomitant with the acquisition of major pathogenicity island SPI-2 (55); in other words, it happened very recently on the timescale of enterobacterial evolution. Nonetheless, the sopD and sopD2 genes have already diverged sufficiently to serve distinct functions in host–pathogen interplay (54, 56) as well as differ with respect to their own transcriptional control (57, 58). Perhaps the selective SgrS-mediated repression of sopD fostered the functional diversification of the two genes. Sequence comparison of the two genes strongly argues that the regulation of sopD vs. nonregulation of sopD2 has been under selective pressure (SI Appendix, Fig. S2). That is, although mutations have accumulated throughout both genes, the first six codons containing the SgrS site have been fully conserved (SI Appendix, Fig. S2) (22). This N-terminal domain is crucial for delivery of SopD through the T3SS and essential for virulence (59). All base changes in this region of sopD or sopD2 were silent in terms of amino acid sequence (SI Appendix, Fig. S2), and this finding includes the conserved crucial C/U discrepancy at position +9 of sopD and sopD2 (Fig. 4). Therefore, the development of this point mutation has enabled selective posttranscriptional control of the two mRNAs by SgrS that maintains the ability of the encoded proteins to be delivered by the T3SS. Generally, gene duplication events play important roles in the evolution of new biological functions (60), and HGT genes show a higher propensity than indigenous genes to undergo duplication (61). In Salmonella, additional effector protein pairs, such as SifA and SifB, resulted from gene duplication (35), and it will be interesting to see whether these pairs are also under selective sRNA-mediated control.
In conclusion, previous global phenotypic and gene expression analyses (18) as well as RNA–protein interaction studies (13) raised the possibility that Hfq-associated sRNAs could directly engage in the regulation of secreted effectors of Salmonella. Our discovery that SgrS regulates SopD synthesis provides proof of such a scenario; it suggests that conserved sRNAs with seemingly unrelated physiological functions constitute a reservoir of regulators that act like conserved transcription factors to tame foreign genes and integrate them into existing regulons. In addition to the HGT genes of the major SPI-1 and SPI-2 regions, Salmonella possesses another 12 pathogenicity islands and many single virulence genes (62); most transcripts from these regions are targets of Hfq. We predict that Hfq governs a large network of posttranscriptional control of HGT genes by either conserved or Salmonella-specific sRNAs that have evolved since Salmonella and E. coli diverged from each other 100–160 million years ago (63).
Experimental Procedures
Bacterial Strains and Growth.
Bacterial strains and their construction details are listed in SI Appendix, Table S2. Strains were grown at 37 °C in LB or on LB plates. Ampicillin (100 μg/mL), kanamycin (50 μg/mL), chloramphenicol (20 μg/mL), and l-arabinose (0.2%) were added where appropriate. Salmonella WT (SL1344) or mutant strains were transformed with plasmids by electroporation. SPI-1– and SPI-2–inducing conditions were as described (13, 18). Briefly, for SPI-1 induction, cultures were inoculated in 5 mL LB containing 0.3 M NaCl in 15-mL Falcon tubes with tightly closed lids. Cultures were incubated vertically with shaking for 12 h at 37 °C.
Oligonucleotides and Plasmids.
Oligonucleotides (DNA and RNA) and plasmids are listed in SI Appendix, Tables S3 and S4. Details on plasmids construction are given in SI Appendix, SI Methods. Target fusions to gfp were constructed as described (34).
Western Blot Analysis and Plate Fluorescence.
Culture samples were taken according to 1 OD600 and centrifuged for 4 min at 16,100 × g at 4 °C, and pellets resuspended in sample loading buffer to a final concentration of 0.01 OD/μL. After denaturation for 5 min at 95 °C, 0.1-OD equivalents of sample were separated on SDS gels. Western blot analyses of GFP and FLAG fusion proteins followed previously published protocols (34).
Northern Blot and Microarray Experiments.
Total RNA was prepared and separated in 5% or 6% (vol/vol) polyacrylamide–8.3 M urea gels (5–10 μg RNA per lane) and blotted as described (18). Membranes were hybridized at 42 °C with gene-specific (32P) end-labeled DNA oligonucleotides in Rapid-hyb buffer (GE Healthcare). Microarray experiments were carried out as described before (30), and SgrS pulse expression was achieved using the pBAD-based pKP12-2 plasmid. Microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession code GSE34851).
In Vitro Translation Assays.
These assays were carried out with modifications as previously described in ref. 40. Briefly, DNA templates carrying a T7 promoter sequence for in vitro transcription were generated by PCR. Primers and sequences of the T7 transcripts are given in SI Appendix, Table S4. T7 templates of gfp fusion mRNAs were amplified from plasmids using a sense primer that adds a T7 promoter to the +1 site of the 5′ UTR and an antisense oligo pZE-T1 122 nt downstream from the gfp stop codon. These transcripts end with the rrnB terminator of the fusion plasmids. RNA was in vitro-transcribed and quality-checked as described in ref. 18. Translation reactions were carried out using PureSystem (PGM-PURE2048C; Cosmo Bio Co., Ltd) according to the manufacturer's instructions. Reactions (10 μL) contained, in addition to 70S ribosomes, mRNA template (40 nM), Hfq (40 nM), and where applicable, full-length SgrS RNA or the 14-mer RNA oligonucleotide. Before addition of PureSystem mix, RNA was denatured for 1 min at 90 °C and chilled on ice for 5 min. Hfq was mixed with mRNA (and sRNA/oligonucleotide) and preincubated for 10 min at 37 °C. PureSystem mix was added, and incubation continued at 37 °C for the time indicated in the figures. Reactions were stopped with 4 volumes ice-cold acetone and kept on ice for 15 min, and proteins were collected by centrifugation (10,000 × g for 10 min at 4 °C). Proteins were quantified by Western blot analysis using a monoclonal GFP antibody.
Supplementary Material
Acknowledgments
We thank Susan Gottesman and Hiroji Aiba for comments on the manuscript, Barbara Plaschke for excellent technical assistance, and Sacha Lucchini for assistance with the transcriptomic analysis. This work was funded by Science Foundation Ireland Grant 08/IN.1/B2104 (to J.C.D.H.'s laboratory) and the Deutsche Forschungsgemeinschaft Priority Programs SPP1258 (Vo875/2-2) and SPP1316 (Vo875/6-1) to J.V.'s laboratory.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 4726 (volume 109, number 13).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119414109/-/DCSupplemental.
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Porwollik S, McClelland M. Lateral gene transfer in Salmonella. Microbes Infect. 2003;5:977–989. doi: 10.1016/s1286-4579(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 3.Galán JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochman H, Soncini FC, Solomon F, Groisman EA. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fass E, Groisman EA. Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol. 2009;12:199–204. doi: 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gal-Mor O, Finlay BB. Pathogenicity islands: A molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen HD, Jewett MW, Groisman EA. Ancestral genes can control the ability of horizontally acquired loci to confer new traits. PLoS Genet. 2011;7:e1002184. doi: 10.1371/journal.pgen.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucchini S, et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarre WW, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 11.Padalon-Brauch G, et al. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papenfort K, et al. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 13.Sittka A, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009;6:266–275. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- 15.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 16.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansong C, et al. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: Implications for virulence and global protein translation. PLoS One. 2009;4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueroa-Bossi N, et al. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol Microbiol. 2006;62(3):838–52. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 20.Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37:5465–5476. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 22.Brumell JH, et al. SopD2 is a novel type III secreted effector of Salmonella typhimurium that targets late endocytic compartments upon delivery into host cells. Traffic. 2003;4:36–48. doi: 10.1034/j.1600-0854.2003.40106.x. [DOI] [PubMed] [Google Scholar]
- 23.Kimata K, Tanaka Y, Inada T, Aiba H. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 2001;20:3587–3595. doi: 10.1093/emboj/20.13.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice JB, Vanderpool CK. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 2011;39:3806–3819. doi: 10.1093/nar/gkq1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang A, et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 28.Vanderpool CK, Gottesman S. The novel transcription factor SgrR coordinates the response to glucose-phosphate stress. J Bacteriol. 2007;189:2238–2248. doi: 10.1128/JB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papenfort K, et al. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuznetsova E, et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem. 2006;281:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 33.Lim S, et al. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 2007;35:1822–1832. doi: 10.1093/nar/gkm060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X, et al. The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Mol Microbiol. 2004;54:1186–1198. doi: 10.1111/j.1365-2958.2004.04344.x. [DOI] [PubMed] [Google Scholar]
- 36.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 37.Papenfort K, et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 38.Maki K, Morita T, Otaka H, Aiba H. A minimal base-pairing region of a bacterial small RNA SgrS required for translational repression of ptsG mRNA. Mol Microbiol. 2010;76:782–792. doi: 10.1111/j.1365-2958.2010.07141.x. [DOI] [PubMed] [Google Scholar]
- 39.Maki K, Uno K, Morita T, Aiba H. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc Natl Acad Sci USA. 2008;105:10332–10337. doi: 10.1073/pnas.0803106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofacker IL. How microRNAs choose their targets. Nat Genet. 2007;39:1191–1192. doi: 10.1038/ng1007-1191. [DOI] [PubMed] [Google Scholar]
- 42.Hao Y, et al. Quantifying the sequence-function relation in gene silencing by bacterial small RNAs. Proc Natl Acad Sci USA. 2011;108:12473–12478. doi: 10.1073/pnas.1100432108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 46.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 47.Guillier M, Gottesman S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balbontín R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol Microbiol. 2010;78(2):380–94. doi: 10.1111/j.1365-2958.2010.07342.x. [DOI] [PubMed] [Google Scholar]
- 49.Peer A, Margalit H. Accessibility and evolutionary conservation mark bacterial small-rna target-binding regions. J Bacteriol. 2011;193:1690–1701. doi: 10.1128/JB.01419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 53.Santiviago CA, et al. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 2009;5:e1000477. doi: 10.1371/journal.ppat.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakowski MA, Cirulis JT, Brown NF, Finlay BB, Brumell JH. SopD acts cooperatively with SopB during Salmonella enterica serovar Typhimurium invasion. Cell Microbiol. 2007;9:2839–2855. doi: 10.1111/j.1462-5822.2007.01000.x. [DOI] [PubMed] [Google Scholar]
- 55.Ochman H, Groisman EA. Distribution of pathogenicity islands in Salmonella spp. Infect Immun. 1996;64:5410–5412. doi: 10.1128/iai.64.12.5410-5412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown NF, et al. Mutational analysis of Salmonella translocated effector members SifA and SopD2 reveals domains implicated in translocation, subcellular localization and function. Microbiology. 2006;152:2323–2343. doi: 10.1099/mic.0.28995-0. [DOI] [PubMed] [Google Scholar]
- 57.Navarre WW, et al. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 58.Xu X, Hensel M. Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect Immun. 2010;78:49–58. doi: 10.1128/IAI.00931-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boonyom R, Karavolos MH, Bulmer DM, Khan CM. Salmonella pathogenicity island 1 (SPI-1) type III secretion of SopD involves N- and C-terminal signals and direct binding to the InvC ATPase. Microbiology. 2010;156:1805–1814. doi: 10.1099/mic.0.038117-0. [DOI] [PubMed] [Google Scholar]
- 60.Andersson DI, Hughes D. Gene amplification and adaptive evolution in bacteria. Annu Rev Genet. 2009;43:167–195. doi: 10.1146/annurev-genet-102108-134805. [DOI] [PubMed] [Google Scholar]
- 61.Hooper SD, Berg OG. Duplication is more common among laterally transferred genes than among indigenous genes. Genome Biol. 2003;4:R48. doi: 10.1186/gb-2003-4-8-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F. So similar, yet so different: Uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett. 2010;305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 63.Mirold S, et al. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J Bacteriol. 2001;183:2348–2358. doi: 10.1128/JB.183.7.2348-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]