Abstract
Important insights into aging have been generated with the genetically tractable and short-lived budding yeast. However, it is still impossible today to continuously track cells by high-resolution microscopic imaging (e.g., fluorescent imaging) throughout their entire lifespan. Instead, the field still needs to rely on a 50-y-old laborious and time-consuming method to assess the lifespan of yeast cells and to isolate differentially aged cells for microscopic snapshots via manual dissection of daughter cells from the larger mother cell. Here, we are unique in achieving continuous and high-resolution microscopic imaging of the entire replicative lifespan of single yeast cells. Our microfluidic dissection platform features an optically prealigned single focal plane and an integrated array of soft elastomer-based micropads, used together to allow for trapping of mother cells, removal of daughter cells, monitoring gradual changes in aging, and unprecedented microscopic imaging of the whole aging process. Using the platform, we found remarkable age-associated changes in phenotypes (e.g., that cells can show strikingly differential cell and vacuole morphologies at the moment of their deaths), indicating substantial heterogeneity in cell aging and death. We envision the microfluidic dissection platform to become a major tool in aging research.
Keywords: lifespan analysis, live-cell imaging, replicative aging, Saccharomyces cerevisiae
Aging is a complex gradual impairment of normal biological function caused by accumulation of molecular damage, finally culminating in death. Investigation of the genetically tractable and short-lived budding yeast Saccharomyces cerevisiae has yielded important insights into general eukaryotic aging: specific genes mediate aging (e.g. SIR2) (1–3) and dietary limitations can increase lifespan (4). S. cerevisiae's replicative aging is considered an important model for aging in mitotically active cells (5, 6), with the replicative lifespan being defined as the number of daughter cells produced by a mother cell before the mother cell ceases dividing (Fig. 1A). Because of asymmetrical inheritance of damage to the mother (7), senescence factors are thought to accumulate in mother cells (8). Comprehensive analyses of age-associated phenotypes are considered to be instrumental in identifying the senescence factors (9, 10).
Fig. 1.
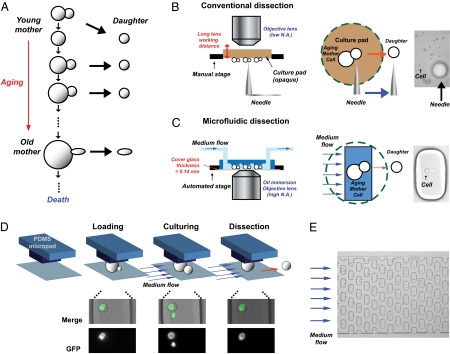
Unique microfluidic method for monitoring the aging process of budding yeast. (A) Schematic illustration of replicative aging. The number of produced buds (daughters) represents the replicative age of the mother cell. Aged cells often increase in size and produce ellipsoidal daughters. (B) Schematic illustration of conventional dissection. A yeast daughter cell is manually removed by a dissection needle (indicated by arrow: typically approximately 10-times larger than cell) from the mother cell. The cells grow on a thick and opaque agar pad, which limits high-resolution microscopic imaging because of the required long-working distance of the objective lens. (C) Schematic illustration of microfluidic dissection. Mother yeast cells are held between a soft PDMS pad and thin cover glass (4 μm distance). This process allows high-resolution fluorescent imaging because high numerical aperture (high N.A.) objectives can be used. Daughter cells are continuously removed by flow of fresh media. (D) Principle of yeast cultivation and microfluidic dissection. Cell loading: Yeast cells are loaded under PDMS micropads. The elastic pads are slightly lifted by the hydrostatic pressure of the cell suspension during loading and hold the cells after a release of the pressure. Cell culturing: Fresh media is continuously provided through the array of micropads. The image shows the vacuole (green) during cell division. Dissection: The media flow (blue arrows) washes daughter cells from their mother cells because of the fact that the daughters’ cell size is smaller than that of the mother cells. Cells expressing Vph1p-GFP served for visualizing the vacuoles. (E) Array of micropads. The chip contains 200 micropads arranged in an array format.
Analyzing the phenotype of replicative aging yeast cells, however, harbors a major technical challenge: continuous budding of the cells causes the original mother cells to be rapidly outnumbered by the exponentially increasing number of daughter cells, and thus makes long-term tracking of the aging cell impossible (Fig. S1). Technologies for studying age-associated phenotypes of replicative aging yeast cells are still very limited (e.g., ref. 11), and until today the prime tool in yeast aging research has been a 50-y-old dissection method (12), in which daughter cells are removed by microscopic micromanipulation with a needle from the larger mother cell on thick opaque culture pads (10, 13) (Fig. 1B). In a laborious and time-consuming manner—one lifespan experiment requiring several days of manual work for removing daughter cells after each mitotic cycle—such dissection allows assessment of the cell lifespan (10) or the isolation of single cells to generate microscopic snapshots of differentially aged cells. This capability has, for example, led to the finding that the morphology of cells changes with age (9, 10, 12). However, because of several constraints, the conventional dissection method does not allow for high-resolution microscopic imaging (e.g., fluorescent imaging), and not for continuous tracking of cells throughout their complete lifespan, which would afford dynamic and essential insights into the phenotype of aging cells.
Unprecedented insights into single cells during aging would be possible with a method that allows for continuous high-resolution microscopic imaging of whole lifespans of yeast cells from their youth through senescence to death. The development of microfluidic devices has raised expectations for their capability to cultivate yeast cells in controlled environments with continuous microscopic observation (14, 15). Unfortunately, none of the currently existing microfluidic devices can be applied for long-term replicative aging studies. Respective devices are limited either by the number of generations over which mother cells can be monitored (typically eight) because of the exponential increase in the number of daughter cells retained in the microfluidic observation chambers (16–18), or by nonideal optical properties imposed by the chip design (19).
In this work, we set out to solve this problem and developed a microfluidic dissection platform with a prealigned single focal plane for long-term live-cell imaging of the complete replicative (and also chronological) lifespan of budding yeast cells. Similar to the classic dissection method, our platform also draws on the fact that the mother is larger than the bud cell (Fig. 1C). Yeast mother cells are trapped under soft elastomer (polydimethylsiloxane, PDMS)-micropad. A continuous medium flowing through the device washes away emerging buds and at the same time ensures a defined and constant environment during the whole aging experiment. This soft elastomer-based “microfluidic dissection platform” allows monitoring of the aging process of single cells from “young” to “death.” In addition to offering the capability of performing lifespan analyses in a less laborious manner, the excellent optical properties of the chip permit in vivo fluorescence measurements during the entire lifespan.
Results and Discussion
Yeast mother cells are trapped under micropads (30 μm × 15 μm) (Fig. 1 C and D) that are arranged in an array-based format (Fig. 1E). The engineered height between the micropad and the glass cover-slide is similar to the diameter of yeast cells (i.e., 4–5 μm), realizing a prealigned single focal plane for long-term live-cell imaging (Fig. 1C). Hydrostatic pressure, applied during the loading of the chip with cells (Fig. S2), lifts up the ∼200 elastic PDMS micropads and allows the cells to pass beneath them. After the release of this pressure, cells are trapped underneath the pads (Fig. 1D, loading) without any detrimental effects on the cells (Fig. S3). After cell loading (described in detail in Fig. S2), a continuous medium flows through the device and (i) washes away emerging buds, which—because of their smaller size—are not held under the micropad (Fig. 1D: culturing, dissection) and (ii) ensures a defined and constant environment during the whole aging experiment. A stable and long-term operation of the chip (> 5 d) is ensured by the overall chip's channel layout.
With this technology, it is possible to monitor, in a fully automated manner without intervention of the experimenter, ∼50 single cells from birth to death (i.e., up to about 60 generations) in a single experiment (Movie S1), despite the fact that occasionally bud cells push neighboring mother cells away from the micropad. This number of cells is comparable with the number of cells observed in conventional lifespan analysis (13). Fig. S4 shows the dynamics of the cell-retention capacity of the chip. We found that the division time of single cells obtained with our device agrees well with findings in another, independent study (20) (71 ± 0.8 min; n = 206 for first to third divisions of wild-type cells at 30 °C in Synthetic Defined Media with the full amino acid complement), indicating that our setup allows for the generation of physiologically correct data. At this point, it is important to note that with our setup starting the experiment with newborn cells is not guaranteed, although considering the typical bud index distribution of an exponentially growing liquid culture (i.e., 80% of the cells have never budded before, 12% once, 6% twice, 3% three times) (21), the majority of loaded cells are still new or recently born cells.
We first asked whether we could use the device to generate classic lifespan data in a simpler and more automated way than with the conventional microdissection method. Therefore, we loaded wild-type cells and recorded the division events and time of individual cells until all cells had entered senescence (after approximately 50–100 h). Furthermore, we used a SIR2 and a FOB1 deletion strain, which are known to show a decreased (2) and increased replicative lifespan, respectively (7, 11, 22). In Fig. 2A, the viability of these strains is shown as a function of the number of buds produced. Consistent with data from conventional dissection analysis (2, 7, 22), the lifespan of the SIR2 deletion mutant is significantly shorter than that of the wild-type strain (medianWT: 21 divisions vs. mediansir2Δ: 11 divisions) and the lifespan of the FOB1 deletion mutant is longer (medianfob1Δ: 32 divisions). A side-by-side comparison of the lifespans obtained through the classic dissection method with the ones generated through our microfluidic section platform (when using identical preculture and culture medium conditions) demonstrates that our experimental system generates very comparable lifespan data (Fig. S5). Moreover, at the same time we overcome other limitations of the classic dissection method: (i) our method effaces the limitations of the agar gel pad required for cell growth during classic dissection and the long-working distance objective lens that severely limits the optical resolution with which cells can be imaged (Fig. 1B); and (ii) it negates the issue of local nutrient depletion and by-product production during the experiment, as we can now realize absolutely constant nutritional conditions (e.g., calorie restriction or noncalorie restriction) throughout the whole experiment through a constant supply of fresh medium.
Fig. 2.
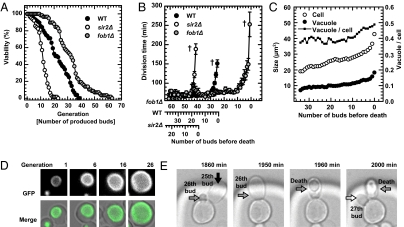
Whole lifespan monitoring of cells. (A) Replicative lifespan analysis with the described platform. Cell viabilities are plotted as a function of number of buds produced (replicate lifespan). The lifespan of the SIR2 gene deletion mutant (sir2Δ, n = 63) is shorter than that of the wild-type (WT, n = 76) and, that of the FOB1 gene deletion mutant (fob1Δ, n = 58) is longer. (B) Division time of WT, sir2Δ, and fob1Δ. The plot shows the progression of the division time of WT, sir2Δ, and fob1Δ when the traces of the individual cells are normalized to the last division. (“†” represents death, error bar: mean ± SEM.) (C) Quantification of vacuole and cell size increase. Near the end of the replicative lifespan the sizes and the ratio of vacuole and cell increase. (D) Visual impression of increase in vacuole and cell size. (Upper) Bright-field images of a tracked cell at the 1st, 6th, 16th, and 26th generation. (Lower) The corresponding vacuole structures with fluorescent imaging using cells expressing Vph1p-GFP. (E) Daughter cells from old mothers can die earlier than the mothers. Black, gray and white arrows indicate the 25th, 26th, and 27th buds from the mother, respectively. The provided times indicate the durations of the cultivation.
In addition to the acquisition of lifespan data, the dynamic progression of the division times of single cells can easily be assessed with our setup. Consistent with earlier reports, we noted that the division time and deviation increased dramatically as the cells approached the end of their lives (12) (100 ± 4.1, 122 ± 6.4, and 151.2 ± 9.5 for wild-type cells at three, two, and one buds before death, respectively) (Fig. 2B). With our platform, it is possible to measure division time very precisely, which has been difficult with the classic dissection method: manual dissection requires a few minutes per cell, and in single experiments, ∼40 cells need to be observed and dissected continuously.
The unprecedented feature of our platform is the possibility for continuous high-resolution imaging of individual yeast cells from birth to death (Movie S1). We exploited this capability to generate novel data on gradual changes in cellular and vacuolar morphology during the complete lifespan of individual cells; the latter was visualized by a GFP-tagged version of VPH1 (a subunit of the vacuolar-ATPase) (Fig. 2 C and D). Consistent with earlier observations made at distinct time points (23), we observed that the surface of the cells becomes more wrinkled with age (12, 24) (Movie S1). In addition, both the cell and vacuole sizes gradually become larger as cells age, with an increase in both sizes during the last three divisions (Fig. 2 C and D), a finding that parallels the earlier observed increase in division time (Fig. 2B). Of interest, the ratio between vacuole size and cell size increases toward the end of the lifespan, potentially indicating higher autophagy activity (25). Exploiting the fact that in some cases bud cells are not immediately removed from the pads, we found that the mother cell typically produced ellipsoidal daughter cells even before the mother's own shape became ellipsoidal (e.g., 25th and 26th buds in Fig. 2E and Movie S1). Of note, these daughter cells died even earlier than the mother (Fig. 2E). Intriguingly, daughter cells of old mothers were sometimes found to be larger than the respective mother cell (Fig. S6 and Movie S2).
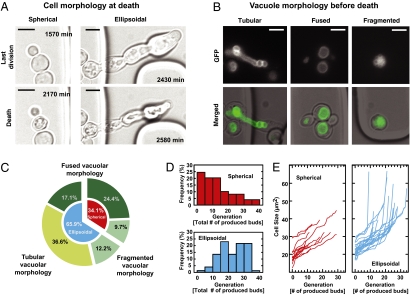
Because we could now track cells during their complete replicative lifespan with continuously recording phenotypic traits, we wanted to explore whether there is heterogeneity in the aging process and death pattern, an insight that remains elusive in studies of yeast cell aging at the population level (11). First, we found two cell morphologies directly before a cell's death (Fig. 3A, Lower), a spherical shape (34.1%) or an ellipsoidal outline (pseudohyphae, 65.9%). Next, we focused on the vacuole morphology and identified three typical vacuole morphologies right before cell death: a tubular shape, a fused appearance, or a fragmented structure (Fig. 3B). Often, completely ruptured vacuoles were observed at the end of cells' life (Movies S1, S3, and S4). As intuitionally expected, the tubular structure was observed only with the ellipsoidal death pattern (Fig. 3 B and C). This type of cell death points to problems in cytokinesis (26, 27), which may also cause failure of vacuole segregation. Usually, after a normal cell division, vacuole structures fuse and form one vacuole (Fig. 3B, Center). However, in about 22% of all cases (Fig. 3C), this fusion fails or is not completed and fragmented vacuolar structures are observed at cell death (Fig. 3B, Right).
Fig. 3.
Age-associated phenotypes. (A) Example images of cell morphology at the last division (Upper) and death (Lower). Cells either show a spherical shape (spherical) or form pseudohyphae (ellipsoidal). (B) Example images of vacuole morphology before death. Cells expressing Vph1p-GFP served for visualizing the vacuolar structure. Vacuoles form either a tubular-like, fused, or fragmented structure. (Scale bars, 5 μm.) (C) Distribution of age-associated phenotypes categorized by cell and vacuole morphologies at the last moment before death. Outer ring, vacuole morphology; inner ring, cell morphology. (D) Age-distribution at the two types of deaths. (Upper) Spherical death pattern (n = 49, median = 12); (Lower) Ellipsoidal death pattern (n = 74, median =23). (E) Dynamic change of the cell size in the two populations. (Left) Cells that show spherical death pattern; (Right) cells that show ellipsoidal death pattern.
With this heterogeneity in morphologies identified, we next asked whether the death types are correlated with any other measures that we obtained. Indeed, we found that cells dying with the spherical death type (Movie S5) die, on average, at a younger age (after 12 buds generated) compared with cells dying the ellipsoidal death (after 23 buds generated) (Fig. 3D), and do so after a sudden increase in their cell sizes (Fig. 3E). This finding implies that the spherical cell death occurs rather at the mid age, suddenly “striking” younger cells, but the ellipsoidal death occurs at rather an older age. This finding indicates that there is indeed heterogeneity in the aging process, resulting in fundamentally different culminations of damage that is ultimately incompatible with life, a fact that underlines the importance of the developed platform for future yeast aging research.
Conclusion
In this article we present a microfluidic dissection platform that allows us to track single yeast cells over their entire replicative (and chronological) lifespans with high-resolution imaging capability, a possibility the yeast-aging research community has long awaited. Not only can this technology replace the tedious manual microdissection methods to determine lifespan data, but it can also be used for high-resolution in vivo fluorescence imaging of aging cells under exactly controlled environmental conditions. This technology thus opens up unique possibilities for future aging research: Its capability for phenotypic tracing is essential (i) to explore the relevance of cell-to-cell heterogeneity in the aging process, and (ii) to address important questions, such as whether certain phenotypes in cellular youth affect old-age behavior, or how damage is asymmetrically inherited by the mother cell and removed from the daughter cell, and how this changes with age. We envision that our technology will enable novel investigations in the quest for molecular mechanisms underlying the aging process and will permit large-scale screens into the aging phenotype for its capability to be easily multiplexed and to allow aging experiments in an unsupervised manner.
Methods
Yeast Strain.
All yeast strains used in this work were in the S288C (BY) background. The SIR2 and FOB1 deletion strains were obtained from the yeast genome deletion collection, and the relevant genotype of the strain for visualization of the vacuole and Msn2 was VMA5-RFP::KANR, VPH1-GFP::HIS3 and HOG1-mCherry:HIS3, and HTA2-CFP with pRS315 MSN2-GFP plasmid, respectively. Yeast cells were prepared in Synthetic Defined Media with 2% glucose or Yeast Peptone Dextrose medium (YPD). Yeast cell cultures were grown overnight to stationary phase and then allowed to resume exponential growth by dilution into fresh growth medium (OD600 = 0.3), and incubation for 3 h at 30 °C before the experiment. The cells were mildly sonicated for 1 min to separate any cell aggregates that could block the microfluidic device. The cells were then introduced into the microfluidic device.
Microfluidic Device.
The microfluidic device is made of PDMS (Sylgard 184; Dow Corning) and produced by replica molding from an SU-8 wafer. The device consists of a main channel (height = 15 μm), which contains an array of micropads with a distance of 4 μm between the PDMs and glass slide, and a side channel (height = 100 μm) as a cell outlet during the loading process (Fig. S2). The microstructures were fabricated by photolithography using negative photoresists (SU-8 2002 and SU-8 10; Microchem). The side channel mold was made by adhesive tape. In a 1:10 wt/wt ratio, the PDMS base and curing agent were mixed, thoroughly stirred, and degassed in a vacuum chamber for 0.5–1 h to remove air bubbles. The degassed mixture was poured onto the SU-8 master mold and cured on a hot plate (65 °C for 1 h and 130 °C for 30 min). The cured PDMS was carefully peeled off the mold. Inlet and outlet connections per holes were punched using a blunt injection needle. Finally, the surfaces of a cover glass and the PDMS mold were subjected to UV irradiation (UV Ozone cleaner PSD-UVT; Novascan) for 6 min to activate the surface for covalent bonding. The mask design required to produce the devices will be made available upon request.
Live-Cell Imaging and Analysis.
The microfluidic device was mounted on the stage of an inverted microscope (Eclipse Ti; Nikon Instruments) equipped with an incubator for cultivation at 30 °C. Yeast cell growth and individual budding events were monitored by time-lapse imaging, successively capturing images every 10 min. The budding events are determined by comparison of two adjacent images, and the moment of cell death was identified by a sudden shrinkage of the cell's body. The hardware-based focusing system (Nikon Instruments; Perfect Focus System) automatically and stably maintained the focus during the whole experiment. LED (pE2; CoolLed) illumination was used as the fluorescent light source. The images were taken with a high numerical aperture (N.A.) oil immersion objective lens (CFI Plan Apo 60×; Nikon; N.A. = 1.4; working distance = 0.13 mm). The exposure times were 1 ms and 50 ms for transmission and GFP images, respectively. Defocused as well as focused transmission images were taken for subsequent segmentation of cells (28). The images were converted into binary images by manual application of the threshold function to ensure proper cell segmentation. The size of segmented cells was measured by the image process function in ImageJ (Wayne Rasband, National Institutes of Health, Bethesda, MD).
Supplementary Material
Acknowledgments
The authors thank Matthias Peter and Uwe Sauer for generously allowing use of the microscope; Dino Di Carlo and Ralf Streichan for helpful discussions; Andreas Hierlemann for generously allowing use of the clean room facility; Reinhard Dechant, Alessandro Baldi, Annina Denoth, and Serge Pelet for helpful discussions and donation of the yeast strain; and Ida van der Klei, Liesbeth Veenhoff, and Yves Barral for excellent comments on the manuscript. This work was supported by an Interdisciplinary Pilot Project and the YeastX project within the Swiss Initiative in Systems Biology (SystemsX.ch), as well as by the NWO-funded Groningen Systems Biology Center for Energy Metabolism and Aging.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113505109/-/DCSupplemental.
References
- 1.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rine J. Cell biology. Twists in the tale of the aging yeast. Science. 2005;310:1124–1125. doi: 10.1126/science.1121310. [DOI] [PubMed] [Google Scholar]
- 4.Fontana L, Partridge L, Longo VD. Extending healthy life span—From yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gershon H, Gershon D. The budding yeast, Saccharomyces cerevisiae, as a model for aging research: a critical review. Mech Ageing Dev. 2000;120:1–22. doi: 10.1016/s0047-6374(00)00182-2. [DOI] [PubMed] [Google Scholar]
- 6.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 8.Henderson KA, Gottschling DE. A mother's sacrifice: What is she keeping for herself? Curr Opin Cell Biol. 2008;20:723–728. doi: 10.1016/j.ceb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell CD, Van Zandycke SM, Quain DE, Smart KA. Replicative ageing and senescence in Saccharomyces cerevisiae and the impact on brewing fermentations. Microbiology. 2000;146:1023–1034. doi: 10.1099/00221287-146-5-1023. [DOI] [PubMed] [Google Scholar]
- 10.Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009;28:1209. doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindstrom DL, Gottschling DE. The mother enrichment program: A genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 13.Medvedik O, Sinclair DA. Caloric Restriction and Life Span Determination of Yeast Cells. Biological Aging: Methods and Protocols. Vol. 371. Springer; 2007. pp. 97–109. [DOI] [PubMed] [Google Scholar]
- 14.Lee PJ, Helman NC, Lim WA, Hung PJ. A microfluidic system for dynamic yeast cell imaging. Biotechniques. 2008;44:91–95. doi: 10.2144/000112673. [DOI] [PubMed] [Google Scholar]
- 15.Taylor RJ, et al. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc Natl Acad Sci USA. 2009;106:3758–3763. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cookson S, Ostroff N, Pang WL, Volfson D, Hasty J. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol Syst Biol. 2005;1:2005.0024. doi: 10.1038/msb4100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falconnet D, et al. High-throughput tracking of single yeast cells in a microfluidic imaging matrix. Lab Chip. 2011;11:466–473. doi: 10.1039/c0lc00228c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowat AC, Bird JC, Agresti JJ, Rando OJ, Weitz DA. Tracking lineages of single cells in lines using a microfluidic device. Proc Natl Acad Sci USA. 2009;106:18149–18154. doi: 10.1073/pnas.0903163106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryley J, Pereira-Smith OM. Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae. Yeast. 2006;23:1065–1073. doi: 10.1002/yea.1412. [DOI] [PubMed] [Google Scholar]
- 20.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanoni M, Vai M, Popolo L, Alberghina L. Structural heterogeneity in populations of the budding yeast Saccharomyces cerevisiae. J Bacteriol. 1983;156:1282–1291. doi: 10.1128/jb.156.3.1282-1291.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Defossez PA, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 23.Egilmez NK, Chen JB, Jazwinski SM. Preparation and partial characterization of old yeast cells. J Gerontol. 1990;45:B9–B17. doi: 10.1093/geronj/45.1.b9. [DOI] [PubMed] [Google Scholar]
- 24.Müller I. Experiments on ageing in single cells of Saccharomyces cerevisiae. Arch Mikrobiol. 1971;77:20–25. doi: 10.1007/BF00407985. [DOI] [PubMed] [Google Scholar]
- 25.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 26.Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon A, et al. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods. 2007;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.