Abstract
Lateral gene transfer (LGT), the acquisition of genes from other species, is a major evolutionary force. However, its success as an adaptive process makes the reconstruction of the history of life an intricate puzzle: If no gene has remained unaffected during the course of life's evolution, how can one rely on molecular markers to reconstruct the relationships among species? Here, we take a completely different look at LGT and its impact for the reconstruction of the history of life. Rather than trying to remove the effect of LGT in phylogenies, and ignoring as a result most of the information of gene histories, we use an explicit phylogenetic model of gene transfer to reconcile gene histories with the tree of species. We studied 16 bacterial and archaeal phyla, representing a dataset of 12,000 gene families distributed in 336 genomes. Our results show that, in most phyla, LGT provides an abundant phylogenetic signal on the pattern of species diversification and that this signal is robust to the choice of gene families under study. We also find that LGT brings an abundant signal on the location of the root of species trees, which has been previously overlooked. Our results quantify the great variety of gene transfer rates among lineages of the tree of life and provide strong support for the “complexity hypothesis,” which states that genes whose products participate to macromolecular protein complexes are relatively resistant to transfer.
Keywords: genome evolution, phylogeny, bacteria, archaea
In Bacteria and Archaea, a variety of mechanisms facilitate the integration of foreign DNA into the genome (1). The adaptive role of this process has been widely documented, and lateral gene transfer (LGT) is now considered one of the main evolutionary forces at work in prokaryotic evolution, allowing acquisition of functions even from distantly related organisms to adapt to changing environments or colonize new ones (2, 3). However, the success of LGT as an evolutionary process impedes the reconstruction of evolutionary patterns: If LGT has been so frequent in the history of life, how can we rely on gene phylogenies to resolve the relationships among species? A widespread opinion states that a tree-like view of relationships among species is incompatible with the actual abundance of LGT (4–6). However, because genes propagate only through cell division or gene transfer, an edge of a gene tree may only represent either vertical (from parent to children) or horizontal (from donor to acceptor) transmission of genes. Identifying the vertical part of the history of life is very challenging, and the approaches to do so have been unsatisfying. The most common method uses universal genes that are a priori considered unlikely to have undergone LGT and sometimes uses ad hoc criteria to remove those that seem to give discordant phylogenetic signal. The combination of the few molecular markers that pass this rigorous screening usually yields a tree of life that is roughly consistent with the original 16S rRNA phylogeny. However, whether based on 16S rRNA or the combination of these few universal genes, this tree can only be interpreted with the prejudice that some genes are relatively immune to LGT. It is also unclear how these few genes could really be seen as representative of complete genomes, which contain hundreds of times more genes (7). Moreover, even slight modifications in the choice of genes, species, or phylogenetic methods give discordant results for key relationships, which suggests that these data are not sufficient to infer reliable phylogenies (8).
Here, we show that LGT itself can be used to reconstruct the history of life. Certain cases of LGT have been described in the literature that provide strong evidence for the existence of several monophyletic groups, [e.g., the massive transfer of mitochondrial genes to the nucleus (9, 10) that demonstrates the unique origin of eukaryotes and the late emergence of amitochondrial eukaryotes, or the transfer of an archaeal tyrosyl-tRNA synthetase in the genomes of animals and fungi that shows the monophyly of opisthokonts (11)]. Similarly, in Bacteria and Archaea, ancestrally transferred genes can be regarded as shared derived characters diagnostic of a clade (Fig. 1), and LGT represents an abundant phylogenetic information that has been largely overlooked. We recently developed Prunier (12), a program that, given a species tree, can efficiently identify LGT in gene phylogenies while taking into account uncertainties in gene tree reconstruction. In simulations, Prunier has been shown to correctly detect most gene transfers and to outperform all other available programs in sensitivity and specificity. Thus, it is now possible to explicitly account for LGT in the phylogenetic reconstruction of a species tree by performing systematic gene tree/species tree reconciliations for all gene families, hence integrating the phylogenetic information contained in both vertical and lateral transmissions of genes. We show that the thousands of gene families that are usually ignored in phylogenetic analysis contain a strong signal for a tree of vertical inheritance and that LGT, far from blurring this tree, carries as-yet-overlooked information on its root.
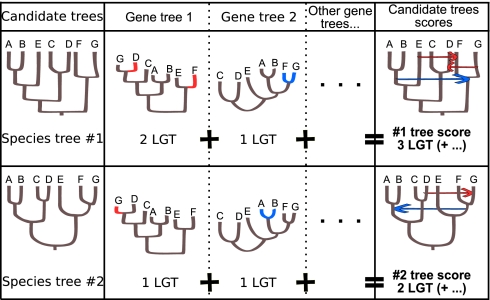
Fig. 1.
LGTs are informative for phylogeny. An LGT can be viewed as a phylogenetic character, and the sharing of an LGT can be viewed as a shared derived character. An objective criterion to select a species tree can be the number of LGTs that need to be invoked in a collection of gene trees. In this illustration, two candidate species trees and two gene trees are presented. Given these species trees, different LGT events are needed to reconcile gene trees. Gene tree 1 has a score of 2 LGTs given species tree 1 and 1 LGT given species tree 2. A global score on all gene trees can be assigned to each candidate species tree. Here, species tree 1 has a score of 3 LGTs, whereas species tree 2 has a score of 2 and is therefore more parsimonious.
Results
We used Prunier to analyze 12,602 single-copy gene family trees distributed in 336 species from 16 bacterial and archaeal phyla (Table S1). We first reconstructed all individual gene trees for each phylum by using a maximum-likelihood approach (Materials and Methods). In most phyla, the level of topological incongruence between gene trees was very high (Table S2), and gene trees reconstructed from universal families were all different from each other. The degree of incongruence among trees was less pronounced in phyla with 20 or fewer representative species (Table S2).
We generated candidate topologies for the “species tree” of each phylum by using a variety of strategies: first, we used 16S and 23S rRNA genes, either alone or combined into a superalignment; second, we combined the information of “universal” protein families in different ways, including various concatenations of gene alignments and supertree approaches, which we will refer to as “phylogenomic” approaches in the rest of the paper (Materials and Methods).
LGT Discriminates Among Phylogenetic Hypotheses.
The number of candidate species trees varies between phyla from one to eight (Fig. 2 and Table S1). In Gammaproteobacteria, all methods yielded a different candidate tree, whereas, in Epsilonproteobacteria, all methods agreed. For each of these candidate trees, we then inferred evolutionary scenarios for all gene trees present in a phylum by using an improved version of Prunier (12) (Materials and Methods and SI Materials and Methods). Prunier aims to resolve all significantly supported differences (i.e., branches with a support value higher than 0.80) between a gene tree and a fully resolved reference tree by invoking a minimal number of LGT events.
Fig. 2.
LGT scores for different candidate species trees. For each of the 16 phyla analyzed, eight candidate species trees (T1–T8) derived from different methods (Materials and Methods and SI Materials and Methods) were tested [T1, concatenated tree (concat); T2, recoded concatenated tree (recod); T3, consensus of trees from universal family (consense); T4, consensus of jackknife replicates (jackknife); T5, super distance matrix tree (SDM); T6, 16S rRNA (16s); T7, 23S rRNA (23s); and T8, 16S + 23S rRNA (16s + 23s)]. Each hypothesis has an associated LGT score that corresponds to the number of LGTs inferred by Prunier in the families under study (Nb fam). Dark blue cells represent trees that are topologically identical to the most parsimonious tree in terms of LGT. Light blue cells correspond to trees that are not statistically different from the most parsimonious tree. Trees with white cells are significantly different from the best tree. Numbers of transfers per hundred branches per family are shown (LGT rate). Chlamydiales-Verruco, Chlamydiales-Verrucomicrobia; Nb sp, number of species.
Within a phylum, different candidate trees typically translate into very different numbers of LGTs in gene families (Fig. 2). In three phyla (Bacteroidetes/Chlorobi, Mollicutes, and Spirochaetes), the differences between candidate species trees and/or the amount of signal from LGT were not sufficient to discriminate among the candidates (Wilcoxon paired test, α = 0.05 with false discovery rate correction for multiple tests). In the 12 remaining phyla (Epsilonproteobacteria having a single candidate species tree), there was a strong statistical signal for one or a few species trees. A general pattern emerges: The candidate trees obtained from 16S rRNA, 23S rRNA, and the combination of these markers (T6–T8) tend to give much higher LGT counts than those obtained from phylogenomic methods (T1–T5). For instance, with the 16S rRNA phylogeny as a reference, the LGT count is significantly higher than with the tree with the best LGT score in 12 of 16 phyla (Fig. 2). All candidate species trees derived from phylogenomic approaches are based on universal, single-copy gene families. Because it may seem circular to evaluate candidate trees based on gene families that were used to reconstruct them, we also evaluated the LGT score of these different phylogenetic hypotheses after removing universal families (i.e., using only “nonuniversal” gene families). The results were remarkably similar, with nonuniversal gene trees always supporting identical or statistically indistinguishable candidate trees from those supported by universal genes. In many phyla, most of the signal to support a candidate tree actually came from these more abundant gene families (Table S3). Although the different phylogenomic methods systematically yield trees with low numbers of LGTs relative to rRNA trees, there does not seem to be a method that consistently outperforms others according to this criterion.
This ability of LGT scores to discriminate among phylogenies is clearly related to the number of transfers observed in the gene families of a phylum. The numbers of transfers identified by Prunier throughout the history of a phylum typically add up to hundreds or thousands, which supports the view that LGT has been abundant in the history of life (Fig. 2). These counts, however, strongly depend on a combination of factors, such as the number of gene families available, the number of species considered, and the phylogenetic signal accumulated since their divergence. Converting them into numbers of LGTs per branch of a gene family tree is necessary to control for these sampling effects. Of course, these estimates of transfer rates depend on factors such as branch length (e.g., species sampling) and cannot be easily compared among phyla, but they can help to depict the magnitude of LGT in the history of different phyla (Fig. 2). The rate of transfers varies among phyla from 5.5% LGTs per branch per family in Lactobacillales to 0.1% in Spirochaetes, with most phyla harboring rates between 2% and 4%. In other words, in a typical Lactobacillales monocopy gene tree, 94.5% of the branches represent vertical descent and 4.5% denote lateral transfers. In Spirochaetes, only 0.1% of the branches of a gene tree correspond to LGT.
Rooting the Species Tree with LGT.
The reconstruction of LGT scenarios depends on the topology of the candidate tree, but because LGTs are time-oriented, it depends also on the position of its root. Because Prunier gives a score for every possible root of an unrooted reference tree, we reported in Fig. 2 only the results for the root position that minimizes the LGT number; however, this means that the minimum LGT score can also provide information on the position of the root of the reference tree. In our previous simulations, we found that the true root was indeed characterized by the best LGT score (12). Thus, we looked whether the LGT score could discriminate among possible roots for each phylum by using a statistical test (Wilcoxon paired test, α = 5%) to compare the score of different roots. In all phyla, several roots have scores statistically equivalent to the best root (Table S4). However, in Gammaproteobacteria, where the number of available gene families was maximal, only 15 of 137 possible roots had scores statistically equivalent to the best. Interestingly, they did not include the one found with the method of the outgroup, where the relatively fast-evolving group of Xanthomonadales and Chromatiales emerge first (Fig. S1). In contrast, the best root according to the LGT score supports the existence of a monophyletic group containing these lineages plus species of Pseudomonadales, Oceanospirillales, Alteromonadales, and Thiotrichales. Only the phylum of Actinobacteria was in the same situation with a large fraction (42 of 59) of possible roots being rejected. These rejected roots included the one generally observed in universal trees, with the early emergence of Rubrobacter and Bifidobacterium (13). In contrast, a root separating Corynebacterineae and Frankia from other Actinobacteria had the best LGT score. It has been shown that the choice of the outgroup can dramatically affect the position of the root of the ingroup when long branches are present (14, 15). The rooting of bacterial phyla is a typical case where long-branch attraction (LBA) can occur because they are by definition distantly related to other phyla. Because the LBA artifact is known to produce trees that are typically unbalanced (16), we used the Colless index (17) to measure the symmetry of trees rooted with the two methods. We observe that when the minimum LGT root differed from the outgroup root, the latter was systematically less balanced (Fig. S2), suggesting that, for several phyla, the currently accepted position of the root may be artifactual. The two phyla for which the difference in numbers of transfers was significant, Gammaproteobacteria and Actinobacteria, correspond to cases where the question of the root should indeed be considered unsettled. In both cases, when these phyla were rooted with an outgroup, the first lineages to emerge displayed branch lengths that are typical of LBA (13, 18), and phylogenomic analyses [using 356 genes for Gammaproteobacteria (19) and 155 for Actinobacteria (20)] failed to resolve the relationships among the so-called “basal” lineages. Because we analyzed phyla in the absence of outgroups, the same LBA artifacts as in universal phylogenies cannot affect our results. Hence, the information brought by LGT can be decisive in finding the position of the root.
Branch-Wise Transfer Numbers and Sequence Evolution.
For each gene family, Prunier identifies the branches of the species tree that has received a gene by transfer. It is thus possible to map all received LGTs on branches of the species tree. We observe a strong universal correlation between branch lengths of the tree of species as deduced from sequence comparisons, and numbers of gene transfer per gene family (significant Spearman's ρ value ranges from 0.59 to 0.87 in different phyla and is 0.61 for all phyla; Table S5). This correlation remains strongly significant (P < 0.0001 with a constant Spearman's ρ ∼ 0.5) when considering only the 75%, 50%, or 25% shortest branches of the species tree. Hence, this effect is not caused by fast-evolving species in which frequent LBA could be interpreted as high LGT rates. Rather, LGTs seem to have accumulated throughout life's history proportionally to sequence divergence, suggesting a relaxed LGT clock. However, other factors such as lifestyle clearly influence LGT rates, as observed, for instance, in the tree of Alphaproteobacteria (Fig. S3): Here, the group of Rickettsiales, which is composed of intracellular parasites, clearly displays LGT numbers that are unexpectedly low with regard to evolutionary rates. This lifestyle in a protected niche is known to cause an acceleration of sequence evolutionary rates associated with genome reduction. Our results suggest that it also allows markedly few opportunities for gene acquisition and/or gene transfer fixation.
LGT Rates in the Tree of Life.
A tree representing the best LGT phylogeny for each phylum is presented in Fig. 3. In this tree, branch lengths represent molecular divergence, whereas branch colors represent transfer rates, expressed in transfers per gene tree containing this branch. The rate of LGT varies widely among branches, from 0% to 31%. Two phyla, Actinobacteria and Gammaproteobacteria, exhibit a number of branches with particularly high rates of transfer. The branch leading to the actinobacterium Rubrobacter xylanophilus is remarkable for both its very high sequence evolutionary rate and its extreme LGT rate. Interestingly, as noted earlier, this species is typically the first to diverge from other Actinobacteria with the outgroup method but not with our approach. Its long branch strongly suggests that the LBA (21) could lead to an overestimation of LGT in gene trees without outgroup and to its early emergence when an outgroup is present (Fig. S1) (13, 18). Still, the majority of gene families in its genome are seen as vertically inherited from the common ancestor of Actinobacteria. Although Prunier takes phylogenetic uncertainty into account, systematic phylogenetic artifacts sometimes yield strongly supported branches that can be interpreted as LGT. Therefore, high rates of transfers coupled with high sequence evolutionary rates should be interpreted with caution. Overall, most of the branches of the tree have relatively low transfer rates, with 90% and 70% of the branches having transfer rates below 10% and 5%, respectively (Fig. 3). Hence, the impact of LGT on the branches of the tree of life is significant but not overwhelming.
Fig. 3.
A prokaryotic tree of life with branch-wise rates of transfers. Within-phyla relationships correspond to the minimum LGT tree, and relationships among phyla are reconstructed from a concatenate of 157 protein alignments after removal of transferred genes (Fig. S1). For each phylum, the root corresponding to the result of the outgroup method is presented, except for Actinobacteria and Gammaproteobacteria for which is shown the minimum LGT root, which was significantly better in terms of LGT score (Table S4; P < 0.05). Branch lengths were estimated on the basis of universal single-copy gene concatenation, and each branch is colored according to the proportion of gene families analyzed where the branch was transferred. Spiro, Spirochaetes; Chlamy, Chlamydiales-Verrucomicrobia; Bacter/Chloro, Bacteroidetes/Chlorobi; Delta, Deltaproteobacteria; Epsilon, Epsilonproteobacteria.
Transfer Rates Among Gene Functional Categories.
We used annotations from the Clusters of Orthologous Groups (COG) database (22, 23) to analyze LGT rates in gene functional categories. We first used a broad definition of functional categories, [i.e., “cellular processes and signaling” (“Cell”), “information storage and processing” (“Info”), and “metabolism” (“Meta”)]. Pooling all phyla together, there was a strongly significant difference between these categories in terms of LGT rates, with Meta gene families exhibiting higher rates of transfer than both Cell and Info gene families did (Kruskal–Wallis test, P value < 0.0001, and Tukey–Kramer honestly significant difference test; Fig. 4). This same pattern was visible and statistically significant in 9 of the 16 phyla (Fig. 4) and confirms what was initially formulated as the “complexity hypothesis” (24), i.e., that genes crucial to the maintenance of genetic information are consistently more resistant to gene transfer than metabolic genes are. We have also found that genes responsible for the establishment of basic cell structures have low transfer rates, similar to informational genes. When considering a finer definition of functional categories, this pattern was confirmed except for two classes, one of Info genes and one of Cell genes, that displayed much higher rates of LGT than expected from their broader category, suggesting that the definition of such broad functional categories can be misleading. These classes are “replication, recombination, and repair” (class L in COG) from the Info category (mean LGT rate = 3.7%) and “defense mechanisms” (class V in COG) from the Cell category (mean LGT rate = 6.2%). A closer inspection of the functional annotation of the genes present in these categories reveals many functions that are expected to be subject to high rates of LGT. For example, class L contains many DNA modification proteins that directly or indirectly promote the integration of foreign DNA in the genome. Similarly, class V contains several transporters as well as restriction-modification and antibiotic-resistance systems that are known to be highly transferred. Remarkably, class V is the class of genes with the highest LGT rate, even compared with Meta genes.
Fig. 4.
LGT rates across different functional categories. The dataset was annotated with the three COG categories: cellular processes and signaling (Cell), information storage and processing (Info), and metabolism (Meta). Average rates of LGT per 100 branches and per family are given for the three categories in corresponding boxes for each phylum and for the pooled dataset (All Phyla). Different colors correspond to classes of statistical equivalence in pairwise tests (Tukey–Kramer test, α = 0.05). Transparent boxes indicate no significant difference between transfer rates of the three categories. Chlamydiales-Verruco, Chlamydiales-Verrucomicrobia.
Discussion
Toward a Comprehensive Map of LGT in Prokaryotic History.
Many studies have aimed at measuring the impact of gene gain in bacterial genomes by using comparisons of gene repertoires or analysis of sequence composition (25). These approaches are known to identify sets of genes that are essentially different from those found with phylogenetic methods, which require that genes are shared among a certain number of species (26). It is possible that the events identified with these methods have quite different, subtler evolutionary consequences (27) because phylogeny detects not only the acquisition of new genes but also, more importantly, the replacement of genes by a homolog from another species. Hence, the LGTs detected with Prunier are those that affect evolutionarily successful gene families and are mainly gene replacements and, to some extent, gain of genes that had no homolog in the recipient genome. The selective pressures governing such gene replacements are poorly understood, and our results provide a guide to test their functional consequences. By searching for LGTs that occurred in the ancestor of a clade, we can identify events that may have played a role in the evolutionary success of these lineages. We detected, for instance, that both the ancestor of Listeria sp. and the ancestor of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis have independently received several genes involved in the biosynthesis of the flagellum. After closer inspection of the genome organization of these species, it appears that these genes are clustered in a region encompassing more than 40 genes mainly involved in flagellar assembly. Several of the corresponding gene families are not present in our dataset because they contain several copies per genome, but their phylogenies attest that they have all been transferred together. Interestingly, these genes have homologs in other Bacillales, where they seem to have been inherited vertically. This finding indicates that they must have been replaced twice independently in the ancestor of Listeria sp. and the ancestor of B. cereus, B. anthracis, and B. thuringiensis, which raises the question of the selective advantage that would explain the repeated acquisition of this flagellum complex.
Joining Pattern and Process in Reconstructing the History of Life.
LGT has shaped the evolution of life in many ways, and the challenge for evolutionary biologists is to reconstruct the history of LGT that makes the history of life. We have shown that an approach that explicitly models LGT can yield a much more complete picture of evolution, because it (i) integrates the phylogenetic information of thousands of gene families, (ii) provides explicit scenarios of evolution for these families, and (iii) recognizes as-yet-unexploited information on the root of the species tree. We show that independent sets of gene families support a same species tree and that most branches (>90%) of a gene tree are compatible with a pattern of vertical inheritance. The parameter (branch support value) used here to detect LGT was chosen based on the results of previous simulations (12), which showed a best compromise for sensitivity and specificity. However, even when changing this parameter, we obtained remarkably similar results, confirming that uncertainties in gene tree reconstruction are indecisive on the choice of a particular candidate topology (example of Actinobacteria in Table S6.). We observe a strong signal that gene functions influence the rate of LGT. In contrast, there was no obvious general relationship between rates of gene transfer and species ecology. The thousands of single-gene histories reconstructed in our analysis each bring information on the evolution of ancestral species that still needs to be decrypted in ecological terms on a case-by-case basis.
Some phyla of interest were not included in this study because of a lack of genomes available in HOGENOM. This is the case of Aquificales. This phylum was shown to present genomes with a significant part from foreign origins (28). It would be interesting to see whether the present approach could identify a rooted tree in this case. However, because these transfers are mainly operational genes from a variety of donor phyla (28), it is likely that the “minimum LGT score criterion” would still identify consistent vertical signal. However, this may not be the case when gene exchanges involve always the same donor and receptor. It was recently proposed that LGTs and, more specifically, gene displacements are more frequent between closely related species (29, 30) and thus could mimic the effect of a shared ancestry (31). At the scale of a bacterial phylum, this phenomenon would, however, have little effect.
Our motivation for using various approaches for the inference of a species tree was not to compare these methods but rather to generate a variety of good candidate trees for evaluation with the LGT score. This process was necessary because of the large dimension of the space of possible species trees. However, our results suggest that phylogenomic methods are systematically superior to strategies based on single genes such as rRNA genes to estimate a species tree. We investigated the reasons for the relatively poor performance of rRNA genes to produce a reasonable species tree. We searched for LGTs in the history of these genes by using Prunier and the best LGT score phylogeny as a reference. None of the gene trees reconstructed from either 16S or 23S rRNA yielded evidence for LGT. Hence, the differences observed in the 16S and 23S gene trees are most probably attributable to uncertainties of phylogenetic reconstruction with relatively short sequences. Because we only tested a limited number of candidate species trees, we cannot exclude that other topologies exist that further optimize the number of LGTs in gene families. However, we observe that independent datasets (universal vs. nonuniversal genes) supported the same species trees, showing that the reference trees supported by the minimum LGT score criterion are probably close to the tree of vertical descent, although a more thorough search and a more complete model of genome evolution, accounting for duplication, transfer, and loss, would probably allow a better exploitation of gene trees. These results highlight the potential of new methodologies that model the biological processes that make gene trees differ from species trees, such as gene transfer, duplication, and loss, to reconstruct the history of life (32).
Materials and Methods
Full list of analyzed genomes, alignments, trees, and LGT scenarios are available at ftp://pbil.univ-lyon1.fr/pub/datasets/DAUBIN/ABBY2011/.
Dataset Description.
We selected homologous families contained in the complete genome database HOGENOM [release version 4, September 2007 (33)] for archaeal and bacterial lineages [according to the National Center for Biotechnology Information taxonomy (34)] that did not cross phylum boundaries and had a minimal number of seven genomes in each taxonomic group. We selected one strain per species as we focused on deep relationships. We ended up with 16 clades (referred to as “phyla” throughout this paper) at different taxonomic levels: two archaeal and 14 bacterial, resulting in 336 selected genomes. An overview of the dataset can be found in Table S1 and more details are available in SI Materials and Methods.
Single-Gene Analysis.
For each phylum, the protein sequences of genes present in one copy per species in at least seven genomes among those selected were extracted from HOGENOM 4 (33), aligned with Muscle [default parameters (35)], and sites were filtered with Gblocks [default parameters (36)]. Then, individual gene trees were built with the maximum-likelihood method implemented in TREEFINDER (37) (WAG + G8 + I + F model).
Candidate Phylogenies.
Five different phylogenomic methods were used to combine the information of universal single-copy genes for each phylum (supermatrix and supertree approaches; SI Materials and Methods). Three other candidate trees were built with rRNA sequences (16S and 23S rRNA; SI Materials and Methods).
Detecting LGTs in Gene Trees Given Candidate Species Trees.
A new version of the program for transfer detection, Prunier (12), was used to detect transfers in the gene trees dataset by using alternately each candidate tree as a reference species tree. Prunier is a phylogenetic method for LGT detection that compares a gene tree with a given reference tree and infers LGT events only for significant topological conflicts between the gene tree and the reference tree. Here, we used a significance threshold of 80% for branch support [Local Rearrangement Expected Likelihood Weights (LR-ELW) supports computed by TREEFINDER (37)] to be considered as significant in LGT detection, and we fixed the “step” parameter to two steps (SI Materials and Methods).
Testing Among Phylogenetic Hypotheses and Choosing the Best Tree.
Prunier infers an LGT scenario for every possible rooting of a given reference tree. For each candidate species tree of a given phylum (unrooted at this step), we computed an LGT score (the sum of the LGT numbers they produce in the gene dataset) for each possible root (2n − 3 possibilities for a tree with n leaves). The root minimizing the number of LGT in genes was elected as the best root for this tree (minimum LGT score criterion). The best among candidate species tree for the phylum was selected with the minimum LGT score criterion, and LGT numbers were used to compare all pairs of rooted candidate species trees by using Wilcoxon tests paired on gene families (see SI Materials and Methods for more details). To compare all different root positions of the best phylum tree, we applied the same procedure: choice of the best root with the minimum LGT score criterion and test between roots with Wilcoxon tests on family-paired LGT numbers. Comparisons of trees and roots were performed with families for which Prunier was able to provide a scenario given the selected parameters (SI Materials and Methods).
Functional Categories Analysis.
We used cross-references between HOGENOM (33) and the COG database (22, 23) to assign COG categories to HOGENOM gene families by using a majority rule. At least one cross-reference was found for 4,401 gene families over 7,060 in the whole dataset. Families with ambiguous category definition (less than 50% of the sequences annotated) were excluded (276 families). Annotated families were gathered into “supercategories”: cellular processes and signaling (Cell; 710 families), information storage and processing (Info; 629 families), and metabolism (Meta; 1,299 families). Another 1,487 families were assigned to a “poorly characterized” function. The comparison of LGT rates between functional categories was performed with Tukey–Kramer honestly significant difference tests (α = 0.05) on a dataset of LGTs retrieved given the best candidate species tree for each phylum, excluding poorly characterized families and families that make Prunier enter the “switch” mode (92 of 12,602 gene trees were excluded; SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank E. Rocha, S. Penel, B. Boussau, G. Szollosi, and all the members of the Bioinformatics and Evolutionary Genomics Group for discussions on the results and comments on the manuscript. We also thank S. Delmotte, B. Spataro, and P. Calvat for their help with using computational resources and the Institut National de Physique Nucléaire et de Physique des Particules (IN2P3) computing center for providing computational resources. This project was supported by the French Agence Nationale de la Recherche (ANR) through Grants ANR-08-EMER-011-03 PhylAriane and ANR-10-BINF-01-01 Ancestrome.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116871109/-/DCSupplemental.
References
- 1.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 2.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 3.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 5.Bapteste E, et al. Do orthologous gene phylogenies really support tree-thinking? BMC Evol Biol. 2005;5:33. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bapteste E, et al. Prokaryotic evolution and the tree of life are two different things. Biol Direct. 2009;4:34. doi: 10.1186/1745-6150-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 9.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 10.Gray MW. Origin and evolution of organelle genomes. Curr Opin Genet Dev. 1993;3:884–890. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Xu Y, Gogarten JP. The presence of a haloarchaeal type tyrosyl-tRNA synthetase marks the opisthokonts as monophyletic. Mol Biol Evol. 2005;22:2142–2146. doi: 10.1093/molbev/msi221. [DOI] [PubMed] [Google Scholar]
- 12.Abby SS, Tannier E, Gouy M, Daubin V. Detecting lateral gene transfers by statistical reconciliation of phylogenetic forests. BMC Bioinformatics. 2010;11:324. doi: 10.1186/1471-2105-11-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pick KS, et al. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 2010;27:1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 16.Moreira D, Le Guyader H, Philippe H. Unusually high evolutionary rate of the elongation factor 1α genes from the Ciliophora and its impact on the phylogeny of eukaryotes. Mol Biol Evol. 1999;16:234–245. doi: 10.1093/oxfordjournals.molbev.a026105. [DOI] [PubMed] [Google Scholar]
- 17.Blum MGB. The mean, variance and limiting distribution of two statistics sensitive to phylogenetic tree balance. Ann Appl Probab. 2006;16:2195–2214. [Google Scholar]
- 18.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 19.Williams KP, et al. Phylogeny of Gammaproteobacteria. J Bacteriol. 2010;192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam MT, Merlo ME, Takano E, Breitling R. Genome-based phylogenetic analysis of Streptomyces and its relatives. Mol Phylogenet Evol. 2010;54:763–772. doi: 10.1016/j.ympev.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- 22.Tatusov RL, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 24.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 26.Ragan MA. On surrogate methods for detecting lateral gene transfer. FEMS Microbiol Lett. 2001;201(2):187–191. doi: 10.1111/j.1574-6968.2001.tb10755.x. [DOI] [PubMed] [Google Scholar]
- 27.Daubin V, Moran NA, Ochman H. Phylogenetics and the cohesion of bacterial genomes. Science. 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- 28.Boussau B, Guéguen L, Gouy M. Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria. BMC Evol Biol. 2008;8:272. doi: 10.1186/1471-2148-8-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creevey CJ, Doerks T, Fitzpatrick DA, Raes J, Bork P. Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS ONE. 2011;6:e22099. doi: 10.1371/journal.pone.0022099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andam CP, Gogarten JP. Biased gene transfer in microbial evolution. Nat Rev Microbiol. 2011;9:543–555. doi: 10.1038/nrmicro2593. [DOI] [PubMed] [Google Scholar]
- 31.Andam CP, Williams D, Gogarten JP. Biased gene transfer mimics patterns created through shared ancestry. Proc Natl Acad Sci USA. 2010;107:10679–10684. doi: 10.1073/pnas.1001418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boussau B, Daubin V. Genomes as documents of evolutionary history. Trends Ecol Evol. 2010;25(4):224–232. doi: 10.1016/j.tree.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Penel S, et al. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009;10(Suppl 6):S3. doi: 10.1186/1471-2105-10-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37(Database issue):D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 37.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.