Abstract
The human genome is densely populated with transposons and transposon-like repetitive elements. Although the impact of these transposons and elements on human genome evolution is recognized, the significance of subtle variations in their sequence remains mostly unexplored. Here we report homozygosity mapping of an infantile neurodegenerative disease locus in a genetic isolate. Complete DNA sequencing of the 400-kb linkage locus revealed a point mutation in a primate-specific retrotransposon that was transcribed as part of a unique noncoding RNA, which was expressed in the brain. In vitro knockdown of this RNA increased neuronal apoptosis, consistent with the inappropriate dosage of this RNA in vivo and with the phenotype. Moreover, structural analysis of the sequence revealed a small RNA-like hairpin that was consistent with the putative gain of a functional site when mutated. We show here that a mutation in a unique transposable element-containing RNA is associated with lethal encephalopathy, and we suggest that RNAs that harbor evolutionarily recent repetitive elements may play important roles in human brain development.
Keywords: genetic disease, long noncoding RNA, long interspersed element 1, medulla oblongata, pediatrics
Short and long interspersed elements (SINEs and LINEs, respectively) constitute the major retrotransposons of higher vertebrate genomes (1). Despite the parallels between their abundance and the evolution of higher cognitive capacities, most of these repetitive elements are still regarded as “junk” DNA. Several copies of retrotransposons inserted into noncoding genomic sequences have evolved either as new regulatory sequences (2, 3) or as a source of nonprotein coding RNAs (ncRNAs), including human microRNAs (miRNAs) (4, 5) and other RNA species, such as the neuronal BC1 (6). In addition, retrotransposons themselves have emerged as specific and transient targets of regulation by small RNAs in both germ cells and somatic cells. Interestingly, the brain is thought to be the major site of RNA expression (7–9).
There is increasing evidence that retrotransposition can induce genetic changes responsible for human diseases, as reviewed by Deininger and colleagues (10). Moreover, we and others have shown that either mutations in or deletion of the noncoding part of the genome can cause disease (11–14). Overall, ncRNA-based regulatory circuits appear central to all complex cellular, physiological, and neurological systems, and in particular, to specific genetic phenomena, including transcriptional and posttranscriptional silencing. However, whether variation in ncRNA that contains a repetitive element can result in pathogenicity remains unknown. In this study, we report that a rare nucleotide variation in a unique transposable element-containing RNA is associated with infantile encephalopathy. This study provides further evidence of how noncoding mutations may contribute to human diseases.
Results and Discussion
Progressive Encephalopathy with Severe Infantile Anorexia Segregates in a Geographic Isolate.
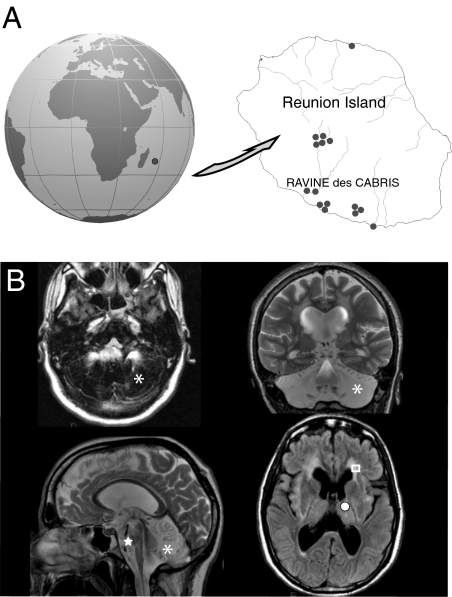
Because of historic, socioeconomic, and geographic constraints (SI Methods), a Caucasian isolate that lives in the southern part of Reunion Island (Fig. 1A), located in the Indian Ocean, presents with a high prevalence of autosomal recessive disorders. Most patients specifically originate from a region named Ravine. In this Ravine isolate, familial recurrence of an extreme phenotype of infantile anorexia led us to suspect autosomal recessive inheritance of the disease (Fig. S1). Inclusion criteria were: (i) infantile anorexia with irrepressible and repeated vomiting during infancy (SI Methods and Table S1) (15), (ii) acute brainstem dysfunction, (iii) severe failure to thrive, and (iv) specific involvement of the posterior fossa upon MRI. Indeed, brain MRI of patients revealed progressive and severe vanishing of the cerebellar white matter and brainstem atrophy, as well as sus-tentorial periventricular white-matter hyperintensities associated with basal ganglia anomalies (Fig. 1B).
Fig. 1.
Progressive encephalopathy associated with vanishing posterior fossa in the Ravine geographic isolate. (A) Distribution of a Caucasian isolate in the Ravine des Cabris region of Reunion Island in the Southern Indian Ocean. Birth locations of patients are indicated by dots. (B) Brain MRI scans of a patient aged 23 y: sagittal T2 (Upper Left), coronal T2 (Upper Right), axial T1 (Lower Right), and axial FLAIR (Lower Left) images, revealing severe atrophy of the pons (indicated by the star), the medulla oblongata, and the spinal cord. Abnormalities of the entire cerebellum are indicated by T2 hyperintensity and T1 hypointensity with a vanishing aspect (indicated by the asterisk). Sus-tentorial periventricular white matter hyperintensities (indicated by the square) are associated with a basal ganglia anomaly (indicated by the solid circle).
Autosomal Recessive Infantile Anorexia Locus Maps to Chromosome 8p22.
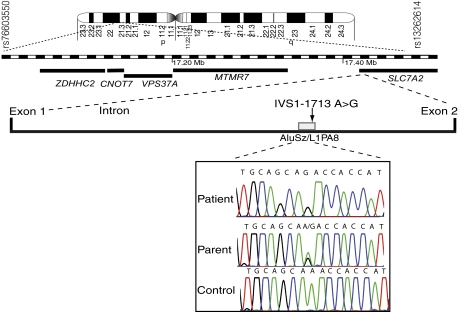
Genome-wide linkage analysis was performed in nine families, comprising 15 patients and 17 unaffected siblings (SI Methods and Fig. S1) and showed linkage to chromosome 8p22 in a 9.5-Mb region (Fig. 2). Homozygosity fine mapping defined a small candidate region between the marker loci D8S1731 and D8S261, which reached maximum cumulative LOD scores of 6.35 and 6.12, respectively, at θ = 0. This region is flanked by rs76603550 and rs13262614 (Figs. S1 and S2). An additional 17 patients from the same population were also typed in this region and were all homozygous for the haplotype (32 patients in total). This region spanned a 400-kb genetic interval, which contains five coding genes (ZDHHC2, CNOT7, VPS37A, MTMR7, and SLC7A2) (Fig. 2). All five genes at the disease locus were regarded as candidates because each was expressed in the brain with discrete and similar patterns (Fig. S2). Sequencing of the exons of those genes was performed in two patients, but no mutations were identified.
Fig. 2.
Mapping of an autosomal recessive infantile anorexia locus. A genome-wide scan using microsatellites mapped the disease locus to chromosome 8p22, and then the candidate region was finely mapped using additional microsatellite markers and SNPs. The minimal region of the disease locus was mapped with selected genetic markers at loci D8S550 and D8S280. All genomic coordinates are relative to the human hg19 assembly, and genes located between markers rs76603550 and rs13262614 are represented as solid lines. Full sequencing of the candidate interval led to the identification of a single-nucleotide variation (indicated by an arrow) in the first intron of the SLC7A2 gene. IVS1-1713 A > G maps within a repeated element, L1PA8, embedded in an AluSz that were identified using Repbase (35) (gray box). An electrophoretogram indicated that the variation is homozygous in patients (AA), heterozygous in parents (AG), and homozygous in ethnically matched controls (GG).
Intronic Nucleotide Variation in a Transposable Element Leads to the Identification of a Long Noncoding RNA.
Complete sequencing of the 400-kb region was then performed on both strands in two patients and two ethnically matched controls. Within the region, we identified 1,725 polymorphisms referenced in the SNP Database (dbSNP) build 132 and five nonreferenced (Dataset S1). The referenced polymorphisms included 1,699 SNPs, 24 indels, and 2 microsatellites. Within the two patients, none of the 1,725 polymorphisms was different from the reference sequence or the controls (Dataset S1). Each of the five nonreferenced variations was intronic SNPs (Dataset S1). One of the five was the same in the homozygous state in the parents as in the patients and was thus disregarded. Genotyping of the remaining four polymorphisms in 1,000 Reunionese controls resulted in identification of IVS1-1713 A > G (position 17,358,053 assembly hg19) as the only rare variation segregating with the disease. Indeed, heterozygosity for the mutant allele was detected in 2% of the Reunionese ancestry-matched population, with no homozygotes in the 1,000 Reunionese chromosome controls. The disease haplotype frequency was 1% with a disease prevalence of 1 in 10,000, which was in accordance with Hardy–Weinberg equilibrium. These data support a founder effect in this Caucasian-admixtured community of Reunion Island.
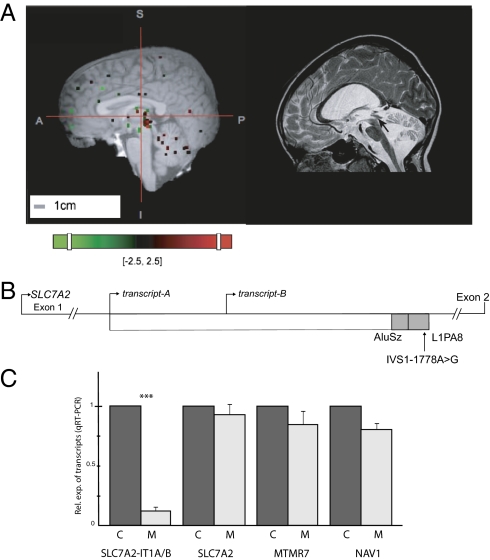
The mutation was located within the 5-kb first intron of SLC7A2 in a degenerated transposable element (Fig. 2). This sequence was composed of the 3′ end of a LINE-1 element, L1PA8, which comprised the mutation and was embedded in a SINE element, AluSz (Fig. 2). Regarding the disease phenotype, SLC7A2 appears to be a good candidate because of the discrete pattern of expression of SLC7A2 in the brain tectal plaque that closely matches the pattern of aqueductal stenosis observed in some Ravine patients (Fig. 3A and Fig. S3A). In the mouse, the SLC7A2 locus is complex because of an overlapping transcript, the CTN-RNA, which under stress conditions serves as a reservoir for SLC7A2 through posttranscriptional cleavage of its repeat elements (16). We thus checked whether dysregulation of SLC7A2 expression or splicing because of the mutation could have occurred. Indeed, exonization mediated by transposed elements is regarded as one of the underlying causes for the high frequency of alternative splicing in human protein-coding genes (17, 18). However, we did not observe ab initio exons, splicing alterations, or a change in the level of expression of SLC7A2 in patients with a combination of in silico analyses, reverse transcription, and real-time quantitative PCR (qRT-PCR) experiments (Fig. S3 B and C, and Table S2). We further determined whether transcriptional activity was associated with the repetitive elements and found a unique expressed sequence tag (AV720089) that comprised the IVS1-1713 A > G mutation. Using 5′- and 3′- RACE experiments, we cloned two unique transcripts, A and B, which were 1,536 bp and 962 bp in length, respectively, and that were colinear to SLC7A2 (Fig. 3B and Fig. S4). Using RT-PCR, we found that transcripts A/B were expressed in several different brain structures (Fig. 3B). However, no homology was found with any known protein, motif, or regulatory motif in either transcript (Table S2). In cross-species sequence comparisons using University of California at Santa Cruz hg19 46-species MultiZ track, we observed the homology-spanning transcript A sequence among all primates. However, the degenerated transposable element spanning the mutation was absent in all placental mammals (Fig. S5). Megablast against the various primate genomes available indicated a homology range between 87% and 97%, with conservation of the A nucleotide, which is the wild-type nucleotide in Reunionese and in all primate genomes (Fig. S5). Thus, the A nucleotide is the ancestral allele.
Fig. 3.
Identification of a noncoding RNA by analysis of intronic nucleotide mutation in a transposed element. (A) Expression of the SLC7A2 gene in brain of control and patient. In the control brain (Left), the 3D planar view of SLC7A2 expression shows reproducible signals with three distinct probes in cerebellum and midbrain. Probes were all located in SLC7A2 on chromosome 8 and span from 17,412,117–17,412,176; 17,419,598–17,421,149; and 17,415,803–17,415,856 (relatively to the human hg19 assembly) (see Methods and the Allen Human Brain Atlas). In the patient brain, the spatial correspondence between images and T1-weighted MR volumes, obtained via a series of assisted registration processes, are consistent with acqueductal stenosis, indicated by an arrow observed on a sagittal T2 plane MRI image from a patient aged 10 y old. (B) Identification of a totally intronic ncRNA. RACE-PCR in human brain led to the identification of two overlapping transcripts: A and B, of 1536-bp and 962-bp in length, respectively. The hg19 coordinates are 17,356,587 and 17,357,161 for the transcription start sites of A and B, respectively, and 17,358,123 for the end of both transcripts. Cloning and sequencing revealed that the rare single nucleotide variation (indicated as an arrow) (hg19 coordinate: 17,358,053), which was found homozygous in patients only, is included in both transcripts. Gray boxes schematize the repetitive elements. Expression of transcripts A/B was evaluated by RT-PCR in adult and in fetal human brain tissues. Vimentin (NM_003380) served as a positive control. Negative controls were tested on each total RNA showing the absence of genomic DNA. (C) Expression of SLC7A2-IT1A/B ncRNA in the brain of patient versus control by qRT-PCR. Postmortem brain RNA extraction was performed similarly in the patient as in the control. Control RNA consisted of commercially available pool of normal brain tissues (Human Total RNA; Clontech). Patient RNA was from a Reunionese female individual, age 23. RNA integrity was ensured using an on-chip capillary electrophoresis, reaching 8.1 and 7.6 for the patient and the control RNA, respectively. Expression of SLC7A2-IT1A/B, SLC7A2, MTMR7, and NAV1 was measured by qRT-PCR, normalized to GAPDH, and shown as a ratio to control. Data are mean ± SD. ***P < 0.01 (n = 3).
Phenotypic Consequences of the Transposable Element-Containing Noncoding Variant.
According to the nomenclature for RNAs, SLC7A2 intronic transcripts A and B were considered to be long noncoding RNA (lncRNA) and named SLC7A2-IT1A and SLC7A2-IT1B, respectively (19, 20). We found that the overall expression of SLC7A2-IT1A/B was specifically reduced more than eightfold in patient brain tissue compared with control, as assessed with qRT-PCR using GAPDH as the internal reference (Fig. 3C). Conversely, the expression of a brain-specific mRNA and the two genes of the mapped locus serving as controls remained unchanged (Fig. 3C).
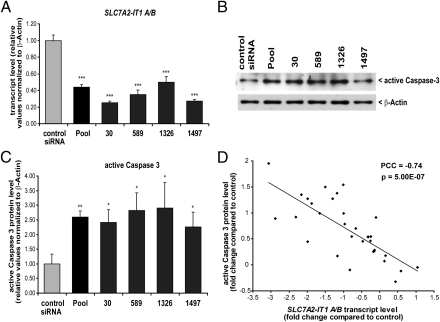
To investigate the function of wild-type SLC7A2-IT1A/B and to mimic the reduced levels in the disease, we performed transient siRNA-based knockdown of SLC7A2-IT1A/B in human neuronal cells (Fig. 4A). We used a set of four SLC7A2-IT1A/B targeting siRNAs, individually as well as pooled, that are directed against different and nonoverlapping sites in the transcripts. Knockdown elicited a significant increase in apoptotic cells as assessed by detection of active Caspase3 (Figs. 4 B and C). A correlation study based on different SLC7A2-IT1A/B knockdown conditions showed a significantly inverse correlation of SLC7A2-IT1A/B transcript levels and active Caspase-3 protein levels (Fig. 4D). Similarly, knockdown of SLC7A2-IT1A/B by antisense oligonucleotides resulted in a significant increase in apoptotic cells as assessed by dual staining of mitochondrial Bax and active Caspase 3 (Fig. S6).
Fig. 4.
RNAi knockdown of SLC7A2-IT1A/B transcripts. Human neuroblastoma Kelly cells were transiently transfected with 10 nM of a nontargeting siRNA pool (control siRNA) or a 10 nM pool of four SLC7A2-IT1-targeting siRNAs (Pool). The targeting siRNAs were further tested individually at 10 nM each. In the figure “30” is A-specific, “589” is specific for A and B transcripts, “1326” is preAluSz-specific, and “1497” is L1PA8 specific. (A) Real-time PCR analysis to estimate SLC7A2-IT1A/B transcript levels. Values were normalized to relative β-actin levels. (B) Detection of active Caspase-3 protein levels by Western blotting. A representative original blot is shown. β-Actin served as loading control. (C) Statistical analysis of Western blotting results. Knockdown of SLC7A2-IT1A/B by ∼40% leads to a more than 2.5-fold elevated active Caspase-3 level. n = 3. (D) Correlation of SLC7A2-IT1A/B transcript levels and active Caspase-3 protein levels. Using different transfection conditions (10 nM up to 100 nM siRNA concentrations for 24 h up to 48 h) SLC7A2-IT1A/B transcript levels were assessed by real-time PCR and correlated to corresponding active Caspase-3 protein levels. PCC, Pearson correlation coefficient; n = 32. *P < 0.05, **P < 0.01, ***P < 0.001.
Potential Structural Effect of the Mutation.
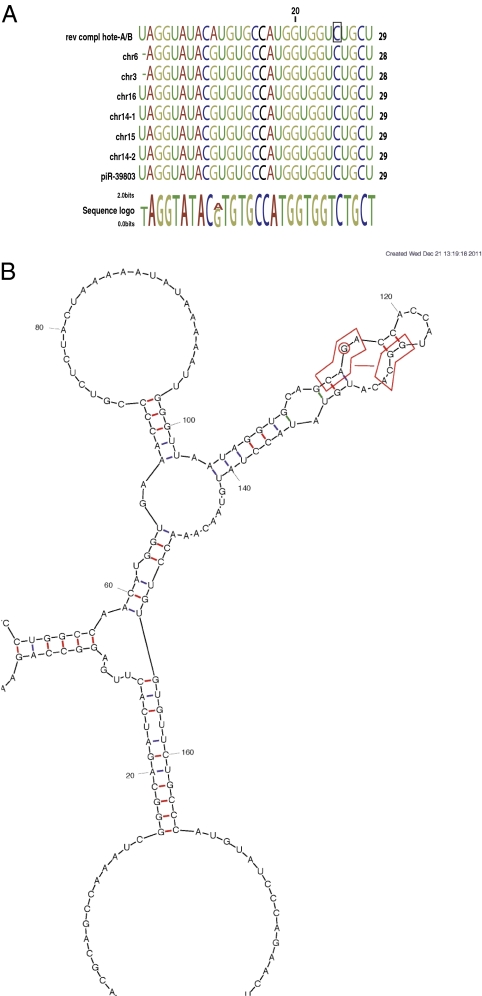
We blasted the SLC7A2-IT1A sequence against the Rfam database and found that the transcript did not correspond to any of the 1,973 ncRNA families. Nonetheless, blasting the mutant sequence of SLC7A2-IT1A against the small RNAs from the comprehensive ncRNA sequence database fRNAdb (<50 nt) identified antisense homology to a Piwi-interacting RNA (piRNA) at the level of the mutation (Fig. 5A). The mutation increased the probability for the mutant SLC7A2-IT1A sequence to de facto form the complementary sequence to a piRNA by 103-fold, reaching an E-value of 4 × 10−6 vs. 0.001 for the wild-type. Based on the threshold values, the homology to the mutant but not the wild-type sequence was significant. Retrotransposon silencing can be mediated by piRNAs (21) and may lead to silencing of the transcript (22). Thus, we hypothesize that the mutated segment within SLC7A2-IT1A/B is a privileged target of piRNAs. According to the recent identification of piRNAs in mammalian brain (23, 24), piRNAs may invade and bind to the complementary mutant sequence, which ultimately leads to the silencing of the transcript (22).
Fig. 5.
(A) Multialignment and sequence logos of reverse complement SLC7A2-IT1A/B (labeled “rev compl SLC7A2-IT1A/B”) and piRNAs. Boxed nucleotide indicates the mutant site. Homology to hsa-piR-39803 piRNAs scattered on different chromosomes is shown. The piRNA sequences were extracted from the piRNA database. (B) Secondary structure as calculated by the program mfold (25) for the RNA fragment comprising the Alu/Line sequence. The same local fold is obtained with or without the mutation A > G for that fragment as well as with the whole ncRNA. The mutated IVS1-1713 A > G is within an internal loop. A slight rearrangement leads to the potential occurrence of a set of non-Watson–Crick pairs, as observed in the internal loop of helix 8 of 7SL RNA of SRP (27). This occurrence is illustrated by a straight line between the two blocks of circled nucleotides, which delineates a potential structural module of the consensus motif 5′-CAGA-3′ and 5′-GGCA-3′.
We further questioned the significance of the mutation on the secondary structure of the lncRNA. Using the mfold program (25), we calculated the 2D structure of the AluSZ/L1PA8 186-bp fragment of SLC7A2-IT1A (Fig. 5B), which was similar to the full-length and mostly unaffected by the mutation. In fact, the mutation occurs in an asymmetrical internal loop of one of the small hairpins of the second three-way junction. Interestingly, upon slight rearrangement of the pairs, the mutated internal loop presents a conserved motif of helix 8 in domain IV of the 7SL RNA of the signal recognition particle (SRP) (i.e., 5′-CAGA-3′ and 5′-GGCA-3′) (Fig. 5B) (26). This latter motif is known to form a tight structural module with very specific binding to the highly conserved SRP protein SRP54 (27). In particular, alignments of 7SL RNAs showed that the third G of the 5′ strand is invariant (28). SRP54 is a key component for interactions with the signal peptide, the SRP receptor, and the binding to two ribosomal proteins, L23a and L35 (26). This observation suggests the intriguing possibility that the recruitment of SRP54 induced by the mutation participates in the observed deleterious effects.
Prompted by the finding that the mutation occurred in a small hairpin, we further studied the structural modularity of SLC7A2-IT1A/B to assess the likelihood that a small RNA is processed from the lncRNA (29). Assessing the SLC7A2-IT1A/B sequence as a source of miRNAs revealed an 82-bp sequence that generated a good triplet-support vector machine candidate that could only be retrieved using the mutant sequence and not the wild-type, consistent with other algorithms used for predicting miRNA precursors (Table S2) (30).
In sum, several hypotheses could explain the effect of the mutation in SLC7A2-IT1A/B, including the possibility of altered posttranscriptional regulation of SLC7A2 as observed with the CTN-RNA in stress conditions (16). From the current insights, we propose to view the A > G mutation as: (i) a mediator of RNA:protein interactions, which may potentially be SRP54; (ii) or as a mediator of RNA:RNA interactions, acting either as a piRNA target or as a miRNA-like sequence. In either scenario, the mutation supports a putative gain of a functional site. A challenge ahead will be to test whether any of these predicted effects of the mutation occurs in the disease context of Ravine encephalopathy.
Concluding Remarks.
In conclusion, the noncoding A > G mutation we identified in Ravine encephalopathy results in significantly decreased levels of expression of the SLC7A2-IT1A/B RNA, which is associated with an increase in cell-death markers. As shown with RNAi experiments, silencing this lncRNA resulted in increased apoptosis, which is consistent with the atrophy and vanishing white matter observed in patients’ brain MRIs. Similarly, the intriguing overlap between SLC7A2 expression and the acqueductal stenosis that we observed may suggest that the spatial expression of SLC7A2-IT1A/B RNA shares regulatory elements with SLC7A2, as has been documented for several ncRNAs and their host genes (31).
Overall, our data unveil the contribution of a mutation in a specific lncRNA in a progressive disorder of the posterior hindbrain involving the medulla oblongata and leading to anorexic behavior. The existence of taxon- and species-specific ncRNAs encompassing repetitive elements suggests the possibility of their involvement in modification of gene regulatory networks that may control development of more complex systems, typically in human-specific brain function (8, 32, 33).
Methods
Patients.
Individuals were included in the cohort upon presentation of distinctive clinical features, such as infantile anorexia with emesis and failure to thrive, which was related to acute brainstem dysfunction and an MRI pattern of specific involvement of the posterior fossa. For each patient, proband, parent, or unaffected sibling, written obtained informed consent was obtained according to the French Ethics Committee, and all procedures were approved by the reviewing board at the Reunion hospital. For each patient, lymphoblastoid cell lines were established by Epstein-Barr virus transformation, and genomic DNA was extracted. The two patients and two healthy individuals used as controls for complete sequencing were unrelated over five generations based on genealogic registry. All controls were ethnically matched individuals, issued from the Centres de Ressources Biologiques (CRB)-DNA bank of Reunion Island.
Linkage Analysis, Sequencing, and Mutation Detection.
For homozygosity mapping, a panel of 400 markers at an average distance of 10 cM was used. A linkage software (M-LINK) was used to calculate two-point LOD scores between the disease phenotype and each marker, under the assumption of a recessive disorder with a mutated allele frequency of 0.01. Amplicons were ∼400–600 bp in length with an average overlap of ∼50 %, and covered the entire 400-kb locus. PCR products were purified with the Qiagen PCR Purification kit, and sequenced directly using the amplification primers and the ABI Big Dye Terminator v1.1 Cycle Sequencing kit. Sequences were determined with an ABI model 3100 Genetic Analyzer (Applied Biosystems), and contigs were assembled using SeqScape v2.1.
Postmortem Specimens.
Brain tissue was obtained following written consent from the next of kin (the parents). The request was only made to parents who had already faced the death of another child because of this disease. Brain tissue was dissected along the cerebellum-pons axis, snap-frozen in liquid nitrogen, and stored at −70 °C. Samples were cut from the snap-frozen postmortem samples on dry ice, transferred immediately to TRIzol solvent (Invitrogen), and total RNA was isolated per the product protocol for quantitative PCR. Control RNA consisted of a pool of normal brain tissues (cerebellum, pons, and medulla oblongata), which were commercially available (Human Total RNA; Clontech). RNA integrity was checked using RNA 6000 Nano Labchips in an Agilent 2100 Bioanalyzer following the manufacturer's protocol.
RNA Extraction and RACE.
For RACE, total RNA was extracted using the RNeasy Mini Kit (Qiagen). One microgram of total RNA was reverse-transcribed according to the protocol provided by the SMART RACE cDNA Amplification kit (Clontech) in a final volume of 100 μL. For 5′-RACE, we performed the first run of amplification according to the protocol provided with the Advantage 2 PCR kit (Clontech) using the primer Rev: 5′-gaatgcggacaaatttgttactac and the used universal primer mixture (UPM) as the adaptor primer, and the second run with 1-μL PCR product from the first run using the nested primer Rev: 5′-agcccatttagacaagtgctatc, with the nested adaptor primer NUPM. The 3′-RACE was essentially the same, except we used the primer Fwd: 5′-gtagtaacaaatttgtccgcattc.
Gene Expression Analyses.
All primer sets are listed (SI Methods). One microgram total RNA was reverse-transcribed with the High Capacity cDNA Archive kit primed with Oligo d(T)16 (Applied Biosystems) in accordance with the supplier's recommendations. Semiquantitative RT-PCR was normalized with respect to vimentin expression in adult and fetal human brain tissues (at 25 PCR cycles, in the exponential phase). A 276-bp cDNA fragment of SLC7A2-IT1A/B mRNA and a 274-bp cDNA fragment of Vimentin mRNA were amplified using the respective primer sets: IT1-A/B-F/-R and VIME1-F/-R. qRT-PCR reactions were performed in triplicate using either a Light Cycler 480 Instrument (Roche Diagnostics) or the GeneAmp 5700 system (Applied Biosystems) according to the manufacturer's instructions. RT-PCR was carried out with M-MLV reverse transcriptase (Applied Biosystems) and the LightCycler 480 DNA SYBR Green I Master reaction mix (Roche Diagnostics). Relative expression was calculated using the Livak (2 -ΔΔCt) method and normalized to control. GAPDH or β-actin served as the control for normalization. SLC7A2, MTMR7, NAV1, GAPDH and β-actin mRNA were quantified using the following primer sets: SLC7A2-E4-F/-R, MTMR7-E7-F/-R, NAV1-E13-F/-R, GAPDH-E3-F/-R, and β-actin-F/-R, respectively. Whole-brain microarray experiments were performed with the same donor (male, aged 24 y, African American, postmortem interval: 23 h) using three distinct probes located on chromosome 8 in SLC7A2 that span 17,412,117–17,412,176, 17,419,598–17,421,149, and 17,415,803–17,415,856 (hg19). Data presented in heat map format are normalized across the entire set of microarray samples. Images and methods are accessible in the Allen Human Brain Atlas (http://www.brain-map.org). SLC7A2 expression profiles were systematically sampled throughout regions of the brain.
Transfection.
SLC7A2-IT1A/B knockdown experiments were performed in Kelly neuroblastoma cells, which were purchased from the German DSMZ, and genotyped as wild type for IVS1-1713 A > G (ACC 355). Stealth siRNAs (LifeTech) were designed to target four different sites of the transcript (SI Methods and Fig. S4). Kelly cells were grown to ∼70% confluency in six-well plates and transfected with either 10–100 nM each siRNA or a pool of the four siRNAs (2.5 nM each) for 24 or 48 h. A commercially available nontargeting siRNA pool (Dharmacon) served as a negative control. Cells were transfected using SilenceMag (SM10500; OZ Bioscience), according to the manufacturer's protocol. Cells were harvested 24 or 48 h after transfection for gene or protein expression analyses. Preparation of protein extracts, Western blotting, and immunodetection were performed as previously described (34). Active Caspase 3 was detected using a monoclonal antibody to Caspase3 (AM08377PU-N, Acris Antibodies; 1:500). Anti–β-actin antibody (MAB1501R; Chemicon) was applied after stripping of the membranes to control for differences in protein loading. Primary antibodies were detected with peroxidase-coupled secondary antibodies and visualized using the Chemi-Glow-West Detection kit (Alpha Innotech). Autoradiographic signals were scanned and quantified using Scion Image software (Scion). The Student's paired t test was applied to reveal statistical significances. P values less than 0.05 were considered significant. To correlate SLC7A2-IT1A/B knockdown with apoptosis, we calculated the fold-change relative to control siRNA transfection for SLC7A2-IT1A/B transcript levels and corresponding active Caspase-3 protein levels per tested condition. The P value based on the Pearson correlation coefficient was calculated.
Sequence Analyses.
The human SLC7A2 sequence was extracted from the Human Genome Project Assembly (GRCh37/hg19) using the transcript ENST00000494857 as a reference from Ensembl. Briefly, the human sequence was analyzed for SNPs using BioMart from Ensembl, repeat sequences (LINEs, SINEs, etc.) using RepeatMasker, and species-specific collection of repeat sequences from the latest Repbase-Update (35). A variety of complementary approaches were used to search for exons and regulatory elements (36): GENSCAN and ExonScan Web Server to identify ab initio exons; Expasy Translate tool to search for potential ORFs checked for homology with BLASTP against Swiss-Prot and PDB-databases; BLASTN (37) against mRNA (expressed sequence tag or cDNA) sequences from GenBank (release 123); the Rfam database (v10.1) (http://rfam.sanger.ac.uk/); tRNAscan-SE (38); the fRNA database (v3.4) (39); and support vector machine-based algorithms to search for real miRNA hairpins (40, 41) (Table S2).
Supplementary Material
Acknowledgments
We thank the DNA bank (Centres de Ressources Biologiques–La Réunion), the families for participating in the study, and the clinicians for the sample collection; J. M. Rozet, C. Huber, A. Rötig, H. Quesneville, H. Roest-Crollius, and S. Boissinot for their contributions; Chris Gordon for helpful proofreading; and F. Darcel, M. L. Jaquemont, and J. F. Lesure for discussions, referring patients, and support. This work was supported by the Institut National de la Santé et de la Recherche Médicale, an Agence Nationale de Recherche EvoDevoMut grant; and Deutsche Forschungsgemeinschaft Grant FA845/2-2 (to M.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Single Nucleotide Polymorphism Database (dbSNP), www.ncbi.nlm.nih.gov/projects/SNP (accession nos. rs76603550, rs77619061, rs80022018, rs79197328, rs76885599, and rs74720879).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111596109/-/DCSupplemental.
References
- 1.Huang CR, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy A, Sela N, Ast G. TranspoGene and microTranspoGene: Transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2008;36(Database issue):D47–D52. doi: 10.1093/nar/gkm949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitelaw E, Martin DI. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 4.Seitz H, et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 5.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Zhong J, et al. Regulatory BC1 RNA and the fragile X mental retardation protein: Convergent functionality in brain. PLoS ONE. 2010;5:e15509. doi: 10.1371/journal.pone.0015509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 9.Satterlee JS, et al. Noncoding RNAs in the brain. J Neurosci. 2007;27:11856–11859. doi: 10.1523/JNEUROSCI.3624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hüttenhofer A, Schattner P, Polacek N. Non-coding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 13.Benko S, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 14.de Pontual L, et al. Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renouil M, et al. Severe anorexia in infants in Reunion: A new autosomal recessive disease? (French) Arch Pediatr. 1999;6:725–734. doi: 10.1016/s0929-693x(99)80354-6. [DOI] [PubMed] [Google Scholar]
- 16.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Krull M, Brosius J, Schmitz J. Alu-SINE exonization: En route to protein-coding function. Mol Biol Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 18.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ Splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 19.Wright MW, Bruford EA. Naming ‘junk’: Human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics. 2011;5:90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vorechovsky I. Transposable elements in disease-associated cryptic exons. Hum Genet. 2010;127:135–154. doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- 21.Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS One. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dharap A, Nakka VP, Vemuganti R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–1109. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee EJ, et al. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer-Eriksson AE, Hainzl T. S-domain assembly of the signal recognition particle. Curr Opin Struct Biol. 2003;13:64–70. doi: 10.1016/s0959-440x(02)00010-6. [DOI] [PubMed] [Google Scholar]
- 27.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 28.Regalia M, Rosenblad MA, Samuelsson T. Prediction of signal recognition particle RNA genes. Nucleic Acids Res. 2002;30:3368–3377. doi: 10.1093/nar/gkf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee MT, Kim J. Self containment, a property of modular RNA structures, distinguishes microRNAs. PLOS Comput Biol. 2008;4:e1000150. doi: 10.1371/journal.pcbi.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone CD, Hannon GJ. Molecular evolution of piRNA and transposon control pathways in Drosophila. Cold Spring Harb Symp Quant Biol. 2009;74:225–234. doi: 10.1101/sqb.2009.74.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 34.Fähling M, et al. Translational regulation of the human achaete-scute homologue-1 by fragile X mental retardation protein. J Biol Chem. 2009;284:4255–4266. doi: 10.1074/jbc.M807354200. [DOI] [PubMed] [Google Scholar]
- 35.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res. 2010;20:311–319. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kin T, et al. fRNAdb: A platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35(Database issue):D145–D148. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hertel J, Stadler PF. Hairpins in a haystack: Recognizing microRNA precursors in comparative genomics data. Bioinformatics. 2006;22:e197–e202. doi: 10.1093/bioinformatics/btl257. [DOI] [PubMed] [Google Scholar]
- 41.Batuwita R, Palade V. microPred: Effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics. 2009;25:989–995. doi: 10.1093/bioinformatics/btp107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.