Abstract
O-linked β-N-acetylglucosamine (O-GlcNAc) is a reversible posttranslational modification found on hundreds of nuclear and cytoplasmic proteins in higher eukaryotes. Despite its ubiquity and essentiality in mammals, functional roles for the O-GlcNAc modification remain poorly defined. Here we develop a combined genetic and chemical approach that enables introduction of the diazirine photocrosslinker onto the O-GlcNAc modification in cells. We engineered mammalian cells to produce diazirine-modified O-GlcNAc by expressing a mutant form of UDP-GlcNAc pyrophosphorylase and subsequently culturing these cells with a cell-permeable, diazirine-modified form of GlcNAc-1-phosphate. Irradiation of cells with UV light activated the crosslinker, resulting in formation of covalent bonds between O-GlcNAc-modified proteins and neighboring molecules, which could be identified by mass spectrometry. We used this method to identify interaction partners for the O-GlcNAc-modified FG-repeat nucleoporins. We observed crosslinking between FG-repeat nucleoporins and nuclear transport factors, suggesting that O-GlcNAc residues are intimately associated with essential recognition events in nuclear transport. Further, we propose that the method reported here could find widespread use in investigating the functional consequences of O-GlcNAcylation.
Keywords: diazirine, glycosylation, metabolism, nucleoporins, posttranslational modification
O-linked β-N-acetylglucosamine (O-GlcNAc) is a common modification of intracellular proteins in metazoa and higher plants (1). Hundreds of O-GlcNAc-modified human nuclear and cytoplasmic proteins have been identified. Modified proteins fall into a variety of functional classes and include transcription factors, ribosomal proteins and translational factors, signaling proteins, cytoskeletal proteins, and components of the nuclear pore complex. Like phosphorylation, O-GlcNAcylation is reversible. Mammalian genomes encode a single O-GlcNAc transferase (OGT) that transfers GlcNAc from the nucleotide sugar donor UDP-GlcNAc to serine or threonine side chains of substrate proteins, and one O-GlcNAcase (nuclear cytoplasmic O-GlcNAcase and acetyltransferase; NCOAT) that hydrolytically removes GlcNAc residues from modified proteins. The O-GlcNAc modification is essential in mammals (2) and O-GlcNAc is critical to cells’ ability to tolerate a variety of forms of stress (3, 4).
While characterization of O-GlcNAc-modification sites has advanced dramatically (5), a comprehensive understanding of the functional consequences of O-GlcNAc modification remains more elusive (6). How does O-GlcNAc affect a protein’s activity, stability, and ability to engage in binding interactions? O-GlcNAc is often found at sites that can be alternatively phosphorylated, leading to a reciprocal relationship between these two modifications. This relationship led to the hypothesis that a key role for O-GlcNAc is to interfere with phosphorylation (7). In fact, experimental evidence suggests that the interplay between these two modifications is complex, involving both negative and positive associations (8–10). Another documented function for O-GlcNAc is to disrupt binding interactions of the modified proteins, presumably by steric interference (11). Less clear is whether the O-GlcNAc modification can impart a novel activity to the modified protein, although recent findings suggest that increased levels of O-GlcNAc can be associated with acquisition of function (12–14).
Selectively observing the cellular behavior of O-GlcNAc-modified proteins remains challenging. Most methods report on the bulk behavior of the protein of interest and do not distinguish among the different posttranslationally modified forms. We reasoned that appending a small photoactivatable crosslinking group to the O-GlcNAc modification would enable selectively induced covalent crosslinking between an O-GlcNAc-modified protein and surrounding molecules. Because the crosslinker is on the O-GlcNAc residue, proteins that lack the modification will not engage in crosslinking and will be rendered essentially invisible in this assay. In this way, even proteins that are O-GlcNAc-modified at substoichiometric levels could be examined.
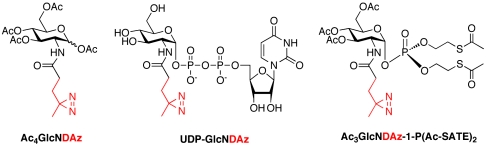
To implement the photocrosslinking approach, we designed an analog of GlcNAc, GlcNDAz, in which the N-acyl substituent was modified to include the diazirine photocrosslinker (Fig. 1). Using a metabolic labeling approach, we induced cultured cells to produce the modified nucleotide sugar donor UDP-GlcNDAz and to transfer GlcNDAz to proteins that are normally O-GlcNAc-modified. Subsequent photoirradiation resulted in the selective covalent crosslinking of O-GlcNDAz-modified proteins. Mass spectrometry analysis of purified complexes revealed crosslinking between O-GlcNAc-modified nucleoporins and nuclear transport factors. These results indicate that the O-GlcNAc modification is intimately associated with the recognition events that occur during nuclear transport.
Fig. 1.
Diazirine-modified GlcNAc derivatives used in this study.
Results
In Vitro Production and Crosslinking of O-GlcNDAz-modified Peptides.
We envisioned that introduction of a photocrosslinking group directly onto an O-GlcNAc residue would enable the covalent crosslinking between the O-GlcNAc modification and proximal molecules. To test this idea, we assessed crosslinking between an O-GlcNDAz-modified peptide and a monoclonal antibody, RL2 (15), that specifically recognizes the O-GlcNAc modification. First, we prepared the diazirine-modified nucleotide-sugar donor UDP-GlcNDAz (Fig. 1) (16) and incubated it with recombinant human OGT and a biotinylated substrate peptide derived from casein kinase II (CKII) (17). The product mixture was analyzed by MALDI mass spectrometry, revealing a mass consistent with transfer of the GlcNDAz residue to the CKII peptide (Fig. S1A). Next, the O-GlcNDAz-modified peptide was incubated with RL2 and UV irradiated to induce photocrosslinking (Fig. S2A). Immunoblot analysis revealed a biotinylated species migrating with an apparent molecular weight identical to that of the light chain of RL2 (Fig. S2B). Observation of the biotinylated species depended on inclusion of the crosslinker (UDP-GlcNDAz) and on UV irradiation. We concluded that this biotinylated species represented the O-GlcNDAz-modified peptide crosslinked to the light chain of the RL2 antibody. These results indicate that the addition of the diazirine modification to GlcNAc does not preclude transfer of the sugar by OGT nor does it abrogate the binding interaction between O-GlcNAc-modified CKII and RL2.
Having established that human OGT is capable of accepting UDP-GlcNDAz, we conducted competition experiments to determine whether OGT would transfer GlcNDAz from UDP-GlcNDAz when UDP-GlcNAc was also available. Lysates prepared from HeLa cells overexpressing human OGT were incubated with UDP-GlcNAc, UDP-GlcNDAz or an equimolar mixture of the two, along with an OGT acceptor peptide (+P2) derived from human α-A crystallin (18) (Fig. S1B). Lysates incubated with UDP-GlcNAc produced a product with the mass (m/z = 1,241) expected for an O-GlcNAc-modified peptide. Lysates incubated with UDP-GlcNDAz produced a product whose mass corresponded to the intact O-GlcNDAz-modified peptide (m/z = 1,309) and an additional product whose mass was consistent with loss of N2 from the O-GlcNDAz-modified peptide (m/z = 1,281). Loss of N2 likely occurs during the mass spectrometry ionization step, a phenomenon we have observed with other diazirine-containing molecules (19). When OGT-containing lysates were incubated with an equimolar mixture of UDP-GlcNAc and UDP-GlcNDAz, masses corresponding to both O-GlcNAc- and O-GlcNDAz-modified peptides were observed. We repeated the experiment with recombinant OGT, rather than lysate, and analyzed the peptide products by HPLC. The O-GlcNAc-modified peptide was the major product (Fig. S1C), but the O-GlcNDAz-modified peptide was also observed. While these data indicate that OGT prefers UDP-GlcNAc over UDP-GlcNDAz, the O-GlcNDAz-modified peptide was readily produced when the two nucleotide sugars were present in comparable amounts.
Cellular Production of UDP-GlcNDAz.
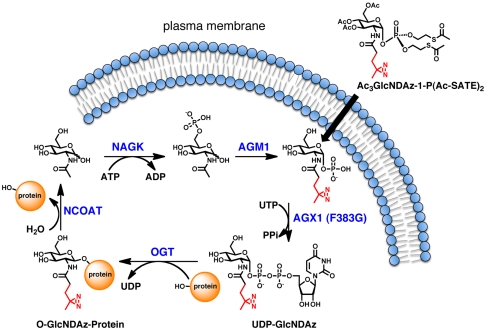
We next focused attention on engineering cells to produce UDP-GlcNDAz, so that O-GlcNDAz-modified proteins could be produced in cells (Fig. 2). In mammalian cells, UDP-GlcNAc can be produced from GlcNAc through three enzymatic steps of the GlcNAc salvage pathway. Hoping to take advantage of the endogenous pathway, we prepared a peracetylated form of GlcNDAz, Ac4GlcNDAz, and added it to the media of cultured cells to test whether the cells could convert this compound to UDP-GlcNDAz. We used high performance anion exchange chromatography (HPAEC) analysis of the lysates to assess UDP-GlcNDAz production in multiple cell lines (HeLa, Jurkat, BJAB K20), but could not detect the photocrosslinking nucleotide sugar (UDP-GlcNDAz). We also failed to observe production of GlcNDAz-1-P, suggesting inadequate conversion by either N-acetylglucosamine kinase (NAGK), which phosphorylates GlcNAc at C6, or N-acetylglucosamine-phosphate mutase (AGM1), which converts GlcNAc-6-P to GlcNAc-1-P. To circumvent this metabolic restriction, we prepared a form of GlcNDAz-1-P in which the hydroxyl groups are peracetylated and the phosphate is protected with two S-acetyl-2-thioethyl (Ac-SATE) groups. We predicted that Ac3GlcNDAz-1-P(Ac-SATE)2 would be capable of diffusing across the plasma membrane, after which the protecting groups would be removed by chemical hydrolysis or the action of intracellular esterases (20). Indeed, HPAEC analysis of HeLa lysates revealed that cells cultured with Ac3GlcNDAz-1-P(Ac-SATE)2 readily accumulated intracellular GlcNDAz-1-P; however, UDP-GlcNDAz was not detected (Fig. S3A). These data suggested that GlcNDAz-1-P is a poor substrate for AGX1 and/or AGX2, the two splice variant isoforms of UDP-GlcNAc pyrophosphorylase.
Fig. 2.
Metabolic labeling strategy for cellular biosynthesis of O-GlcNDAz-modified proteins. Cellular GlcNAc can be converted to UDP-GlcNAc via three enzymatic steps catalyzed by N-acetylglucosamine kinase (NAGK), N-acetylglucosamine-phosphate mutase (AGM1), and UDP-GlcNAc pyrophosphorylase (AGX1 or AGX2). HeLa cells expressing AGX1(F383G) and cultured with Ac3GlcNDAz-1-P(Ac-SATE)2 efficiently produced UDP-GlcNDAz.
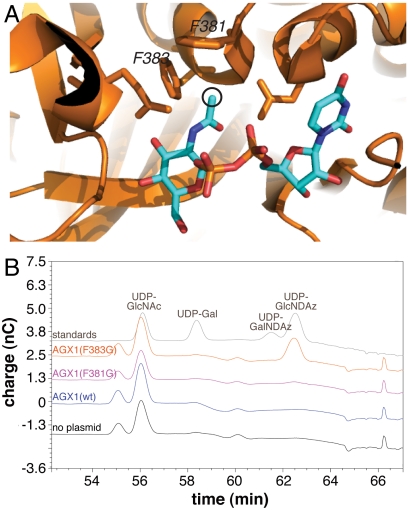
Examination of the X-ray crystal structure of human AGX1 (PDB code 1JV1) (21) revealed that the N-acetyl group of the bound UDP-GlcNAc resides in a compact hydrophobic pocket bounded by two phenylalanine residues (Fig. 3A; F381 and F383). We hypothesized that the hydrophobic pocket was too small to accommodate the N-acyl diazirine substituent and designed mutants of AGX1 (F381G and F383G) predicted to have expanded binding pockets. HeLa cells were transiently transfected with plasmids encoding wild-type AGX1, AGX1(F381G), or AGX1(F383G), and cultured in media containing Ac3GlcNDAz-1-P(Ac-SATE)2. HPAEC analysis of the lysates showed that cells transfected with mutant AGX1(F383G) efficiently produced a metabolite whose mobility matched that of synthetic UDP-GlcNDAz (Fig. 3B). The identity of UDP-GlcNDAz was confirmed by UV irradiating both the lysate and the UDP-GlcNDAz standard compound and comparing their HPAEC patterns (Fig. S3B). In addition, the UDP-GlcNDAz peak from the AGX1(F383G)-expressing lysate was collected and analyzed by MALDI-TOF mass spectrometry, revealing the expected mass (Fig. S3C). HeLa cells transfected with wild-type AGX1 did not produce UDP-GlcNDAz, indicating that UDP-GlcNDAz production was not simply due to AGX1 overexpression (Fig. 3B). Furthermore, UDP-GlcNDAz production occurs in a time-dependent manner (Fig. S3D). Thus, expression of AGX1(F383G) permits conversion of GlcNDAz-1-P to UDP-GlcNDAz in cells, likely by expanding the enzyme’s binding pocket to accommodate the unnatural sugar. Notably, cells expressing AGX1(F383G) and cultured in media containing Ac4GlcNDAz did not produce UDP-GlcNDAz, confirming that conversion of GlcNDAz to GlcNDAz-1-P is inefficient (Fig. S3E).
Fig. 3.
Structure-guided mutagenesis of AGX1 results in efficient UDP-GlcNDAz production. (A) The X-ray crystal structure of human AGX1 (PDB code 1JV1)(21) reveals that F381 and F383 surround the N-acetyl group of UDP-GlcNAc. Black circle indicates position where the unnatural alkyl diazirine substituent is attached in UDP-GlcNDAz. (B) HPAEC-PAD analysis of lysates from HeLa cells transiently transfected with DNA encoding wild-type AGX1, AGX1(F381G), AGX1(F383G), or no vector and cultured with Ac3GlcNDAz-1-P(Ac-SATE)2. Chromatogram of standard nucleotide sugars is shown at the top.
Cellular Production of O-GlcNDAz-modified proteins.
We wished to investigate whether HeLa cells producing UDP-GlcNDAz could incorporate GlcNDAz into O-GlcNDAz-modified proteins. We reasoned that UV irradiation would result in covalent conjugation of the O-GlcNDAz-modified proteins to neighboring molecules. Thus, irradiation of an O-GlcNDAz-modified protein should increase the protein’s apparent molecular weight, as assessed by immunoblot analysis. To test this idea, we examined the known interaction between O-GlcNAc-modified proteins and the O-GlcNAc-recognizing antibody, RL2. We transfected HeLa cells with AGX1(F383G) and cultured them in media containing Ac3GlcNDAz-1-P(Ac-SATE)2. Cell lysates were incubated with RL2. After UV irradiation, RL2 was precipitated by protein G beads. The elute from the beads was analyzed by immunoblot, using antibodies that recognize two known O-GlcNAc-modified proteins, transcription factor SP1 and nucleoporin NUP153 (Fig. S4). In both cases, we observed distinct higher molecular weight complexes only when the cells were both transfected with AGX1(F383G) and cultured in media containing Ac3GlcNDAz-1-P(Ac-SATE)2. This result strongly suggested that the cells produce O-GlcNDAz-modified proteins and that O-GlcNDAz-modified proteins can be selectively photocrosslinked to neighboring molecules.
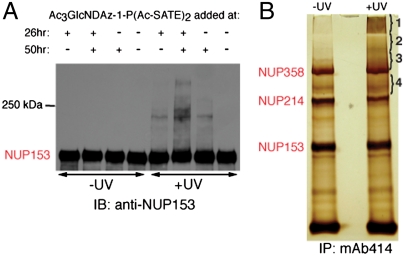
Next, we investigated whether O-GlcNDAz-modified proteins could be crosslinked to endogenous binding partners in live cells. Again, HeLa cells expressing AGX1(F383G) were cultured with Ac3GlcNDAz-1-P(Ac-SATE)2. Cultured cells were irradiated, lysed, and analyzed by immunoblot using mAb414, which recognizes nucleoporins containing phenylalanine-glycine (FG) repeats, including NUP62, NUP153, NUP214, NUP358 (Fig. S5A). In addition to bands corresponding to the nucleoporins, additional mAb414 reactivity at higher molecular weights was observed only when cells were transfected with AGX1(F383G) and cultured with Ac3GlcNDAz-1-P(Ac-SATE)2. Adding Ac3GlcNDAz-1-P(Ac-SATE)2 at two separate time points enhanced formation of the crosslinked species. While the higher molecular weight species on the mAb414 immunoblot were not well resolved, distinct photocrosslinked complexes could be observed by probing with antibodies specific for an individual nucleoporin, NUP153 (Fig. 4A) or NUP62 (Fig. S5B). Taken together, these results support the idea that crosslinking is occurring through a diazirine-modified O-GlcNAc residue. In addition, we found similar effects on nucleoporin crosslinking when we introduced AGX1(F383G) and Ac3GlcNDAz-1-P(Ac-SATE)2 into T84 intestinal epithelial cells (Fig. S5C and S5D), indicating that the engineering strategy is not cell line-specific.
Fig. 4.
FG-repeat nucleoporins are crosslinked through O-GlcNDAz residues. (A) HeLa cells transiently expressing AGX1(F383G) were cultured with Ac3GlcNDAz-1-P(Ac-SATE)2 (added 26 and/or 50 h after transfection). Cells were irradiated with 365 nm light, then lysed. Lysates were analyzed by immunoblot using a rabbit anti-NUP153 antibody. (B) HeLa cells stably expressing AGX1(F383G) were cultured with Ac3GlcNDAz-1-P(Ac-SATE)2. Irradiated and nonirradiated cells were lysed and lysates were immunoprecipitated with mAb414. Immunoprecipitates were separated by SDS-PAGE and visualized by silver staining. Indicated regions of the UV-irradiated sample were analyzed by mass spectrometry.
FG-repeat Nucleoporins Interact with Nuclear Transport Factors.
Our next goal was to identify proteins that were covalently conjugated to FG-repeat nucleoporins via O-GlcNDAz directed crosslinking. To accomplish this, we prepared HeLa cells that stably expressed AGX1(F383G) and cultured the cells with Ac3GlcNDAz-1-P(Ac-SATE)2. These UDP-GlcNDAz-producing cells were UV irradiated and lysed. We conducted an immunoprecipitation of the lysate using mAb414. The immunoprecipitate was separated by SDS-PAGE and visualized by silver-staining (Fig. 4B). The banding pattern was compared to an identical lysate that was not UV irradiated. We identified molecular weight ranges in which significant silver staining was observed in the UV-irradiated sample, but not the nonirradiated sample. These regions were excised from the gel and subjected to in-gel trypsin digest, followed by high performance liquid chromatography tandem mass spectrometry (HPLC/MS/MS) analysis to identify tryptic peptides. The Mascot search engine was used to analyze the mass spectrometry data and develop a candidate list of proteins present in the immunoprecipitate (Table S1). As expected, the putative crosslinked regions contained nucleoporins recognized by mAb414: NUP153, NUP214, and NUP358. In addition, peptides corresponding to several known nuclear transport factors were identified: exportin-1 (CRM1), transportin-1 (TNPO1), transportin-2 (TNPO2), importin subunit β1 (KPNB1), and nuclear RNA export factor 1 (NXF1). The remaining lower confidence hits also corresponded to proteins (histone 2B and γ-catenin) known to enter the nucleus.
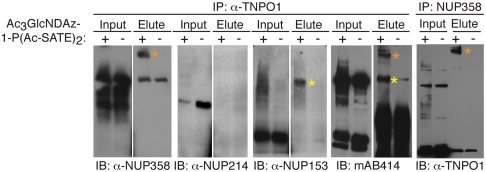
To validate the mass spectrometry data, we used a reciprocal immunoprecipitation strategy. UDP-GlcNDAz-producing HeLa cells were UV irradiated, then lysed and immunoprecipitated with an anti-TNPO1 antibody to isolate TNPO1 along with any covalent complexes that contained TNPO1. The immunoprecipitate was analyzed by immunoblot using mAb414, as well as antibodies specific for the individual nucleoporins, NUP153, NUP214, and NUP358. As predicted, the NUP153 and NUP358 antibodies each recognized a higher molecular weight species, consistent with crosslinking between TNPO1 and the nucleoporins. In addition, mAb414 recognized two high molecular weight species, whose apparent molecular weights matched those of the putative crosslinked complexes recognized by anti-NUP153 and anti-NUP358 (Fig. 5). Further confirmation of TNPO1 crosslinking to NUP358 came from a reverse immunoprecipitation experiment. We immunoprecipitated the crosslinked HeLa cell lysate with anti-NUP358 and analyzed the immunoprecipitate with anti-TNPO1 immunoblotting. The anti-NUP358 immunoprecipitate showed clear evidence of a covalent TNPO1-NUP358 complex (Fig. 5, Right). Taken together, these results demonstrate that FG-repeat nucleoporins (namely NUP358 and likely NUP153) engage in interactions with the nuclear transport factor TNPO1 and that an O-GlcNAc residue is located at or near to the interaction site. We did not obtain evidence for crosslinking between TNPO1 and NUP214, which could reflect a lack of interaction between these two proteins or the fact that NUP214 crosslinked complexes were poorly visualized on the immunoblot (see input lanes in Fig. 5).
Fig. 5.
Transportin-1 interacts with NUP153 and NUP358. HeLa cells stably expressing AGX1(F383G) were cultured with or without Ac3GlcNDAz-1-P(Ac-SATE)2. After UV irradiation, the cells were lysed and the lysates were immunoprecipitated with the indicated antibodies. Input lysates and immunoprecipitates were analyzed by immunoblots, as indicated. Two distinct crosslinked species were identified: NUP358-TNPO1 (orange asterisk) and NUP153-TNPO1 (yellow asterisk).
Effects on O-GlcNAc Metabolism.
Our metabolic labeling method relies on the introduction of an unnatural metabolite, Ac3GlcNDAz-1-P(Ac-SATE)2, and the overexpression of a mutant form of a metabolic enzyme, AGX1. Thus, we were concerned that this strategy could result in perturbations to the normal O-GlcNAc patterns in the cell. Cells stably transfected with the AGX1(F383G)-encoding plasmid produced dramatically increased levels of AGX1 (Fig. S6A), which might be expected to lead to an increased pool of UDP-GlcNAc/UDP-GlcNDAz. We found that the effect of AGX1(F383G) overexpression on the UDP-GlcNAc level was slight (Fig. 3B), although AGX1(F383G) overexpression does lead to the added appearance of UDP-GlcNDAz. Overall, the amount of UDP-GlcNAc/UDP-GlcNDAz in the engineered cells is about twofold higher than the amount of UDP-GlcNAc in normal cells.
We also examined whether NCOAT, the hydrolytic enzyme that removes O-GlcNAc, was capable of recognizing diazirine-modified sugar. We prepared lysates from normal HeLa cells and from HeLa cells transiently transfected with a plasmid encoding NCOAT. These lysates were incubated with artificial substrates, p-nitrophenyl-β-d-GlcNAc (pNP-GlcNAc) or p-nitrophenyl-β-d-GlcNDAz (pNP-GlcNDAz). We observed significant hydrolysis of pNP-GlcNAc, which dramatically increased with NCOAT overexpression, while hydrolysis of pNP-GlcNDAz was barely detectable with either lysate (Fig. S7). This result implies that O-GlcNDAz residues are resistant to removal by NCOAT and suggests that O-GlcNDAz-modified proteins likely accumulate in cells. Despite effects on the UDP-GlcNAc/UDP-GlcNDAz pool and the resistance of O-GlcNDAz to NCOAT removal, when we probed the lysates from HeLa cells expressing AGX1(F383G) and cultured with Ac3GlcNDAz-1-P(Ac-SATE)2 using RL2, we saw no significant alterations in O-GlcNAcylation (Fig. S6B). This result suggests that our metabolic labeling method does not dramatically perturb the normal O-GlcNAcylation pattern in these cells.
Discussion
We describe a metabolic engineering approach to selectively incorporate crosslinkers into O-GlcNAc residues on nuclear and cytoplasmic proteins. Our initial, in vitro experiments demonstrated that a diazirine-modified form of GlcNAc could be transferred by OGT and recognized by an O-GlcNAc-specific antibody, RL2. These results indicated that the relatively small diazirine modification did not abrogate GlcNAc recognition by OGT or by RL2. The ability of OGT to tolerate N-acyl modifications to GlcNAc has been shown previously (22) and is consistent with modeling of the OGT-UDP-GlcNAc complex based on the recently reported OGT structure (23).
To incorporate GlcNDAz into O-GlcNAc-modified proteins, we initially hoped to exploit the GlcNAc salvage pathway. However, we discovered at least two metabolic barriers to the conversion of GlcNDAz to UDP-GlcNDAz. First, GlcNDAz is not efficiently converted to GlcNDAz-1-P. We did not conduct experiments to distinguish whether the metabolic barrier is at the phosphorylation step that produces 6-phosphosugar, or in the mutase reaction that converts the 6-phosphosugar to the 1-phosphosugar (or both). Instead, we bypassed the initial barrier by directly delivering a protected form of GlcNDAz-1-P to cells. The second barrier occurs in the pyrophosphorylase step that converts the 1-phosphosugar to the UDP-sugar. Here we used structure-guided mutagenesis to produce a mutant form of AGX1 that tolerates the diazirine modification. Thus, using an approach that combines chemistry and genetic engineering, we induced mammalian cells to produce both UDP-GlcNDAz and O-GlcNDAz-modified proteins. Normally, the O-GlcNAc modification is removed from proteins through the action of the O-GlcNAcase, NCOAT. We found that NCOAT had reduced activity toward an artificial GlcNDAz-containing substrate, suggesting that, in cells, the O-GlcNDAz modification could be more long-lived than O-GlcNAc, and could even be effectively permanent. Thus, our method may cause some perturbations to normal O-GlcNAc patterns, even though no dramatic differences were observed (Fig. S6B).
Metabolic barriers to metabolism of hexosamine analogs have also been observed in efforts to incorporate chemical reporters into O-GlcNAc-modified proteins. An azide-modified GlcNAc analog, GlcNAz, was inefficiently metabolized to the corresponding UDP-GlcNAc analog in 293T cells, but AGX2 overexpression enabled UDP-GlcNAz production, suggesting the presence of only a single metabolic barrier (24). This same report demonstrated that the metabolic barrier could be circumvented by providing cells with an azide-modified GalNAc analog (GalNAz) that was efficiently metabolized to UDP-GalNAz, and further converted to UDP-GlcNAz by the action of the UDP-galactose 4′-epimerase (GALE). More recently, Pratt and coworkers examined metabolism of both azide-modified hexosamines and their alkyne-modified counterparts (25). Their results confirmed the metabolic interconversion of azido-sugars, but indicated that the alkynyl-sugars did not efficiently interconvert. Similar to the alkynyl case, we did not observe any production of UDP-GalNDAz in our experiments, suggesting that GALE may not readily interconvert the larger, diazirine-modified UDP-sugars. Thus, the hexosamine salvage pathway appears to be sensitive to the exact nature of the N-acyl modification and metabolism of any novel analog should be examined carefully. For the experiments described here, the lack of UDP-GalNDAz production is desirable since it is expected to yield more selective incorporation of the diazirine into only GlcNAc-containing glycans.
We demonstrated the utility of O-GlcNDAz crosslinking by using this technique to covalently crosslink nuclear pore proteins and nuclear transport factors. The mammalian nuclear pore is a 120 MDa complex assembled from about 30 different nucleoporins, each present in multiple copies (26). Transport in and out of the nucleus is regulated by the pore complex: small molecules can diffuse through the pore, while larger molecules are escorted through by karyopherins (27). Karyopherin-cargo complexes translocate through the center of the pore by a poorly understood mechanism that likely involves interactions between karyopherins and the unstructured, FG-repeat nucleoporins that project into the pore. In metazoa, FG-repeat nucleoporins are highly O-GlcNAc-modified, but the functional significance of these modifications has remained unclear. Early work showed that wheat germ agglutinin (WGA), which binds O-GlcNAc, can block transport through the nuclear pore (28), but this finding may reflect the steric bulk of WGA rather than a specific role for O-GlcNAc. More recently, experiments conducted in Caenorhabditis elegans offered an opportunity to investigate the significance of the O-GlcNAc modification, since loss of ogt-1 (the C. elegans ortholog of OGT) is not lethal. Hanover and coworkers examined the behavior of several transcription factors in the ogt-1 deletion strain and found that neither their nuclear localization nor their kinetics of transport were affected (29). Nonetheless, the O-GlcNAc residues may have a more pronounced role in mammals, where OGT is essential.
We carried out O-GlcNDAz-based crosslinking experiments to gain information about the direct binding partners of O-GlcNAc-modified FG-repeat nucleoporins. Notably, we obtained evidence for direct interactions between TNPO1 and both NUP153 and NUP358, a finding consistent with previous experiments (30–32). However, our cell-based crosslinking method enables additional conclusions. First, our data indicate that the observed interactions occur in a normal cellular environment, since the crosslinking event was triggered by irradiation of intact cells. Second, interactions occur at normal protein expression levels since our experiments involved crosslinking of endogenous proteins. Third, the formation of the covalent complex suggests the proteins interact directly, and not through a third party. Finally, because crosslinking occurs through the photoaffinity label on the GlcNAc, we conclude that there is an O-GlcNAc at or near the interaction site.
While our experiments do not directly address the question of whether the interactions between FG-repeat nucleoporins and nuclear transport factors are O-GlcNAc-dependent, these recognition events do share important features with typical glycan-mediated interactions. First, glycan-mediated interactions are typically low-affinity, with rapid off-rates (33). Similarly, interactions between FG-repeat nucleoporins and nuclear transport factors typically display nanomolar to micromolar equilibrium dissociation constants (34). Indeed, efficient nuclear transport requires that these interactions be transient, allowing cargo to be efficiently transported through the pore and not retained at a specific site. Second, both cell surface glycan-mediated interactions (35) and FG-repeat nucleoporin-nuclear transport factor interactions are typically multivalent (34, 36), which may enhance interaction specificity. Thus, the use of photocrosslinking groups offers an important strategy to covalently trap these transient and multivalent interactions (37).
In summary, we report a general method for identifying the interaction partners of O-GlcNAc-modified proteins and showed that this method can be applied in at least two cell lines. In the experiments presented here, we used antibodies against endogenous proteins to isolate crosslinked complexes for further characterization. In the absence of suitable antibodies, epitope-tag could be appended to O-GlcNAcylated proteins to enable efficient analysis. We predict that this photocrosslinking approach could be applied to many of the other hundreds of proteins known to be O-GlcNAc-modified (5).
Methods
Synthesis of GlcNDAz Compounds.
Synthesis of Ac4GlcNDAz and UDP-GlcNDAz have been described (16, 38). Ac3GlcNDAz-1-OH was produced by selective deprotection of Ac4GlcNDAzAc3GlcNDAz-1-P(Ac-SATE)2 was synthesized by phosphitylation of Ac3GlcNDAz-1-OH with bis(S-acetyl-2-thioethyl) N,N-diisopropylphosphoramidite (20) and subsequent oxidation with mCPBA. p-nitrophenyl-β-d-GlcNDAz (pNP-GlcNDAz) was prepared by standard methods. Analytical data for Ac3GlcNDAz-1-P(Ac-SATE)2 and pNP-GlcNDAz are presented in Fig. S8.
HPAEC-PAD Analysis of GlcNDAz-containing Metabolites.
HeLa cells were transiently transfected with pCMV6-XL5-AGX1(F383G). After 43 h, cells were transferred to serum-free DMEM media containing low glucose (1.0 g/L). Ac4GlcNDAz, Ac3GlcNDAz-1-P(Ac-SATE)2, Ac4GlcNAc, or DMSO (vehicle) were added to achieve a final concentration of 100 μM. After 5 h, cells were harvested and lysed in 75% ethanol by sonication and centrifuged at 20,000 × g. Supernatant was dried, resuspended in 40 mM sodium phosphate buffer (20–60 μL per million cells), and filtered through Amicon® Ultra centrifugal filter unit (Millipore, 10,000 MWCO). Filtrates were analyzed by HPAEC (ICS-3000 system, Dionex) with CarboPac™PA1 (Dionex) and pulsed amperometry detector (PAD) (39, 40).
Crosslinking of Cellular O-GlcNDAz Proteins.
HeLa cells were transiently transfected with mutant AGX1(F383G). Culture medium was replaced with serum-free, low-glucose DMEM 26 h after transfection. Ac3GlcNDAz-1-P(Ac-SATE)2 (100 μM final concentration) was added at 26 h and/or 50 h after transfection. Cells were harvested 20 h after final addition of Ac3GlcNDAz-1-P(Ac-SATE)2 and washed with DPBS. Cells were resuspended in DPBS and irradiated with UV light (365 nm, UVP, XX-20BLB lamp) while on an ice bath; control cells were kept on ice in dark. A 5% CuSO4 pentahydrate aqueous solution was used to filter out longer wavelength light. Cells were lysed by RIPA buffer, and analyzed by SDS-PAGE and immunoblot.
Identification of Nucleoporin Interaction Partners.
For immunoprecipitation of nucleoporins with mAb414, HeLa cells stably transfected with AGX1(F383G) were used. Ac3GlcNDAz-1-P(Ac-SATE)2 (100 μM final concentration) was added 0 and 24 h after the medium was changed to serum-free, low-glucose (1.0 g/L) DMEM. Twenty h later, cells were harvested, UV irradiated, as described above, then lysed in an immunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 2.0 mM EDTA, 1 mM DTT, 1 mM PMSF, 1× protease inhibitor cocktail). After rotating the lysate with mAb414 (0.5 μL/mg lysate) overnight at 4 °C, the resulting solution was mixed with protein G sepharose (50 μL) for 4 h at 4 °C. After washing beads five times with the immunoprecipitation buffer, proteins were eluted with 50 μL of 2× loading dye containing 5 mM DTT. Eluted samples were resolved by 5% SDS-PAGE, and silver staining was performed using the SilverQuest™ Silver staining kit (Invitrogen). Indicated regions of the gel were excised, destained, and in-gel digested with trypsin. Extracted peptides were analyzed by HPLC/MS/MS analysis.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Yuh Min Chook and Beatriz Fontoura for advice and discussions; Katharine Ullman, Mary Dasso, and Brian Burke for antinucleoporin antibodies; John Hanover, Suzanne Walker, and David Vocadlo for plasmids; Junmei Zhang for mass spectrometry analysis; Masato Kato and Ilmin Kwon for recombinant OGT; and Shunzi (Susan) Li, Kathlynn Brown, and Tanya Leavy for peptide synthesis. This work was supported by a Welch Foundation grant (I-1686) to J.J.K, an National Institutes of Health grant (GM66047) to C.R.B., and a Howard Hughes Medical Institute fellowship of the Life Sciences Research Foundation to M.B. J.J.K. is an Alfred P. Sloan Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114356109/-/DCSupplemental.
References
- 1.Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 2.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akimoto Y, Hart GW, Hirano H, Kawakami H. O-GlcNAc modification of nucleocytoplasmic proteins and diabetes. Med Mol Morphol. 2005;38:84–91. doi: 10.1007/s00795-004-0264-1. [DOI] [PubMed] [Google Scholar]
- 4.Lazarus BD, Love DC, Hanover JA. O-GlcNAc cycling: Implications for neurodegenerative disorders. Int J Biochem Cell B. 2009;41:2134–2146. doi: 10.1016/j.biocel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Hart GW. Glycomic approaches to study GlcNAcylation: Protein identification, site-mapping, and site-specific O-GlcNAc quantitation. Clin Proteomics. 2008;4:5–13. [Google Scholar]
- 6.Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc Code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 7.Hart GW, et al. O-linked N-acetylglucosamine: the “yin-yang” of Ser/Thr phosphorylation? Nuclear and cytoplasmic glycosylation. Adv Exp Med Biol. 1995;376:115–123. [PubMed] [Google Scholar]
- 8.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: Site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci USA. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smet-Nocca C, et al. Identification of O-GlcNAc sites within peptides of the Tau protein and their impact on phosphorylation. Mol Biosyst. 2011;7:1420–1429. doi: 10.1039/c0mb00337a. [DOI] [PubMed] [Google Scholar]
- 10.Rexach JE, et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Özcan S, Andrali SS, Cantrell JE. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;1799:353–364. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gewinner C, et al. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–3572. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 13.Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 14.Dentin R, Hedrick S, Xie J, Yates III J, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 15.Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bond MR, et al. Metabolism of diazirine-modified N-acetylmannosamine analogues to photo-cross-linking sialosides. Bioconjug Chem. 2011;22:1811–1823. doi: 10.1021/bc2002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 18.Leavy TM, Bertozzi CR. A high-throughput assay for O-GlcNAc transferase detects primary sequence preferences in peptide substrates. Bioorg Med Chem Lett. 2007;17:3851–3854. doi: 10.1016/j.bmcl.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bond MR, Whitman CM, Kohler JJ. Metabolically incorporated photocrosslinking sialic acid covalently captures a ganglioside-protein complex. Mol Biosys. 2010;6:1796–1799. doi: 10.1039/c0mb00069h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefebvre I, et al. Mononucleoside phosphotriester derivatives with S-acyl-2-thioethyl bioreversible phosphate-protecting groups: Intracellular delivery of 3′-azido-2′,3′-dideoxythymidine 5′-monophosphate. J Med Chem. 1995;38:3941–3950. doi: 10.1021/jm00020a007. [DOI] [PubMed] [Google Scholar]
- 21.Peneff C, et al. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 2001;20:6191–6202. doi: 10.1093/emboj/20.22.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross BJ, Kraybill BC, Walker S. Discovery of O-GlcNAc transferase inhibitors. J Am Chem Soc. 2005;127:14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]
- 23.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyce M, et al. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci USA. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaro BW, Yang Y-Y, Hang HC, Pratt MR. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc Natl Acad Sci USA. 2011;108:8146–8151. doi: 10.1073/pnas.1102458108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chook YM, Süel KE. Nuclear import by karyopherin-βs: Recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanover JA, et al. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutten S, Wälde S, Spillner C, Hauber J, Kehlenbach RH. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J Cell Sci. 2009;122:1100–1110. doi: 10.1242/jcs.040154. [DOI] [PubMed] [Google Scholar]
- 32.Lau CK, et al. Transportin regulates major mitotic assembly events: From spindle to nuclear pore assembly. Mol Biol Cell. 2009;20:4043–4058. doi: 10.1091/mbc.E09-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr Opin Chem Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Terry LJ, Wente SR. Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Euk Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramachandraiah G, Chandra NR, Surolia A, Vijayan M. Computational analysis of multivalency in lectins: structures of garlic lectin-oligosaccharide complexes and their aggregates. Glycobiology. 2003;13:765–775. doi: 10.1093/glycob/cwg095. [DOI] [PubMed] [Google Scholar]
- 36.Bednenko J, Cingolani G, Gerace L. Importin β contains a COOH-terminal nucleoporin binding region important for nuclear transport. J Cell Biol. 2003;162:391–401. doi: 10.1083/jcb.200303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka Y, Bond MR, Kohler JJ. Photocrosslinkers illuminate interactions in living cells. Mol Biosyst. 2008;4:473–480. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka Y, Kohler JJ. Photoactivatable crosslinking sugars for capturing glycoprotein interactions. J Am Chem Soc. 2008;130:3278–3279. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- 39.Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: Sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 40.Yu S-H, Bond MR, Whitman CM, Kohler JJ. Metabolic labeling of glycoconjugates with photocrosslinking sugars. Methods Enz. 2010;478:541–562. doi: 10.1016/S0076-6879(10)78026-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.