Abstract
Mutations in the human mitochondrial genome are implicated in neuromuscular diseases, metabolic defects, and aging. An efficient and simple mechanism for neutralizing deleterious mitochondrial DNA (mtDNA) alterations has unfortunately remained elusive. Here, we report that a 20-ribonucleotide stem-loop sequence from the H1 RNA, the RNA component of the human RNase P enzyme, appended to a nonimported RNA directs the import of the resultant RNA fusion transcript into human mitochondria. The methodology is effective for both noncoding RNAs, such as tRNAs, and mRNAs. The RNA import component, polynucleotide phosphorylase (PNPASE), facilitates transfer of this hybrid RNA into the mitochondrial matrix. In addition, nucleus-encoded mRNAs for mitochondrial proteins, such as the mRNA of human mitochondrial ribosomal protein S12 (MRPS12), contain regulatory sequences in their 3′-untranslated region (UTR) that confers localization to the mitochondrial outer membrane, which is postulated to aid in protein translocation after translation. We show that for some mitochondrial-encoded transcripts, such as COX2, a 3′-UTR localization sequence is not required for mRNA import, whereas for corrective mitochondrial-encoded tRNAs, appending the 3′-UTR localization sequence was essential for efficient fusion-transcript translocation into mitochondria. In vivo, functional defects in mitochondrial RNA (mtRNA) translation and cell respiration were reversed in two human disease lines. Thus, this study indicates that a wide range of RNAs can be targeted to mitochondria by appending a targeting sequence that interacts with PNPASE, with or without a mitochondrial localization sequence, providing an exciting, general approach for overcoming mitochondrial genetic disorders.
The mtDNA of mammals encodes 13 proteins of the electron transport chain, 22 tRNAs, and 2 rRNAs (1, 2). The majority of mitochondrial proteins and some noncoding RNAs are encoded in the nucleus and imported into mitochondria (3, 4). Whereas the pathways of protein translocation into mitochondria have been well studied, the mechanisms for nucleus-encoded RNA import into mitochondria, however, are not well understood, even though mitochondrial RNA import is a universal process (5, 6). Recent detailed sequencing analysis of the mitochondrial transcriptome indicates that the collection of mitochondrial RNAs is more complex than previously thought, including an enrichment of several nucleus-encoded tRNAs and other noncoding RNAs (7), indicating that these RNAs are actively imported into mitochondria.
A subset of nucleus-encoded tRNAs is imported into the mitochondria of almost every organism (5, 8). The number of imported tRNAs ranges from one in yeast to all in trypanosomes. Recent mitochondrial transcriptome analysis suggests that this tRNA collection is broader in mammals than previously thought (7) as several unsuspected nucleus-encoded tRNAs are imported and processed to mature forms. In addition to in vitro evidence for a protein-only RNase P enzyme within mitochondria (9), the nucleus-encoded RNA components of RNase P and MRP enzymes are also imported into mitochondria and function in mitochondrial (mt)DNA replication and transcription (10–12). The 5S ribosomal RNA translocates into mitochondria, assembles with ribosomes (13, 14), and may function in mtRNA translation, although the exact function of the 5S ribosomal RNA within mitochondria has yet to be firmly established. Finally, specific nucleus-encoded microRNAs have been isolated from mitochondria (7, 15). Thus, a broad spectrum of nucleus-encoded RNAs localize to mitochondria, although the functions for many of these diverse RNAs in mitochondria have not yet been determined.
The nucleus-encoded RNAs in the mitochondrion have potentially diverse import pathways, but the details of these pathways and import mechanisms are still being revealed (16). tRNAs in every organism are imported by a variety of pathways, some of which require cytoplasmic chaperones (17). The 5S ribosomal RNA is imported from the cytosol with the assistance of two proteins, matrix-localized rhodanese and mitochondrial ribosomal protein L18 (14). The ribosomal protein L18 serves as a conduit or chaperone to facilitate the association of the imported 5S ribosomal RNA with mitochondrial ribosomes. And the mammalian polynucleotide phosphorylase (PNPASE) enzyme localizes to the mitochondrial intermembrane space (18–20) where it may function as an RNA receptor to augment translocation of RNAs into the mitochondrial matrix (6, 21).
Specific mutations in mtDNA have been implicated in muscular and neuronal diseases and in the decline of organ function with aging (22, 23). Despite a significant need, there are currently no effective treatments for deleterious mtDNA alterations. DNA import into mitochondria to repair mtDNA alterations has been difficult and low in efficiency, so approaches at mitochondrial repair have exploited import pathway mechanisms (24). For example, allotopic expression of a subset of mitochondrial genes (recoded mtDNA expressed from the nucleus) has been explored to partially correct the effects of deleterious mutations for some mitochondrial genes, including tRNAs (25–27). However, this approach has been limited because imported mitochondrial proteins fail to assemble correctly in respiratory complexes. The import of RNA has been restricted to tRNAs from different species and requires additional foreign factors. We recently identified a unique approach to target nucleus-encoded RNAs to mitochondria, using the 20-ribonucleotide stem-loop sequence of H1 RNA, the RNA component of the RNase P enzyme that regulates its import (6). When appended to a nonimported RNA, the H1 RNA import sequence, designated RP, enables the fusion transcript to be imported into isolated mitochondria (6).
Two longstanding models of human mtDNA disease are cytoplasmic hybrids (cybrids) derived from samples from patients with myoclonic epilepsy with ragged red fibers (MERRF) and mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) (28–31). These cybrid lines harbor an A8344G (mt-tRNAAAALys) mutation for MERRF and an A3243G (mt-tRNAUURLeu) mutation for MELAS, both of which cause inefficient mtRNA translation, which results in defective cell respiration. We demonstrate here that the mitochondrial defects in these mutant cybrid cells can be partially rescued by targeted import of allotopically encoded wild-type tRNAs, using the RP import signal and, for corrective tRNAs, a mitochondrial localization signal, derived from the 3′-untranslated region (UTR) of human mitochondrial ribosomal protein S12 (MRPS12), which targets the mRNA to the mitochondrial outer membrane (32). We also show that the RP import sequence is also capable of importing much larger, mitochondrial protein-encoding mRNAs in vivo, broadening the approach to target defects in all mtDNA alterations.
Results
H1 RNA Import Sequence Regulates Mitochondrial Import of mt-tRNA Precursors.
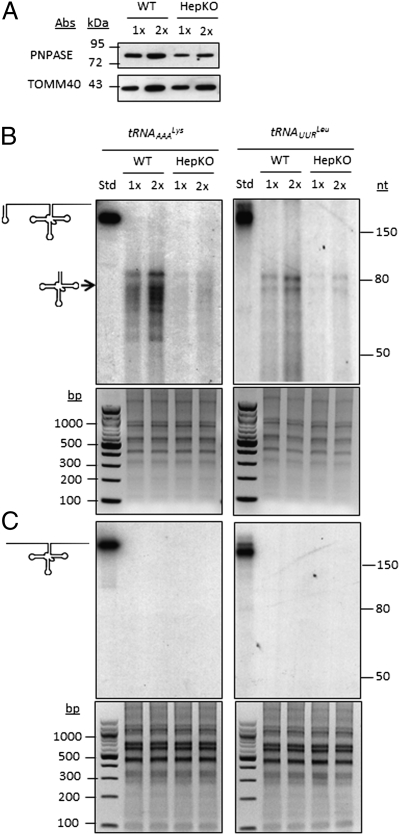
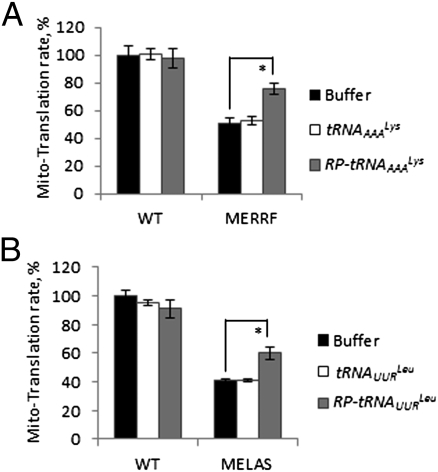
Initially, we determined whether corrective, in vitro synthesized mitochondrial tRNA (mt-tRNA) precursors could be imported into isolated mitochondria and, if so, whether they were processed into mature mt-tRNAs. The mt-tRNAAAALys precursor contains 67 and 74 ribonucleotides, and the mt-tRNAUURLeu precursor contains 93 and 76 ribonucleotides that are cleaved from the 5′ and 3′ transcript ends, respectively, during mt-tRNA maturation (2). In engineering the imported mt-tRNAs, the 5′ end of each mt-tRNA precursor contained or lacked the 20-nt stem-loop sequence of H1 RNA that directs the import of this RNA component of the RNase P enzyme (6); this sequence is designated the RP sequence (6). Engineered tRNAs were then added to import assays that used mouse liver mitochondria isolated from wild-type or a liver-specific “knockout” (designated HepKO) of Pnpt1, the gene encoding for PNPASE (6) (Fig. 1A). The abundance of PNPASE in the HepKO mitochondria was decreased to approximately one-third the level present in WT mitochondria. Residual PNPASE expression in the HepKO liver is from heterogeneous liver elements and from the AlbCRE recombinase strain, which is hepatocyte specific but deletion incomplete (33). Only mt-tRNA precursors with the appended RP sequence were efficiently imported into isolated mitochondria, and import was markedly impaired to approximately one-sixth to one-eighth the level seen in WT mitochondria with reduced PNPASE expression (Fig. 1 B and C). Importantly, the 5′ and 3′ mt-tRNA precursor sequences were removed inside the mitochondria to yield mature 60- to 80-ribonucleotide mt-tRNAs (Fig. 1B). To examine whether the imported mt-tRNAs could rescue defective mtRNA translation, in vitro import was combined with mitochondrial in organello protein synthesis studies. The A8344G mutation (mt-tRNAAAALys) in MERRF and the A3243G mutation (mt-tRNAUURLeu) in MELAS cause a substantial reduction in mtRNA translation (28, 29). A statistically significant increase in the steady-state abundance of total translated mitochondrial polypeptides was observed in both MERRF and MELAS cells following incubation with the mt-tRNA precursors containing the RP import sequence, but not with mt-tRNA precursors lacking RP (Fig. 2 A and B and Fig. S1). These data indicate that the RP sequence enabled PNPASE-dependent mt-tRNA precursor import into isolated mitochondria and that the imported mt-tRNA precursors were processed and functioned in at least partially correcting defective mtRNA translation.
Fig. 1.
H1 RNA import sequence regulates mitochondrial import of mt-tRNA precursors. (A) Hepatocyte-specific Pnpt1 knockout (HepKO) in 6-wk-old mice (6). Immunoblot from 6-wk-old WT and HepKO mouse livers shows ∼50% reduction in PNPASE expression. (B) Radiolabeled mt-tRNA precursors with (Upper) a 5′ H1 20-ribonucleotide predicted stem-loop sequence (designated RP, marked with loop in schematic) were in vitro transcribed and incubated with WT or HepKO liver mitochondria. Nonimported RNA was digested with added nuclease, followed by RNA isolation, separation on a urea acrylamide gel, and autoradiography. Import reactions were repeated with 1× and 2× amounts of mt-tRNA. (Lower) Loading control showing equivalent amounts of mitochondria used in the imports, as revealed by total mitochondrial nucleic acids separated on an agarose gel. (C) As in B, but the mt-tRNA precursor lacks the 5′ H1 20-ribonucleotide predicted stem-loop sequence.
Fig. 2.
Import mt-tRNA precursors with the RP sequence partially rescue the translation defect of isolated MERRF and MELAS mutant mitochondria. (A) mt-tRNA precursors with or without RP were imported into isolated WT or MERRF mitochondria from cybrid lines for 2 min at RT, followed by an additional 5 min with rNTP supplementation. Following RNase A digestion of the nonimported mt-tRNA, mitochondria were pelleted and resuspended in an in organello translation buffer with radiolabeled methionine and cysteine for 30 min at 37 °C. The autoradiograms are shown in Fig. S1. Individual lanes were quantified, and total radioactivity was calculated and normalized to total protein amounts. The WT control in the presence of assay buffer was set at 100%. n = 3 independent experiments. *P < 0.01 with Student's t test. (B) As in A with WT and MELAS cybrid cell lines. n = 3 independent experiments. *P < 0.01 with Student's t test.
The RP Sequence Directs Import of mt-tRNAs into Mitochondria in Vivo.
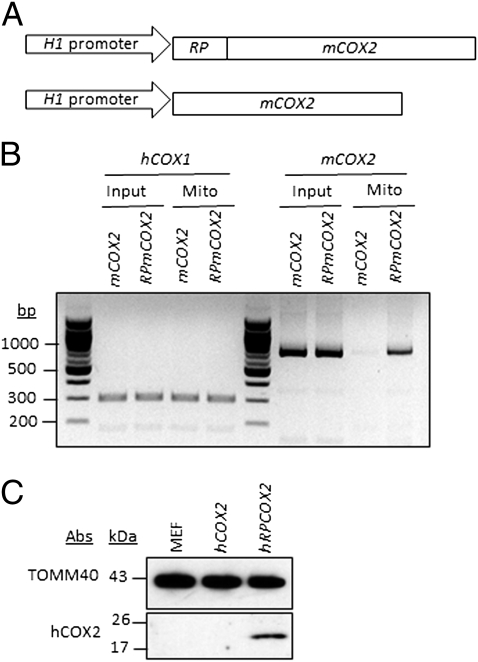
To determine whether the RP import sequence functions in vivo, a mouse cytochrome oxidase 2 (mCOX2) mtRNA was used for import into human cells, because the sequence of mCOX2 differs significantly from that of human COX2 (hCOX2) (1, 2). The mCOX2 gene, with or without the added 5′ RP import sequence, was placed under the control of the H1 promoter (Fig. 3A) and constructs were introduced into HeLa cells via transient transfection. Two days after transfection, mitochondria were isolated and subjected to digitonin treatment (100 μg/1 mg of mitochondrial protein) in the presence of nuclease to generate mitoplasts as a means to determine whether the mCOX2 RNA was indeed imported into the mitochondrial matrix. The presence of the mCOX2 RNA in the mitochondrial matrix was examined by RT-PCR. Only the mtRNA fusion transcript containing the RP import sequence directed the mCOX2 transcript into the mitochondrial matrix with an import efficiency of ∼5–10%, indicating that the RP import sequence is required and functions in vivo (Fig. 3B). To examine whether the imported mtRNA is translated, hCOX2 expression constructs, with or without the RP import sequence, were generated and stably introduced into mouse embryonic fibroblasts, because the monoclonal COX2 antibody is specific for human COX2 protein. Cells expressing RP-hCOX2, but not hCOX2, nucleus-encoded mtRNA showed mitochondrial transcript import (Fig. S2 A and B) and hCOX2 protein translation within mitochondria (Fig. 3C), indicating that the RP import sequence also is required and functions with coding mtRNAs in vivo. hCOX2 localization was also verified within mitochondria (Fig. S2 B and C). hCOX2 protein was trypsin resistant, similar to inner membrane protein TIMM23 (Fig. S2B), in contrast to outer membrane protein TOMM40. In contrast to soluble PNPASE, hCOX2 assembled in the membrane because the protein was recovered in the pellet fraction after carbonate extraction, similar to the control, TOMM40 (Fig. S2C). Thus, hCOX2 is imported and assembles into the mitochondrial inner membrane. The data also show that the RP sequence enables mitochondrial import and processing of RNAs much larger (683 ribonucleotides) than tRNAs (60–80 ribonucleotides), providing a broader therapeutic potential. Thus, the RP sequence can potentially be used in general strategies to target large RNAs for import into mitochondria.
Fig. 3.
In vivo import of mitochondrial-coded COX2 into mitochondria, using the RP sequence. (A) Diagrams of mCOX2 expression vectors. (B) Mitochondria were isolated from HeLa cells expressing mCOX2 or RP-mCOX2. Mitoplasts were made with digitonin, followed by treatment with nuclease. RNA was then isolated from total cell lysates (Input) or from nuclease-treated mitoplasts (Mito) and analyzed by primer-specific RT-PCR. hCOX1 is a control for total and mitochondria-isolated RNAs. (C) Mitochondria were isolated from mouse embryonic fibroblasts stably expressing hCOX2 or RP-hCOX2. hCOX2-specific expression was analyzed by Western blot from isolated mitochondria.
Functional Rescue of Mitochondrial tRNA Mutants.
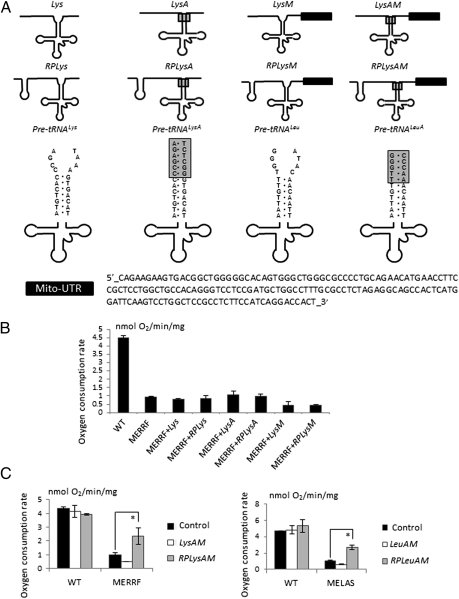
The in vivo rescue of function in mitochondria with mt-tRNA mutations has proven challenging (5). The usual processing of nucleus-encoded tRNA precursors occurs inside the nucleus (34, 35). When stably expressed from inside the nucleus, mt-tRNA precursors with the fused RP sequence did not rescue the respiratory defect of MERRF or MELAS cells (Fig. 4 A and B). Instead, the RP and 5′ mt-tRNA presequences were cleaved inside the nucleus (Fig. S3A). Moving the RP import signal to the 3′ end of the mt-tRNA presequence was also ineffective as both 5′ and 3′ mt-tRNA presequences were cleaved in the nucleus (Fig. S3B). To stop the cleavage of mt-tRNA presequences with the RP sequence inside the nucleus, several ribonucleotides adjacent to the aminoacyl stem of the mt-tRNA were replaced, creating LysA, RPLysA, LeuA, and RPLeuA (Fig. 4A). The ribonucleotides adjacent to tRNA stems are usually unpaired; when the aminoacyl stem is extended by eliminating the mismatch caused by unpaired ribonucleotides, the cleavage rates of tRNA presequences are significantly reduced (36). When LysA, RPLysA, LeuA, and RPLeuA were expressed in mammalian cells, an increase in unprocessed mt-tRNA precursors was detected (Fig. S3C). However, these nucleus-encoded mt-tRNA precursors still failed to rescue the MERRF or MELAS respiration defect (Fig. 4B).
Fig. 4.
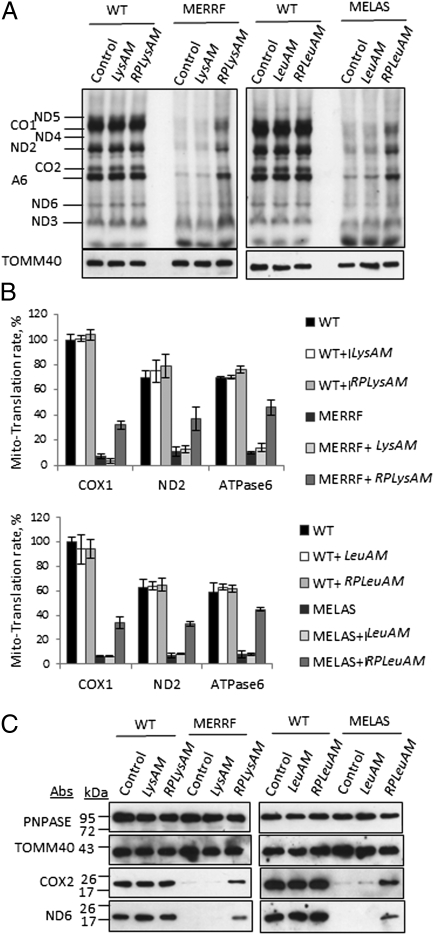
Three elements are required for mt-tRNA precursors encoded in the nucleus to rescue mt-tRNA respiratory defects in vivo. (A) Schematic of the mt-tRNA precursors generated for the in vivo rescue assay. The single step loop at the 5′ of the second row structures is the H1 RNA import sequence, RP. The shaded box indicates ribonucleotides that were changed to make tRNA precursors less susceptible to processing in the nucleus. The solid box is the 3′-UTR of MRPS12 that localizes RNA to the vicinity of mitochondria (32). (B) tRNALys precursors lacking one or two of the three elements do not rescue the MERRF respiratory defect. (C) tRNALys or tRNALeu precursors with all three elements rescue respiration in MERRF and MELAS cells. n = 3 independent experiments. *P < 0.01 with Student's t test.
We reasoned that the mt-tRNA precursors might not localize near the mitochondria and, therefore, the RP sequence could not function as an import signal, as it does with isolated mitochondria in vitro. Consistent with this reasoning is the known requirement for regional localization of cellular mRNAs. It was originally assumed that mRNAs associated only with the endoplasmic reticulum, but now it has been shown that different mRNAs can be targeted to different subcellular locations where translation occurs locally (37). Mitochondrial mRNAs, mainly of prokaryotic origin, are targeted to the mitochondrion by a 3′-UTR (38, 39). As an example, the mRNA of human mitochondrial ribosomal protein S12 (MRPS12) is targeted to the proximity of mitochondria through the microtubule network (32). Targeting is mediated by elements within 154 ribonucleotides of its 3′-UTR (32). To test whether the 3′-UTR of MRPS12 assists in the import of mt-tRNA precursors into mitochondria in vivo, we fused the MRPS12 3′-UTR to the 3′ end of stabilized and RP-containing mt-tRNA precursors. In total, eight expression constructs were generated, including LysM (mt-tRNAAAALys precursor with MRPS12 3′-UTR), RPLysM (mt-tRNAAAALys precursor with RP and MRPS12 3′-UTR), LysAM (mt-tRNAAAALys precursor with the extended stem and MRPS12 3′-UTR), RPLysAM (mt-tRNAAAALys precursor with RP, the extended stem, and MRPS12 3′-UTR), LeuM (mt-tRNAUURLeu precursor with MRPS12 3′-UTR), RPLeuM (mt-tRNAUURLeu precursor with RP and MRPS12 3′-UTR), LeuAM (mt-tRNAUURLeu precursor with the extended stem and MRPS12 3′-UTR), and RPLeuAM (mt-tRNAUURLeu precursor with RP, the extended stem, and MRPS12 3′-UTR) (Fig. 4A).
Stable polyclonal transfectants with the various tRNA chimeras in MERRF and MELAS cells were made and cell respiration was measured with an XF24 Extracellular Flux Analyzer (Seahorse Biosciences). When all three elements (the extended stem, the RP sequence, and MRPS12 3′-UTR) were present, the mt-tRNA precursors rescued MELAS and MERFF respiration defects (∼2.5-fold increase); otherwise, no rescue was detected (Fig. 4C). Expression of mt-tRNA precursors in vivo did not have a significant effect on the respiration of wild-type cybrid cells, suggesting little perturbation of other cellular functions (Fig. 4C). To evaluate whether the rescue of respiration is from a correction in mtRNA translation by imported wild-type mt-tRNAs, an in vivo mitochondrial translation assay was performed with MERRF and MELAS cells expressing different versions of the mt-tRNA precursors. Consistent with the respiration results, MERRF and MELAS cells showed a substantial reduction in the synthesis of mitochondrion-encoded proteins compared with the wild-type cybrid cells. Only when the mt-tRNA precursors with all three elements were expressed, did mitochondrial protein synthesis recover (threefold to sixfold increase) (Fig. 5 A and B). A complete recovery was not expected, as the mutant mt-tRNAs were still present in the mitochondria and likely competed with the imported wild-type mt-tRNAs during mtRNA translation. Stable mitochondrial protein levels in MERRF and MELAS cells were also examined. Consistent with the increase in respiration and in vivo translation results in the mutant cells expressing the mt-tRNA precursors containing all three elements, the levels of mitochondrial-encoded COX2 and ND6 proteins were markedly increased (Fig. 5C). To assess the assembly and activity of the respiratory chain complexes, in-gel activity assays for complex I were performed, using blue-native gel electrophoresis. In the MELAS and MERRF cells expressing the mt-tRNA precursors containing all three elements, the activity of complex I increased to 30–40% of the WT level (Fig. S4). These results show that for nucleus-encoded mt-tRNA precursors to be imported into and adequately function within mitochondria, an extended stem is required for a subpopulation of these precursors to escape the nucleus without the presequences being processed. Also, a mitochondria localization sequence such as MRPS12 3′-UTR is required, probably for the precursor to traffic to the proximity of mitochondria. And finally, an RNA import sequence, in this case RP, is required for the mitochondria to internalize the mt-tRNA precursor.
Fig. 5.
Rescue of respiration is due to restoration of mitochondrial translation. (A) Analysis of mitochondrial translation in vivo with stable rescue cell lines. Mitochondrial translated proteins were separated by SDS/PAGE and visualized by autoradiography. (B) Quantification of specific bands on gels from A. Autoradiogram counts were normalized to protein amounts and expressed relative to WT control samples. (C) Steady-state levels of nucleus-encoded and mitochondrial-encoded proteins in WT, MERRF, and MELAS cells. TOMM40 and PNPASE also serve as loading controls.
Discussion
Defects in mtDNA are implicated in a plethora of human conditions, including neurodegenerative and cardiovascular diseases, muscular disorders, and the process of aging (22, 23). Despite a significant need, there are currently no effective treatments for deleterious mtDNA alterations. Our results show that these disorders can potentially be treated using targeted mitochondrial RNA import. We show that the RP import sequence mediates import of not only mitochondrial tRNA precursors but also much larger nucleus-expressed mRNAs into the mitochondrial matrix. Disease-causing mutations and deletions in the mitochondrial genome seem to be fairly equally distributed along the mitochondrial DNA (5, 22, 23) and this approach may generalize to rescue them all.
Up to now, experimental approaches using mitochondrial RNA import to rescue mitochondrial function have been restricted to correcting the defects caused by mitochondrial tRNA mutations. These efforts require the introduction of nonnative tRNAs with foreign protein factors or the transfer of a large multisubunit aggregate into cells, which is of low efficiency and difficult to reproduce in desirable disease-relevant settings (27, 40, 41). Our approach, however, has no such restriction; rationally engineered human mitochondrial tRNAs and mRNAs can both be efficiently targeted and functional. The fusion RNA presequences are encoded in the nuclear genome and can be imported into mitochondria where they are processed, restore translation, and are degraded via normal pathways in the mitochondrion. Importantly, these modified RNAs do not appear to have negative effects on other cellular processes, as wild-type cell metabolism is not affected. In vivo, the efficient import also depends on how well the RP import sequence is protected from processing in the nucleus and cytosol and whether the RNA precursors can gain access to the vicinity of mitochondria. We have shown that even for precursors that are normally processed in the nucleus and have no access to the vicinity of mitochondria, there are avenues to manipulate the processing and trafficking, although it is interesting that, at least for allotopically expressed COX2 transcripts, a mitochondrial localization sequence is not required for RP sequence-PNPASE–mediated import.
The approach we have described here can be adapted for importing different kinds of RNA into mammalian mitochondria. The RNA sequences can be altered to inhibit processing steps in maturation in the nucleus without affecting the function of the mature RNAs in the mitochondria. For RNAs sequestered elsewhere in the cell, a mitochondria localization sequence from the 3′-UTR of MRPS12 can be appended to redirect the RNA precursor to the mitochondrion. Thus, this approach may generalize to mtDNA mutations in mt-tRNAs, mt-rRNAs, and protein-encoding mtRNAs as well as to heteroplasmic mtDNA populations, where ribozymes can be targeted (42). Thus, this rational transcript engineering approach may represent a unique therapeutic opportunity for a wide range of diseases caused by mutations in the mitochondrial genome for which current effective therapies are lacking.
Materials and Methods
Cell Culture, Transfection, and Transduction.
Mammalian cell lines were maintained in DMEM growth medium supplemented with 10% FBS and 2% l-glutamine. MERRF and MELAS cybrid lines (kindly provided by Carlos Moraes, University of Miami Miller School of Medicine) were maintained in DMEM growth medium supplemented with 10% FBS, 2% l-glutamine, and 0.5 mg/mL uridine. Transient transfections were performed using Bio-T reagent (Bioland Scientific). In transfections with COX2 constructs, the calcium phosphate uptake method was used. Retroviral supernatants were produced by transient transfection of the 293T Phoenix packaging cell line, after which the cells were bulk infected and selected in puromycin.
Biochemical Assays with Mitochondria and Additional Methods.
Detailed methods are listed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Carlos Moraes at the University of Miami Miller School of Medicine for kindly providing the MELAS and MERRF cybrid cell lines. This work was supported by grants from the National Institutes of Health (GM061721, GM073981, CA90571, and CA156674), the American Heart Association (0640076N), the California Institute of Regenerative Medicine (RS1-00313 and RB1-01397), and the Broad Stem Cell Research Center at University of California, Los Angeles.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116792109/-/DCSupplemental.
References
- 1.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Entelis NS, Kolesnikova OA, Martin RP, Tarassov IA. RNA delivery into mitochondria. Adv Drug Deliv Rev. 2001;49:199–215. doi: 10.1016/s0169-409x(01)00135-1. [DOI] [PubMed] [Google Scholar]
- 5.Alfonzo JD, Söll D. Mitochondrial tRNA import—the challenge to understand has just begun. Biol Chem. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duchêne AM, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 9.Holzmann J, et al. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Chang DD, Clayton DA. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- 11.Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 12.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: Multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 13.Smirnov A, et al. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. RNA. 2008;14:749–759. doi: 10.1261/rna.952208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smirnov A, Entelis N, Martin RP, Tarassov I. Biological significance of 5S rRNA import into human mitochondria: Role of ribosomal protein MRP-L18. Genes Dev. 2011;25:1289–1305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kren BT, et al. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sieber F, Duchêne AM, Maréchal-Drouard L. Mitochondrial RNA import: From diversity of natural mechanisms to potential applications. Int Rev Cell Mol Biol. 2011;287:145–190. doi: 10.1016/B978-0-12-386043-9.00004-9. [DOI] [PubMed] [Google Scholar]
- 17.Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem. 2011;80:1033–1053. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- 18.Rainey RN, et al. A new function in translocation for the mitochondrial i-AAA protease Yme1: Import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HW, et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen HW, Koehler CM, Teitell MA. Human polynucleotide phosphorylase: Location matters. Trends Cell Biol. 2007;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Shimada E, Koehler CM, Teitell MA. PNPASE and RNA trafficking into mitochondria. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 23.Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci USA. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyriakouli DS, Boesch P, Taylor RW, Lightowlers RN. Progress and prospects: Gene therapy for mitochondrial DNA disease. Gene Ther. 2008;15:1017–1023. doi: 10.1038/gt.2008.91. [DOI] [PubMed] [Google Scholar]
- 25.Manfredi G, et al. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 26.Karicheva OZ, et al. Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic Acids Res. 2011;39:8173–8186. doi: 10.1093/nar/gkr546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolesnikova OA, et al. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- 28.Masucci JP, Davidson M, Koga Y, Schon EA, King MP. In vitro analysis of mutations causing myoclonus epilepsy with ragged-red fibers in the mitochondrial tRNA(Lys)gene: Two genotypes produce similar phenotypes. Mol Cell Biol. 1995;15:2872–2881. doi: 10.1128/mcb.15.5.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schon EA, Koga Y, Davidson M, Moraes CT, King MP. The mitochondrial tRNA(Leu)(UUR) mutation in MELAS: A model for pathogenesis. Biochim Biophys Acta. 1992;1101:206–209. [PubMed] [Google Scholar]
- 30.Kishnani PS, et al. Acute pancreatitis in an infant with lactic acidosis and a mutation at nucleotide 3243 in the mitochondrial DNA tRNALeu(UUR) gene. Eur J Pediatr. 1996;155:898–903. doi: 10.1007/BF02282842. [DOI] [PubMed] [Google Scholar]
- 31.Shoffner JM, et al. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 32.Russo A, et al. cis-acting sequences and trans-acting factors in the localization of mRNA for mitochondrial ribosomal proteins. Biochim Biophys Acta. 2008;1779:820–829. doi: 10.1016/j.bbagrm.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 34.Ceballos M, Vioque A. tRNase Z. Protein Pept Lett. 2007;14:137–145. doi: 10.2174/092986607779816050. [DOI] [PubMed] [Google Scholar]
- 35.Frank DN, Pace NR. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, et al. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA. 1997;3:175–185. [PMC free article] [PubMed] [Google Scholar]
- 37.Donnelly CJ, Fainzilber M, Twiss JL. Subcellular communication through RNA transport and localized protein synthesis. Traffic. 2010;11:1498–1505. doi: 10.1111/j.1600-0854.2010.01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marc P, et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sylvestre J, Margeot A, Jacq C, Dujardin G, Corral-Debrinski M. The role of the 3′ untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol Biol Cell. 2003;14:3848–3856. doi: 10.1091/mbc.E03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahata B, Mukherjee S, Mishra S, Bandyopadhyay A, Adhya S. Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science. 2006;314:471–474. doi: 10.1126/science.1129754. [DOI] [PubMed] [Google Scholar]
- 41.Kolesnikova OA, et al. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- 42.Val R, et al. Organelle trafficking of chimeric ribozymes and genetic manipulation of mitochondria. Nucleic Acids Res. 2011;39:9262–9274. doi: 10.1093/nar/gkr580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.