Abstract
General transcription factor TFIIH, previously described as a 10-subunit complex, is essential for transcription and DNA repair. An eleventh subunit now identified, termed Tfb6, exhibits 45% sequence similarity to human nuclear mRNA export factor 5. Tfb6 dissociates from TFIIH as a heterodimer with the Ssl2 subunit, a DNA helicase that drives promoter melting for the initiation of transcription. Tfb6 does not, however, dissociate Ssl2 from TFIIH in the context of a fully assembled transcription preinitiation complex. Our findings suggest a dynamic state of Ssl2, allowing its engagement in multiple cellular processes.
Keywords: DNA repair, general transcription factor, RNA polymerase II, TFIIK, transcription pre-initiation complex, yeast
RNA polymerase II requires a set of general transcription factors for promoter recognition and the initiation of transcription (1). The largest of the general factors, TFIIH, comprises nearly a dozen subunits, with a total mass in excess of 500 kDa, and harbors three catalytic activities, two helicases and a protein kinase. TFIIH tends to dissociate upon isolation into subcomplexes that perform essential roles in other cellular processes. A “core” complex of Tfb1, Tfb2, Ssl1, Tfb4, Tfb5, and the 5′ to 3′ DNA helicase Rad3 (see Table 1 for human nomenclature) is required not only for transcription but also for DNA repair (2–5). A kinase complex, also known as TFIIK, comprising Tfb3, the cyclin Ccl1, and the cyclin-dependent kinase Kin28, phosphorylates the C-terminal domain (CTD) of RNA polymerase II (6–8), and is also required in metazoans for cell cycle control (9). An additional, dissociable component, the 3′ to 5′ helicase Ssl2, unwinds DNA around the transcription start site, and is also required both for DNA repair and for mRNA transactions.
Table 1.
Proteins identified by LC-MS/MS analysis of TFIIH
| Yeast | Human | MW | number of peptides |
| 1 Ssl2 | XPB | 95k | 23 |
| 2 Rad3 | XPD | 90k | 22 |
| 3 Tfb1 | p62 | 73k | 21 |
| 4 Tfb2 | p52 | 59k | 12 |
| 5 Ssl1 | p44 | 52k | 10 |
| 6 Tfb3 | Mat1 | 38k | 10 |
| 7 Ccl1 | CyclinH | 45k | 13 |
| 8 YOR352w(Tfb6) | 39k | 12 | |
| 9 Tfb4 | p34 | 37k | 6 |
| 10 Kin28 | Cdk7 | 35k | 6 |
| 11 Tfb5 | Tfb5 | 8k | 3 |
Ssl2 was originally discovered as a suppressor of a stem-loop inserted in the 5′-untranslated region of yeast HIS4 mRNA (10), suggesting a possible role of Ssl2 in mRNA transactions or translation initiation. Genetic and biochemical studies revealed that TFIIH associates with the DEAD box RNA helicase Dbp5 and may be recruited to a nascent ribonucleoprotein particle (RNP) in an early step of mRNA export from the nucleus (11). Further evidence for the involvement of Ssl2 in mRNA export came from the finding that Saccharomyces cerevisiae ssl2-rtt and S. pombe ptr8-1 mutations (PTR8 is the Saccharomyces pombe homolog of SSL2) cause nuclear accumulation of poly(A)+ mRNA (12). Moreover, Ptr8 genetically interacts with Tho2, a component of the transcription/export (TREX) complex (12).
We report here on a finding made in the course of TFIIH isolation from yeast that may underlie the multifunctional roles of Ssl2.
Results
Identification of Tfb6 as Subunit of Yeast TFIIH.
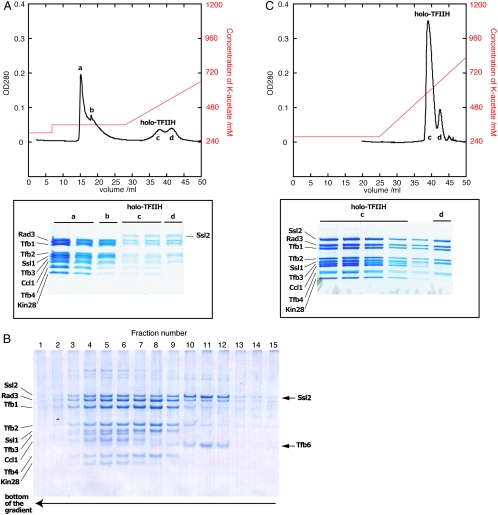
Affinity purification of TFIIH from yeast by means of a TAP-tag on Tfb3, followed by chromatography on UnoQ, gave a series of protein peaks containing the previously described 10-, 9-, 7-, and 6-subunit complexes (refs. 13,14; Fig. 1A). The 10-subunit complex, referred to as holo-TFIIH, constituted only about 10% of the total purified protein, at a yield of less than 0.5 mg from 5 kg of yeast. Mass spectrometry of the holo-TFIIH peak identified the ten previously reported subunits of TFIIH, as well as peptides encoded by an uncharacterized open reading frame, YOR352w (Table 1, Dataset S1). BLAST searches revealed 92% sequence similarity to putative homologues in fungi, and 19% sequence identity and 45% similarity to the human nuclear mRNA export factor isoform 5 (http://mips.helmholtz-muenchen.de/genre/proj/yeast). The protein product of YOR352w would have an expected molecular weight of 39 kDa, and so may have escaped detection in the past due to comigration with Ccl1 on SDS-PAGE (Fig. 1A, Lower).
Fig. 1.
Deletion of TFB6 enables the isolation of holo-TFIIH (A) Elution profile from UnoQ. Wild type yeast (5 kg) from 400L culture was lysed and purified on an IgG column. The eluate was loaded onto a UnoQ (Upper) and analyzed by SDS-PAGE (Lower). Four peaks were observed, corresponding to the 10-subunit (lacking Ssl2) (a), 7-subunit (lacking Ssl2 and TFIIK) (b), 11-subunit (holo-) (c), and 8-subunit (lacking TFIIK) TFIIH (d). (B) Peak c in (A) was subjected to glycerol gradient sedimentation and analyzed by SDS-PAGE. Approximately 40% of Ssl2 was dissociated from TFIIH and formed a dimer with Tfb6 (fractions 10–12). (C) TFIIH was purified from 2.5 kg (200L culture) of the TFB6-deletion strain of yeast as in (A). One major peak corresponding to 11-subunit TFIIH was observed with a minor peak corresponding to 8-subunit TFIIH (lacking TFIIK).
To investigate the association of the YOR352w protein with TFIIH, the UnoQ peak of holo-TFIIH was further fractionated on a glycerol gradient (Fig. 1B). Approximately 40% of the Ssl2 sedimented more slowly than TFIIH (fractions 4–9 in Fig. 1B), along with a 39-kDa protein (fractions 10–12 in Fig. 1B). Mass spectrometry identified the 39-kDa protein as the product of YOR352w, which we refer to as Tfb6. The apparent association of Tfb6 with Ssl2 is in keeping with genome-wide proteomic studies demonstrating an interaction of the two proteins (15, 16). Interaction was confirmed by affinity purification by means of a TAP tag on Tfb6 (16). Tfb6 may be regarded as an eleventh subunit of yeast TFIIH, with a tendency to dissociate from the complex.
Tfb6 Facilitates Dissociation of Ssl2 from TFIIH.
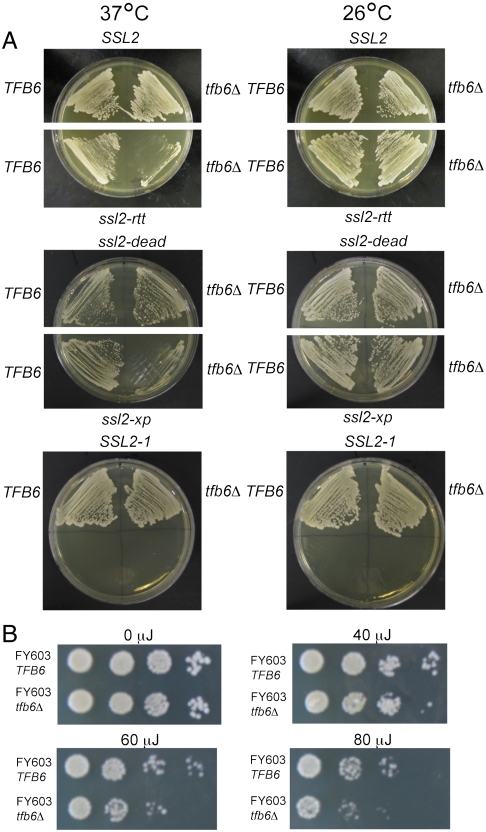
The disruption of TFB6 did not affect the cell growth, but it conferred temperature-sensitive growth when combined with the ssl2-rtt or ssl2-xp alleles (Fig. 2A). The ssl2-rtt mutation causes nuclear accumulation of poly(A)+ mRNA (12). The mutant allele ssl2-xp encodes a protein resembling the mutated XPB protein from UV-sensitive human cells and confers UV (10). The tfb6Δ strain was slightly more sensitive to UV light than wild type (Fig. 2B), suggesting a possible role in DNA repair.
Fig. 2.
Synthetic temperature-sensitive phenotype and UV sensitivity (A) Temperature-sensitive growth of four SSL2 mutants and wild type cells, all bearing deletions of TFB6. Cells were streaked on YPD plates and incubated at 26 °C or 37 °C. The ssl2-rtt allele carries the single amino acid substitution E556K. The ssl2-dead allele contains mutations V490A and H491D. The ssl2-xp allele has a 3′-truncation of the gene that eliminates 94 C-terminal amino acid residues. The SSL2-1 allele carries the single amino acid substitution W427L. The SSL2-1 allele was isolated as a dominant suppressor of a stem-loop inserted in the 5′-untranslated region of HIS4 (10). The ssl2-rtt, ssl2-dead and SSL2-1 mutations stimulate Ty1 transposition (18). (B) Yeast cultures grown in YPD were diluted and plated onto YPD and incubated at room temperature for 30 min. Uncovered plates were then irradiated with UV light (254 nm) at the indicated doses followed by incubation in the dark for 3 d.
The dissociation of Ssl2 as a complex with Tfb6 raised the question whether Ssl2 would still tend to dissociate in the absence of Tfb6. When TFIIH was purified from a tfb6Δ strain of yeast by affinity and UnoQ chromatography, as described above for the wild type, one major peak of 10-subunit TFIIH was obtained, along with a minor peak corresponding to this complex lacking TFIIK (Fig. 1C). The yield of the 10-subunit complex was about 8 mg from 2.5 kg of yeast, a 20-fold increase over that obtained from wild type (Figs. 1 A and C). The deficiency of Ssl2 in previous preparations of TFIIH (14) may therefore be explained by a propensity to dissociate as a heterodimer with Tfb6.
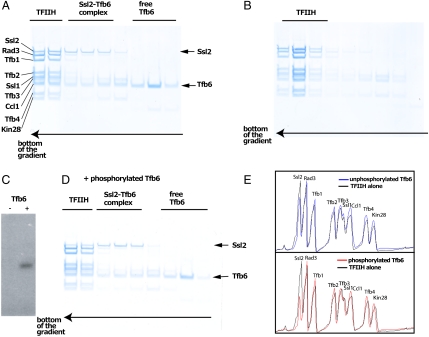
Direct evidence for a destabilizing role of Tfb6 came from adding the protein back to tfb6Δ-TFIIH. Tfb6 expressed and purified from bacteria was added to 10-subunit TFIIH, followed by sedimentation in a glycerol gradient. Some 25% of the Ssl2 appeared in slower sedimenting fractions along with Tfb6 (Fig. 3A), whereas tfb6Δ-TFIIH remained intact in the absence of Tfb6 (Fig. 3B).
Fig. 3.
Tfb6 facilitates the dissociation of Ssl2 from TFIIH (A–B) TFIIH was incubated with (A) or without (B) recombinant Tfb6 and subjected to glycerol gradient sedimentation. The fractions were analyzed by SDS-PAGE. (C) Tfb6 was treated with TFIIK in vitro and analyzed by SDS-PAGE. An autoradiograph of the gel is shown, with a negative control in the left lane. (D) TFIIH was incubated with phosphorylated Tfb6, subjected to glycerol gradient sedimentation, and analyzed by SDS-PAGE as in (A). (E) SDS-PAGE in (A) and (C), scanned and plotted with (Lower) or without (Upper) phosphorylation of Tfb6.
Dissociation of Ssl2 from TFIIH by Tfb6 is Regulated by Phosphorylation.
Mass spectrometry identified four phosphorylation sites (i.e., Thr71, Thr84, Ser104, and Ser105) in Tfb6 from 11-subunit TFIIH (Dataset S2). Thr71 and Ser105 were likely phosphorylated by cyclin-dependent kinases (CDK) because the phosphorylated residues are followed by prolines (TP and SP). None of the four sites was phosphorylated in free Tfb6 purified from a strain of yeast bearing TAP-tagged Tfb6, suggesting a relationship between association of Tfb6 with TFIIH and phosphorylation. To investigate this possibility, bacterially expressed Tfb6 was phosphorylated by TFIIK in vitro (Fig. 3C) and combined with tfb6Δ-TFIIH, followed by sedimentation in a glycerol gradient (Fig. 3D). Approximately 70% of Ssl2 was dissociated from TFIIH by the phosphorylated Tfb6 (Fig. 3D), whereas only about 25% of Ssl2 was dissociated by the unphosphorylated protein (Figs. 3 A and E). Tfb6-TFIIH interaction may be regulated, at least in part, by phosphorylation.
Roles of Tfb6 in Transcription.
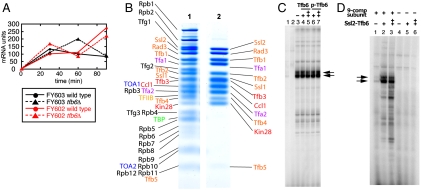
The tfb6Δ strain was indistinguishable from wild type in the kinetics of induction of GAL7 by galactose (Fig. 4A). By contrast, a tfb5Δ strain was previously shown to be defective in the expression of GAL7 (4), demonstrating a requirement for TFIIH at this promoter, so Tfb6 is evidently dispensable for transcription, at least at GAL7. We also investigated the possible involvement of Tfb6 in preinitiation complex assembly and transcription in vitro by reconstitution with RNA polymerase II, tfb6Δ-TFIIH, and the additional required general transcription factors (Fig. 4 B and C). The set of proteins lacking Tfb6, tfb6Δ-TFIIH proved capable of assembly in a complete preinitiation complex (Fig. 4B) and also of the initiation of transcription (Fig. 4C). The addition of Tfb6, even in 15-fold excess over tfb6Δ-TFIIH, had no effect on transcription initiation (Fig. 4C). The lack of effect is noteworthy, suggesting that Tfb6 fails to dissociate Ssl2 from TFIIH in the context of a fully assembled preinitiation complex. More direct support for this idea came from adding the Ssl2-Tfb6 complex (isolated from yeast by affinity purification by means of a TAP-tag on Ssl2) back to the 9-subunit complex lacking Ssl2 and Tfb6 [fractions (a) in Fig. 1A] in transcription assays. Whereas the 9-subunit complex did not support transcription (lane 1 in Fig. 4D), activity was restored by the Ssl2-Tfb6 complex (lanes 2–3 in Fig. 4D).
Fig. 4.
Effects of Tfb6 on transcription (A) GAL7 mRNA levels were quantified by qPCR. Galactose was added to a final concentration of 2% at zero time and the cells were harvested at the times indicated. (B) The reconstituted PIC was subjected to glycerol gradient sedimentation and was analyzed by SDS-PAGE. Lane 1, isolated PIC with 82-bp HIS4 promoter DNA; lane 2, isolated TFIIH with TFIIE. (C) In vitro run-off transcription assay. HIS4 promoter DNA (5 pmol) was mixed with 7.5 pmol of TFIIB, 7.5 pmol of TFIIA, 5 pmol of TBP, 7.5 pmol of TFIIE, 3 pmol of tfb6Δ-TFIIH, and 2 pmol of RNA polymerase II-TFIIF complex. The assay was performed with 15 pmol (lanes 4, 6) or 45 pmol (lanes 5, 7) of unphosphorylated or phosphorylated Tfb6 (p-Tfb6). Lanes 1 and 2 show the negative control of omitting Ssl2 and TBP from transcription, respectively. Arrows indicate the two major promoter-specific transcripts. (D) Reconstituted transcription activity of 9-subunit TFIIH and Ssl2-Tfb6 heterodimer. The transcription assay was performed with (lanes 1–3) or without (lanes 4–6) the 9-subunit TFIIH lacking Ssl2, titrated with 4 pmol (lanes 2, 5) or 8 pmol (lanes 3, 6) of the Ssl2-Tfb6 heterodimer. The assay was performed under the same conditions as in (C).
Discussion
The propensity of Ssl2 to dissociate from yeast TFIIH (14, 17), viewed in the past as no more than an impediment to the isolation of holo-TFIIH, now takes on physiological significance. Our findings suggest a pathway, centered on Ssl2 and Tfb6, connecting transcription initiation with other functions of TFIIH: Tfb6 enters with TFIIH in a transcription preinitiation complex, and Ssl2 melts the promoter DNA to create a transcription bubble; transcription initiation within the bubble is accompanied by action of the TFIIK kinase component of TFIIH upon both Tfb6 and the RNA polymerase II CTD; phosphorylation of Tfb6 provokes the dissociation of an Ssl2-Tfb6 complex, which goes on to perform additional roles.
Our findings have significant technical implications as well. Previously, the isolation of TFIIH gave a low yield of the complete complex, and the purified protein was heterogeneous, due in large part to the dissociation of Ssl2 (14). Only about 10% of the purified protein was in the form of holo-TFIIH, while the remaining 90% lacked Ssl2. We now find that knockout of TFB6 prevents the dissociation of Ssl2, leading to the isolation of complete 10-subunit TFIIH in a yield more than 20-fold greater than from wild type yeast. This technical advance opens the way to definitive biochemical and structural studies, not only of TFIIH, but of the entire RNA polymerase II transcription machinery.
Experimental Procedures
Yeast Methods.
Strains ssl2-rtt (DG1775) and wild type (DG1772) were provided by Drs. T. Tani and D. Garfinkel, respectively (10, 12, 18). To create the tfb6Δ strain, we replaced the gene encoding YOR352w (TFB6) with KanMX using a PCR-mediated strategy in strains CB010 (MATa pep4∷HIS3 prb1∷LEU2 prc∷HISG can1 ade2 trp1 ura3 his3 leu2-3,112 cir-o RPB3∷TAP∷KlTRP1 GAL+ RAF+ SUC+), FY602 (S288c MATa leu2Δ1 ura3-52 trp1Δ63 his3Δ200 lys2-128∂), FY603 (S288c MATa leu2Δ1 ura3-52 trp1Δ63 his3Δ200 lys2-128∂), DG1775 (MATa ura3-167 his3-D200 trp1-hisG leu2-hisG Ty1-270his3-AI Ty1-588neo Ty1-146[tyb1∷lacZ] ssl2∷TRP1 (ssl2-rtt/pRS416)) and DG1772 (MATa ura3-167 his3-D200 trp1-hisG leu2-hisG Ty1-270his3-AI Ty1-588neo Ty1-146[tyb1∷lacZ] SSL2∷TRP1 (SSL2/pRS416)). The replacement of TFB6 was verified by PCR. Tfb3-TAP-tagged and Tfb4-TAP-tagged strains were obtained as described before (19).
Protein Purification.
S. cerevisiae harboring the TAP-tagged Tfb3 or TAP-tagged Tfb4 was grown in 200L or 400L YPAD. The yeast culture was harvested at OD 6.0 and lysed by disruption with glass beads in buffer (400) (50 mM Hepes (pH 7.6), 1 mM EGTA, 5% glycerol, 0.1% 3-(decyldimethyl-ammonio) propanesulfonate (Sigma), with the mM concentration of potassium acetate in parentheses). The cell extract was stirred with 100 mM ammonium sulfate and 0.2 % PEI for 1 h, centrifuged, and the supernatant loaded onto an IgG column. After incubation with TEV protease for 15 h at 4 °C, TFIIH was eluted with buffer (300). The eluate was further purified on UnoQ (Bio-Rad) and/or a glycerol gradient (10–40% v/v). Thus, a nonnative C-terminal calmodulin-binding tag was retained on Tfb3 or Tfb4.
For the expression of recombinant Tfb6, the Escherichia coli Rosetta2 (DE3) strain (Stratagene) was transformed with the pRSFDuet vector (Novagen) harboring the TFB6 gene fused to a sequence encoding a C-terminal His6-tag, grown in 2xYT at 30 °C, and induced with 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) for 3 h. The cells were lysed by sonication, and the soluble fraction was purified by Ni2+-affinity chromatography (GE Healthcare) and gel filtration Superdex 75 (GE Healthcare).
Yeast TFIIF was TAP-tagged on the C-terminus of Tfg2. Cells were grown in 100L YPAD to OD 9.0 and harvested. A whole-cell lysate was generated by bead beating in buffer TEZ (200)(50 mM Tris (pH 7.5), 1 mM EDTA, 10 uM ZnCl2, 0.15% 3-(decyldimethyl-ammonio) propanesulfonate (Sigma), 3 mM DTT, protease inhibitors with the mM concentration of ammonium sulfate in parentheses). The cell extract was stirred with 0.25% PEI overnight, centrifuged, and the supernatant loaded onto an IgG column. TFIIF was washed with TEZ (500) and TEZ (25), and was eluted using TEZ (200) following incubation with TEV protease for 15 h at 4 °C. The eluate was further purified on a Heparin column. The purified TFIIF and yeast Pol II were mixed in a 1.6∶1.0 molar ratio and were dialyzed into 10 mM Tris (pH 7.5), 10 uM ZnCl2, 150 mM potassium acetate, 5% glycerol, and 10 mM DTT, and the Pol II-TFIIF complex was purified on Superose 6.
Full-length Toa1 (residues 1–286) and Toa2 (residues 1–122) subunits of S. cerevisiae TFIIA were subcloned into the pRSFDuet vector (Novagen) and were overexpressed separately in Rosetta 2(DE3) cells (Novagen). TFIIA was reconstituted with refolded Toa1 and Toa2 essentially as described (20). Full-length S. cerevisiae TBP (residues 1–240) was subcloned into pRSFDuet vector and overexpressed in Rosetta 2(DE3) cells. TBP was purified by Heparin and SP-Sepharose chromatography. His6-tagged full-length S. cerevisiae TFIIB (residues 1–345) was purified as described (21). Full-length Tfa1 (residues 1–482) and Tfa2 (residues 1–328) subunits of S. cerevisiae TFIIE were subcloned into pRSFDuet and coexpressed in Rosetta 2(DE3) cells. The cells were lysed by sonication, and the soluble fraction was purified using Ni2+-affinity chromatography, Mono-Q chromatography, and gel filtration through Superdex 75.
Binding Assay.
Tfb6 (0.8 nmol) and TFIIH (0.2 nmol) were incubated in 300 mM potassium acetate, 20 mM Hepes (pH 7.6), 2 mM magnesium acetate, 1 mM EDTA, 5 mM DTT and 5%(v/v) glycerol at 4 °C. The protein mixture was loaded onto a 10–40%(v/v) glycerol gradient and centrifuged for 6 h at 60,000 rpm (Beckman SW60 rotor). The fractions were analyzed by SDS-PAGE and staining with Coomassie blue.
Phosphorylation of Tfb6.
For radioactive labeling of Tfb6, 1 ug/ml of Tfb6 was treated with 1 ug/ml TFIIK in 20 mM Hepes (pH 7.6), 7.5 mM magnesium acetate, and 100 mM potassium acetate, 10 mM DTT, 5% glycerol, 0.1% 3-(decyldimethylammonio) propanesulfonate (Sigma), 100 μM ATP, and 0.033 μCi/μL of [γ-32P] ATP. Reactions were stopped by adding EDTA and analyzed by SDS-PAGE.
PIC Assembly.
To obtain HIS4 promoter DNA (-92 to -11), the complementary oligonucleotides were synthesized (Integrated DNA Technologies) and annealed. 0.5 nmol of the promoter DNA was mixed with 0.75 nmol of TFIIB, 0.75 nmol of TFIIA, 0.5 nmol of TBP, 0.7 nmol of TFIIE, 0.3 nmol of TFIIH in a 40 μL of buffer (500) (20 mM Hepes (pH 7.6), 5 mM DTT, 2 mM magnesium, and 5% glycerol, with the mM concentration of potassium acetate in parentheses). The mixture was dialyzed into buffer (300), buffer (220), buffer (150), and then combined with 0.25 nmol of RNA polymerase II-TFIIF complex. The mixture was further dialyzed into buffer (100) and buffer (80), and was loaded onto a 10–40% (v/v) glycerol gradient containing 20 mM Hepes (pH 7.6), 5 mM DTT, 2 mM magnesium acetate, 80 mM potassium acetate and centrifuged for 9 h at 40,000 rpm (Beckman SW60 rotor).
In Vitro Transcription Assay.
To obtain the dsDNA template, HIS4 promoter (-104 to +98) was amplified with EcoRV site at both ends and was cloned into pDrive (Qiagen). XL10-gold strain (Stratagene) harboring the plasmid was grown and the plasmid was isolated using Plasmid Maxiprep (Qiagen). The plasmid was treated with EcoRV and the linear template was isolated on a 2% agarose gel. Thus, nonnative ATC and GAT were retained at 5′ and 3′ ends, respectively. Five pmol of dsDNA template was mixed with 7.5 pmol of TFIIB, 7.5 pmol of TFIIA, 5 pmol of TBP, 7.5 pmol of TFIIE, 3 pmol of TFIIH, and 2 pmol of RNA polymerase II-TFIIF complex with or without Tfb6 in a 7 μL of buffer A (50 mM Hepes (pH 7.6), 300 mM potassium acetate, 5 mM DTT, and 5% glycerol). Thirteen μL of buffer B [50 mM Hepes (pH 7.6), 2 mM magnesium sulfate, 30 mM potassium acetate, and 10 mM DTT] was added and then incubated for 15 min at room temperature. Transcription was initiated by adding 20 μL of buffer B containing 1.6 mM ATP, 1.6 mM GTP, 1.6 mM CTP, 40 μM UTP, 10 mM Mg-acetate, 1 unit of RNase out, 0.5 μCi of [α-32P] UTP, and stopped after 45 min by adding 185 μL of stop buffer [10 mM Tris, 300 mM sodium acetate (pH 5.5), 5 mM EDTA, 0.7% SDS, 0.1 mg/mL glycogen, 0.013 mg/ml of proteinase K]. Transcripts were recovered and analyzed as described (22) with minor modifications.
RNA Analysis.
For the galactose induction time course experiment, strains FY602 and FY603 were grown in YPR (1% yeast extract, 2% peptone, 2% raffinose) at 30 °C to an OD600 of 0.5. Galactose was added to a final concentration of 2% and the cells were harvested at the indicated times. Total RNA was isolated with RNeasy (Qiagen). The levels of mRNA were quantified relative to ACT1 by RT-qPCR.
Mass Spectrometry.
TFIIH peptides were analyzed by nanoLC MS on a LCQ Deca XP+ ion trap (Thermo Fisher Scientific). The LC was an Eksigent nano2D with an in-house packed reversed phase C18 column (150 μM ID) and the electrospray interface was a Nanomate (Advion). The peptides were identified by searching the MS/MS spectra against a yeast protein database using SEQUEST on a Socerer platform (SageN). The static modification of propionamide (Cys) and variable modifications of oxidation (Met) were used.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Drs D. Garfinkel and T. Tani for the gift of yeast strains, and colleagues for critical reading of the manuscript. This research was supported by NIH Grants GM36659 and AI21144 to R.D.K. K.M. was supported by the JSPS, the Kanae Foundation, and the Uehara Memorial Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201448109/-/DCSupplemental.
References
- 1.Conaway RC, Conaway JW. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 2.Svejstrup JQ, et al. Different forms of TFIIH for transcription and DNA repair: Holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 3.Chang WH, Kornberg RD. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell. 2000;102:609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 4.Ranish JA, et al. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet. 2004;36:707–713. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- 5.Giglia-Mari G, et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 6.Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 7.Svejstrup JQ, Feaver WJ, Kornberg RD. Subunits of yeast RNA polymerase II transcription factor TFIIH encoded by the CCL1 gene. J Biol Chem. 1996;271:643–645. doi: 10.1074/jbc.271.2.643. [DOI] [PubMed] [Google Scholar]
- 8.Takagi Y, et al. Revised subunit structure of yeast transcription factor IIH (TFIIH) and reconciliation with human TFIIH. J Biol Chem. 2003;278:43897–43900. doi: 10.1074/jbc.C300417200. [DOI] [PubMed] [Google Scholar]
- 9.Roy R, et al. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 10.Gulyas KD, Donahue TF. SSL2, a suppressor of a stem-loop mutation in the HIS4 leader encodes the yeast homolog of human ERCC-3. Cell. 1992;69:1031–1042. doi: 10.1016/0092-8674(92)90621-i. [DOI] [PubMed] [Google Scholar]
- 11.Estruch F, Cole CN. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell. 2003;14:1664–1676. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuki F, et al. Participation of XPB/Ptr8p, a component of TFIIH, in nucleocytoplasmic transport of mRNA in fission yeast. Genes Cells. 2007;12:35–47. doi: 10.1111/j.1365-2443.2006.01032.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons BJ, et al. Subunit architecture of general transcription factor TFIIH. Proc Natl Acad Sci USA. 2012;109:1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feaver WJ, et al. Dual roles of a multiprotein complex from S.cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 15.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 16.Brooks MA, et al. Systematic bioinformatics and experimental validation of yeast complexes reduces the rate of attrition during structural investigations. Structure. 2010;18:1075–1082. doi: 10.1016/j.str.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Svejstrup JQ, Feaver WJ, LaPointe J, Kornberg RD. RNA polymerase transcription factor IIH holoenzyme from yeast. J Biol Chem. 1994;269:28044–28048. [PubMed] [Google Scholar]
- 18.Lee BS, et al. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics. 1998;148:1743–1761. doi: 10.1093/genetics/148.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borggrefe T, Davis R, Bareket-Samish A, Kornberg RD. Quantitation of the RNA polymerase II transcription machinery in yeast. J Biol Chem. 2001;276:47150–47153. doi: 10.1074/jbc.M109581200. [DOI] [PubMed] [Google Scholar]
- 20.Ranish JA, Lane WS, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2009;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayre MH, Tschochner H, Kornberg RD. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J Biol Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.