Abstract
Initiation factor 2 (IF2) is a key factor in initiation of bacterial protein synthesis. It recruits initiator tRNA to the small ribosomal subunit and facilitates joining of the large ribosomal subunit. Using reconstituted translation system of Escherichia coli and optical tweezers, we directly measure the rupture force between single ribosomal complexes and mRNAs for initiation complexes in the presence and the absence of IF2. We demonstrate that IF2 together with codon recognition by initiator tRNA increases the force required to dislocate mRNA from the ribosome complexes; mRNA stabilization by IF2 required the presence of a joined 50S subunit, and was independent of bound guanine nucleotide. IF2 thus helps lock the 70S ribosome over the start codon during initiation, thus maintaining reading frame. Our results show how mRNA is progressively stabilized on the ribosome through distinct steps of initiation.
Keywords: force measurement, single-molecule translation
Translation initiates by assembly of elongation-competent ribosomes positioned over the correct start codon. Initiation in bacteria is a multistep process. The small (30S) ribosomal subunit forms a preinitiation complex (PIC), containing mRNA, initiation factors and fMet-tRNAfMet, which recruits the large (50S) ribosomal subunit. The 50S subunit recruitment facilitates initiation factor displacement, resulting in elongation-competent ribosome with initiatior tRNA positioned in the peptidyl-tRNA site (P site) of the ribosome. Initiation is guided and regulated by three initiation factors: IF1, IF2 and IF3. IF1 and IF3 work in concert to prevent premature joining of initiation subunits, safeguarding ribosome from initiation with incorrect start codons and tRNA (1). IF2 is a GTPase and central initiation factor. Upon IF2 and tRNA binding, IF2 facilitates 50S subunit joining that promotes rapid hydrolysis of GTP by IF2, which in turn stimulates IF2 release from the ribosome (1).
IF2 guides major structural rearrangements on the ribosome during initiation (2, 3). Cryo-EM and single-molecule experiments have shown that 50S subunit joins in a rotated, unlocked conformation with respect to the 30S subunit in the presence of IF2 and nonhydrolyzable GTP. Upon GTP hydrolysis, the 50S ribosomal subunit rotates clockwise by approximately 6° relative to the 30S subunit, thus putting ribosome into nonrotated, classical conformation. In this process, IF2 undergoes large conformational changes itself. In 30S PICs, the tRNA-binding domain of IF2 protrudes toward the shoulder of the 30S subunit, whereas the GTP-binding domain interacts on the top of the 30S spur, near helix 14 of the 16S rRNA. After 50S subunit joining, IF2 rearranges, with the GTPase domain moving near the large subunit sarcin-ricin loop and GTPase-associated center, while tRNA-binding domain moves toward the beak of the small subunit. After GTP hydrolysis, GTP-binding domain moves away from the GTPase center by approximately 10 Å toward ribosomal protein L6 and the tRNA-binding domain is shifted toward the beak, pivoting by approximately 20° around the GTPase domain along the intersubunit interface. During this process, IF2 guides initiator tRNA position and conformation. In the 30S PIC, initiator tRNA is located in an intermediate conformation in which the anticodon stem of the tRNA is located in the P site and bent toward the mRNA, while the elbow and acceptor end are displaced toward the exit site (E site). Upon assembly of an elongation-competent 70S initiation complex, the initiator tRNA shifts to the classical P/P conformation (3).
The ribosome must establish and maintain the correct translational reading frame during initiation. Both the Shine-Dalgarno (SD) sequence and initiator tRNA-start codon interactions are central to the establishment of reading frame. Yet the mechanism for how reading frame is preserved during initiation remains unclear. Here we use single-molecule force methods to measure directly the mechanical stability of the ribosome-mRNA complexes along the initiation pathway in Escherichia coli. We show that IF2 and 50S subunits work together to establish and preserve open reading frame during late steps in initiation.
Results
Experimental Design.
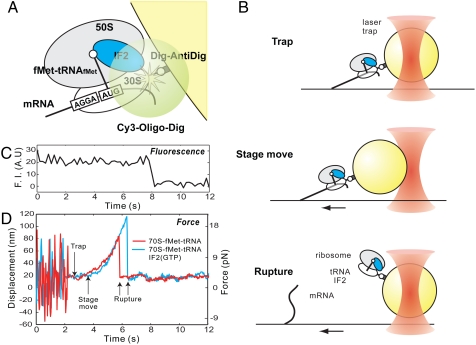
Initiation complexes were assembled on a model mRNA comprised of the first 57 nucleotides (nts) of T4 gene 32 mRNA containing SD sequence, located upstream of the AUG initiation codon with covalently linked biotin on the 5′ end. The initiation complexes assembled over model mRNA were immobilized on the surface of the coverslip via streptavidin–biotin interactions (4). The 30S ribosomal subunits in the experiment contain a genetically engineered meta-stable hairpin extension at the helix 44 of the 16S rRNA (4). Previous bulk and single-molecule fluorescence experiments demonstrated functionality of labeled ribosomal subunits (4, 5). A 22 nt DNA oligonucleotide complementary to the hairpin insertion labeled with 5′ Cy3 and 3′ digoxigenin was annealed to the hairpin. This molecular handle allows bead attachment to the initiation complexes. Upon initiation complex formation, 1-μm polystyrene beads covered with covalently linked antidigoxigenin antibodies were tethered to the initiation complexes via antibody–digoxigenin interactions; therefore, initiation complexes were tethered between the surface of the coverslip and the bead (Fig. 1A). The Cy3 fluorescence from the attached DNA oligonucleotide was used to validate the presence of a single tether between the bead and the coverslip surface (Fig. 1C). Upon observing a single Cy3 photobleaching event, the tethered bead was captured by an optical trap. After bead capture, the coverslip surface was moved at the constant speed to displace the beads from the center of the optical trap. The bead displacement generated constantly increasing force linearly proportional to the bead displacement. The force was increased until the complex ruptured (Fig. 1B), which was registered as a precipitous return of the bead to the center of the optical trap. The amplitude of the return movement is linearly proportional to the exerted force during mRNA-ribosome complex rupture and was used to calculate applied force (Fig. 1D). The resulting rupture force was calculated using standard methods by observing the mode of the Gaussian distribution of multiple single-molecule rupture events.
Fig. 1.
Experimental design. (A) Immobilization scheme. Ribosomal complexes were assembled on biotinylated mRNA and immobilized on the surface of the coverslip via biotin-streptavidin interactions. Meta-stable hairpin insertion at helix 44 of the 16S rRNA was used to dual-label small ribosomal subunits with Cy3 and digoxigenin. The digoxigenin handle was used to secure attachment of the 1 μm polystyrene beads covered with complimentary antibodies. (B) Surface-bound beads were captured by an optical trap; the glass surface was then moved at 100 nm/s, causing bead displacement from the center of the optical trap. Bead dislocation generated a returning force linearly proportional to the displacement. Force was increased until the complex ruptured. (C) Single photobleaching events of the Cy3-labeled ribosomes were used to ensure presence of the single molecular link between surface of the coverslip and the bead. (D) Time-position traces. Prior to capturing, the bead fluctuates around attachment point. Bead capture suppresses fluctuations. The stage then moves the bead out of the trap center. The force was increased until the complex ruptured, the rupture of the initiation complex is registered a precipitous return of the bead to the trap center.
tRNA, Not IF2, Is a Major Determinant of Stability of the Early Initiation Complexes.
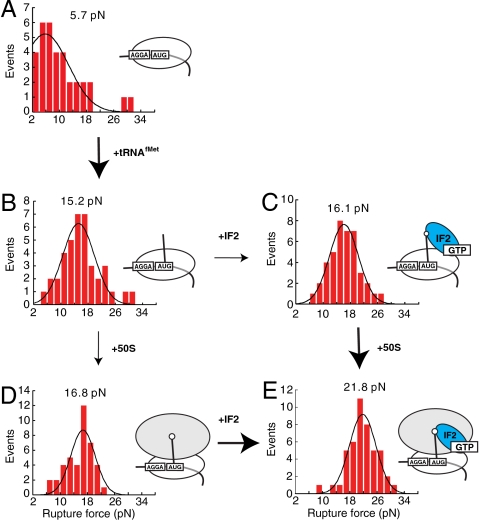
Stability of ribosome-mRNA interactions within 30S initiation complexes was measured in presence of initiator tRNA and IF2. Naked 30S-mRNA complexes were ruptured at 5.7 pN (Fig. 2A). Addition of the initiator tRNA increased the mean rupture force to 15.2 pN (Fig. 2B). Further addition of IF2∶GTP to the 30S∶tRNA complexes did not affect complex stability because the rupture force was measured to be 16.1 pN (Fig. 2C). Thus, IF2 does not affect the stability of mRNA-ribosome interactions in PICs. However adding tRNA to the 30S-mRNA drastically increased rupture force by 10.5 pN (Fig. 2 A and B).
Fig. 2.
The rupture force distribution during early initiation steps. Each distributions fitted by Gaussian function have peak rupture force displayed above fitting curve. (A) 30S∶mRNA. (B) 30S∶tRNAfMet∶mRNA. (C) 30S∶IF2(GTP)∶fMet-tRNAfMet∶mRNA. (D) 70S∶fMet-tRNAfMet∶mRNA. (D) 70S∶IF2(GTP)∶fMet-tRNAfMet∶mRNA complexes. In the absence of IF2 50S subunit joining does not affect initiation complex stability. The presence of IF2 increases stability of the initiation complex by 5 pN upon large subunit joining. The arrow and bold arrow indicate no change and larger change in rupture force respectively.
IF2 and tRNA Cooperatively Stabilize Late Initiation Complexes.
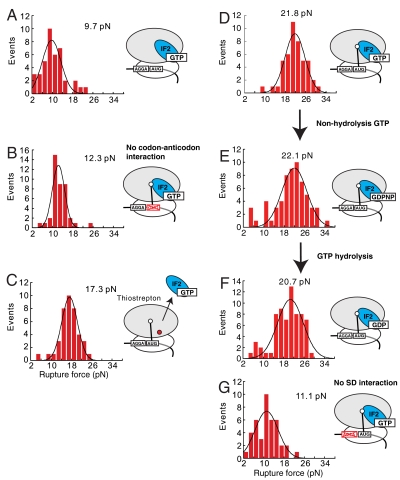
To probe the effect of 50S subunit joining on mRNA rupture force, ribosome-mRNA interactions were measured for 70S initiation complexes containing mRNA and P-site bound initiator fMet-tRNAfMet. The rupture force distribution was centered at 16.8 pN, (Fig. 2D) in agreement with previously observed 15.2 pN rupture force of the 70S ribosomes with initiator tRNA in the P site, independently determined by using a similar setup (6). Addition of IF2 to this complex (70S∶tRNA) increased the rupture force by 5–21.8 pN (Fig. 2E). Stabilization of the ribosome–mRNA interactions by IF2 required the presence of initiator tRNA bound to a correct start codon. Binding of the IF2 to the 70S ribosomes lacking initiator tRNA (70S∶IF2 complexes) did not affect complex stability, yielding a rupture force similar to that as with naked 70S ribosomes [9.7 (Fig. 3A) and 10.6 pN, ref. 6, respectively]. Replacement of the AUG start codon for a noncognate GUG in the 70S∶IF2(GTP)∶fMet-tRNAfMet complexes decreased the rupture force by 9.5 pN from 21.8 to 12.3 pN (Fig. 3B). This change is greater than contributions of initiator tRNA in the P site in 70S complexes of 4.6 pN (6) and therefore IF2 contributes of 5 pN. Thus stabilization of mRNA on the ribosome requires correct codon-anticodon interactions between initiator tRNA and mRNA.
Fig. 3.
The rupture force distribution fitted by Gaussian function for following ribosome complexes. (A) 70S∶IF2(GTP)∶mRNA. (B) 70S∶IF2(GTP)∶fMet-tRNAfMet∶noncognate mRNA. (C) 70S∶IF2(GTP)∶fMet-tRNAfMet∶mRNA in the presence of thiostrepton. (D) 70S∶IF2(GTP)∶fMet-tRNAfMet∶mRNA. (D) 70S∶IF2(GDPNP)∶fMet-tRNAfMet∶mRNA. (F) 70S∶IF2(GDP)∶fMet-tRNAfMet∶mRNA. (G) 70S∶IF2(GTP)∶fMet-tRNAfMet∶no-SD mRNA The initiation complexes were assembled in the excess of the tRNA, IF2 and corresponding small molecules.
Large Subunit and IF2 Interplay.
Joining of the 50S subunit in the presence of tRNA and IF2 increases the mRNA rupture force by 5 pN; 30S-mRNA complexes ruptured at 16.1 pN (Fig. 2C) whereas 70S-mRNA complexes ruptured at 21.8 pN (Fig. 2E). However, in the presence of only tRNA 50S subunit joining did not affect mRNA stability (Fig. 2 B and D). Thus, stabilization of mRNA on the ribosome by IF2 requires the presence of the 50S ribosomal subunit. Overall, upon IF2 and tRNA binding followed by formation of 70S complexes, the rupture force increases by 5.7–21.8 pN. Interestingly, large-subunit joining in the absence of the tRNA and IF2 also lead to the 5 pN increase in rupture force [5.7 pN (Fig. 2A) and 10.6 pN, ref. 6, respectively], whereas overall stabilization of the mRNA-ribosome interactions was only 6 pN upon 70S formation. These results suggest that maximal stabilization of mRNA on the ribosome requires large subunit joining to occur via tRNA- and IF2-dependent pathways.
To reveal the relationship between IF2 and 50S subunit in mRNA stabilization, we measured the rupture force of 70S∶IF2∶tRNA complexes in the presence of thiostrepton. Thiostrepton is a cyclic oligopeptide antibiotic that binds near the GTPase center and blocks the function of the G factors involved in translation. It has been shown to inhibit late initiation steps (7–9). The rupture force for 70S-IF2-tRNA complexes in the presence of thiostrepton was decreased by 5 pN (Fig. 3C). Thus, proper interaction between IF2 and the 70S ribosomal particle is required for full stabilization of the mRNA-ribosome interaction during initiation.
GTP Hydrolysis by IF2 Does Not Affect mRNA-Ribosome Complex Stability.
To assess the role of GTP hydrolysis by IF2 in the stabilization of mRNA-ribosome interactions, we measured the rupture force in the presence of GTP, GDP, and a nonhydrolysable GTP analog GDPNP. The 70S initiation complexes were assembled on mRNA in the presence of the IF2 and fMet-tRNAfMet. Saturating concentrations of IF2 (1 μM) ensured IF2 occupancy on the ribosome. The rupture forces were found to be independent of guanine nucleotide and were 21.8 pN for 70S∶tRNA∶IF2 (GTP), 20.7 pN for 70S∶tRNA∶IF2 (GDP), and 22.1 pN for 70S∶tRNA∶IF2 (GDPNP) complexes (Fig. 3 D–F).
IF2 Stabilization Is Independent of the SD Interaction.
Our prior studies showed that interactions between the SD sequence in mRNA and 70S ribosome add approximately 11 pN to the mRNA complex stability (6). To test a relationship between SD and IF2 stabilization of initiation complexes, we measured forces required for complex disruption on mRNA lacking a SD sequence. 70S-tRNAfMet complexes lacking SD sequence ruptured at 4.8 pN (figure 3h in ref. 6). The addition of the IF2 to the ribosomal complexes resulted in increase of the complex stability by 6.3 pN to 11.1 pN, similar to the increase of 5 pN observed on SD-containing mRNA upon IF2 binding (figure 3h in ref. 6 and Fig 3G). Similar to the factorless 70S complexes, the 70S∶tRNAfMet∶IF2 complexes assembled on SD-lacking mRNA required 10.7 pN less force compared to the 70S∶tRNAfMet∶IF2 assembled on SD-containing mRNA. The observed decrease in complex stability is the same as previously observed SD contribution of approximately 11 pN into the mRNA-ribosome complex stability (6). IF2 therefore stabilizes mRNA on the ribosome independently from SD interaction.
Discussion
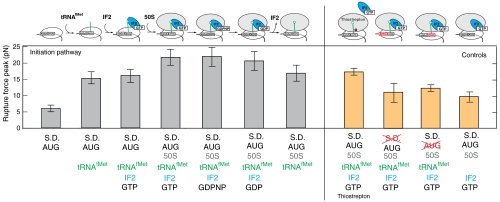
The results presented here demonstrate how the ribosome establishes and maintains mRNA interactions during initiation. We have determined the rupture force for mRNA-ribosome interactions for each distinct step during initiation (Fig. 4). First, mRNA is recruited to the 30S subunits via SD interaction, then IF2 binds to the ribosome and recruits initiator tRNA. The 30S∶IF2∶initiator tRNA complex then facilitates rapid 50S subunit binding, with subsequent hydrolysis of GTP by IF2 and dissociation of the factors to create an elongation-competent ribosome. The strength of interactions between mRNA and the ribosome increases along this pathway. In the 30S-mRNA complex, mRNA is transiently bound to the ribosome, and a relatively weak force of 5.7 pN is required to dislocate mRNA from the ribosome. Upon initiator tRNA binding, the force required to dislocate mRNA rises dramatically to 16.1 pN due to the codon-anticodon interactions in the P site. In the 30S PIC, IF2 binding does not further increase the mRNA rupture force. Subsequent 50S subunit joining in the presence of IF2 further increases the rupture force by 5 pN. This results in a total fourfold increase in the final 70S initiation complex stability comparing to the initial 30S∶mRNA complex. This observed increase in mRNA-ribosome stability required the successful establishment of the codon-anticodon interactions between initiator tRNA and AUG codon of mRNA. Omission of the tRNA or replacement of the start codon with noncognate GUG codon decreased the rupture force to that required for rupturing naked 70S-mRNA complexes.
Fig. 4.
The summary of the stability of the mRNA-ribosome interactions along the initiation pathway (gray bars) and for 4 control experiments (orange bars). Error bars represent the standard deviation of Gaussian fitting for each ribosome complexes. Complex compositions are shown at the bottom of panel.
The results presented here suggest that mRNA position on the ribosome, and thus the open reading frame, is established and secured in a series of steps that increase the strength of ribosome-mRNA interaction, with SD, tRNA, IF2 and large subunit contributing equally by approximately 5 pN to the mRNA stability on the ribosome. Stabilization of mRNA by IF2 and 50S subunit was independent of the SD interaction. Thus later initiation steps, including initiator tRNA recruitment and large-subunit joining, are independent checkpoints in the establishment of the open reading frame. These results demonstrate that SD sequences play a role in the initial establishment of the open reading frame. However, the subsequent securing of the open reading frame is SD independent and is determined by the interplay between IF2, tRNA and ribosomal subunit.
The 50S subunits together with IF2 play an active role in stabilizing the mRNA-ribosome interaction. The 50S subunit binding improves stability of the mRNA complexes by 5 pN and requires IF2 to lock the mRNA on the ribosome. In the absence of 50S subunits, IF2 has no effect on the complex stability. Single-molecule fluorescence studies demonstrated that 50S subunits join 30S PICs in a rotated (ratcheted) conformation relative to the small subunit (5). Upon GTP hydrolysis, 50S subunits rotate clockwise, putting the 70S ribosome into a nonrotated (nonratcheted) conformation. Interestingly, no difference in the displacement force was found between IF2 containing complexes before and after GTP hydrolysis. Thus, it is indicated that ribosome interacts similarly with mRNA in ratcheted and nonratcheted conformations in the presence of IF2.
Translation embodies a strategy of progressive investment in stronger mRNA binding during initiation to establish the reading frame. The complex of IF2 bound to the 70S ribosomal particle after GTP hydrolysis is a final intermediate of initiation and has maximal stability for mRNA-ribosome interaction. Upon dissociation of IF2, the elongation-competent ribosome is destabilized by 5 pN. A site tRNA binding adds 10 pN of mRNA stability to compensate for the loss of IF2’s contribution, which drops to 16 pN upon peptide bond formation (6). Thus, the ribosome maintains a minimum rupture force of > 15 pN during elongation to ensure reading frame maintenance but allow processivity and ribosome movement during translation. Understanding the interplay of ribosome conformation, mRNA context, and elongation will require additional single-molecule investigation.
Materials and Methods
Reagents and Supplies.
30S ribosomal subunits and 70S ribosomes employed in this study have a 23 nucleotide extension in helix 44 of the 16S rRNA (4, 6). The DNA oligonucleotide labeled with Cy3 at the 5′ end and digoxigenin at the 3′ end (Cy3-AAAGGGAGATCAGGATATAAAG-digoxigenin) and 5′-biotinylated T4 gene32 derived mRNAs were synthesized by Japan Bio Services Co., Ltd. Carboxylate microspheres (1 μm in diameter; Polysciences, Inc.) were coated with polyclonal antibody to digoxigenin (Acris Antibodies) as previously described (10).
Sample Preparation for Rupture Force Measurements.
Mutant E. coli ribosomes were labeled by incubation with twofold molar excess of 5′ Cy3 and 3′ digoxigenin-modified oligonucleotide at 30 °C for 30 min. Initiation complexes were assembled by incubation of ribosomal particles with 5′-biotinylated mRNA and appropriate factors [tRNA, IF2, guanine nucleotides (Sigma) and ribosomal subunits] at 37 °C for 5 min. tRNA, IF2, guanine nucleotides, and 50S subunits were added in twofold excess to the 30S ribosomal particles. The initiation complexes were flash-frozen in liquid nitrogen and stored at -80 °C. Model mRNA (CAACCUAAAACUUACACACCCGGUAAGGAAAUAAAAAUGUUUAAACGUAAAUCUACU, start codon shown in bold) was based on the on T4 phage g32 mRNA and contained 5′ UTR with UAAGGA SD sequence, AUG start site followed by six elongator codons.
A flow cell was assembled from a coverslip and a glass slide separated by double-sided tape. The coverslips were treated with an oxygen plasma asher prior to use. Ribosomes were immobilized by stepwise delivery of following solutions: (i) 3 mg/mL biotinylated BSA (11) followed by a wash with Polymix buffer [50 mM Tris-OAc, pH 7.5, 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 5 mM Mg(OAc)2, 5 mM β-mercaptoethanol, 5 mM putrescine, 1 mM spermidine, and 1% (wt/vol) glucose]; (ii) 0.33 mg/mL streptavidin (Invitrogen) followed by a wash with Polymix buffer; (iii) 30S or 70S ribosomal complexes followed by a wash with Polymix buffer containing 0.5 mg/mL β-casein (Sigma); and (iv) anti-digoxigenin-coated beads followed by a wash with Polymix buffer containing 0.5 mg/mL β-casein and O2 scavenging system [25 mM glucose, 50 units/mL glucose oxidase (Sigma) and 50 units/mL catalase (Sigma)] and supplemented with appropriate factors [1 μM tRNA, 1 μM IF2, 2 mM guanine nucleotides, 500 nM 50S subunits and 10 μM thiostrepton (Sigma)] where required.
Rupture Force Measurements.
The microscopy system equipped with optical tweezers was as described previously (6). The trap stiffness (0.18 pN/nm) was calculated as described previously (6). The Cy3 fluorescence was used to validate presence of the single tether between bead and the glass surface. Upon observing a single Cy3 photobleaching event, the tethered bead was captured by an optical trap and external load was applied at a rate of 18 pN/s by moving a piezostage at 100 nm/s. This resulted in dislocation of the bead from the center of the optical trap. Upon rupture of the ribosome complex, the bead rapidly returned to the center of the optical trap. The rupture force was calculated by multiplying trap stiffness to the displacement distance. The rupture force distributions were fitted to a single Gaussian. All experiments were performed at room temperature (approximately 23 °C).
Acknowledgments.
This work was supported by Japan Science and Technology Agency (S.U.) and National Institutes of Health Grants GM51266 and GM073999 (to J.D.P.). This work was also supported by Grants-in-Aid for Young Scientists (Startup; Grant 21870008 to R.I.), Scientific Research on Priority Areas (Grant 22020006 to R.I.), and Scientific Research (B) (Grant 21370065 to T.F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006;25(11):2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myasnikov AG, et al. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol. 2005;12(12):1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- 3.Simonetti A, et al. Structure of the 30S translation initiation complex. Nature. 2008;455(7211):416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 4.Dorywalska M, et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33(1):182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17(7):793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uemura S, et al. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446(7134):454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- 7.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. A quantitative kinetic scheme for 70S translation initiation complex formation. J Mol Biol. 2007;373(3):562–572. doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25(4):519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta AD, Finer JT, Spudich JA. Use of optical traps in single-molecule study of nonprocessive biological motors. Methods Enzymol. 1998;298:436–459. doi: 10.1016/s0076-6879(98)98039-9. [DOI] [PubMed] [Google Scholar]
- 11.Itakura S, et al. Force-generating domain of myosin motor. Biochem Biophys Res Commun. 1993;196(3):1504–1510. doi: 10.1006/bbrc.1993.2422. [DOI] [PubMed] [Google Scholar]