Abstract
Crimean–Congo hemorrhagic fever virus (CCHFV), a virus with high mortality in humans, is a member of the genus Nairovirus in the family Bunyaviridae, and is a causative agent of severe hemorrhagic fever (HF). It is classified as a biosafety level 4 pathogen and a potential bioterrorism agent due to its aerosol infectivity and its ability to cause HF outbreaks with high case fatality (∼30%). However, little is known about the structural features and function of nucleoproteins (NPs) in the Bunyaviridae, especially in CCHFV. Here we report a 2.3-Å resolution crystal structure of the CCHFV nucleoprotein. The protein has a racket-shaped overall structure with distinct “head” and “stalk” domains and differs significantly with NPs reported so far from other negative-sense single-stranded RNA viruses. Furthermore, CCHFV NP shows a distinct metal-dependent DNA-specific endonuclease activity. Single residue mutations in the predicted active site resulted in a significant reduction in the observed endonuclease activity. Our results present a new folding mechanism and function for a negative-strand RNA virus nucleoprotein, extend our structural insight into bunyavirus NPs, and provide a potential target for antiviral drug development to treat CCHFV infection.
Keywords: structure biology, virology
Crimean–Congo hemorrhagic fever virus (CCHFV) is a tick-borne virus and the causative agent of Crimean–Congo hemorrhagic fever (CCHF), a severe human disease that occurs in over 30 countries in Asia, the Middle East, Southeastern Europe, and Africa (1, 2) and results in high mortality. There is currently no vaccine against CCHF and available therapeutic interventions are limited. The high pathogenicity of CCHFV has led to it being considered as a severe public health threat and a potential bioterrorism agent in the wider world. It is therefore classified as a World Health Organization biosafety level 4 pathogen, limiting fundamental investigations in the laboratory (2).
CCHFV is a negative-sense single-stranded RNA [(−)ssRNA] virus with a three-segmented genome and belongs to the genus Nairovirus within the family Bunyaviridae (2). The Bunyaviridae is the largest negative-sense viral family and comprises more than 350 species, including many significant human pathogens, such as Rift Valley fever, Crimean–Congo hemorrhagic fever, hanta, and sandfly fever viruses. The bunyavirus genomes consist of small (S), medium (M), and large (L) RNA segments, which encode a viral nucleocapsid protein (NP), glycoprotein precursor, and polymerase proteins, respectively (3). According to previous studies on the function of nucleoproteins in bunyaviruses, some of these NPs may be able to recognize specific viral RNA sequences (4–6), but mostly bind to single-stranded RNA (ssRNA) in a nonspecific way (7, 8). Because the Bunyaviridae family includes hundreds of different genuses and the nucleoproteins of each genus show little homology or other features in common, the addressing of the exact function and mechanism of each group of nucleoproteins case by case is necessary for our understanding on bunyaviruses replication and assembly. To our knowledge, the only NP structure reported to date in the Bunyaviridae family is the Rift Valley fever virus nucleoprotein (9, 10), which shows weak binding affinity with RNA and displays a conformational change before oligomerization into a ribonucleoprotein (RNP) complex (9). Previous structures of NPs from other families, such as the influenza virus (Orthomyxovidae) (11, 12), rabies virus (Rhabdoviridae) (13), vesicular stomatitis virus (VSV) (Rhabdoviridae) (14), and borna disease virus (BDV) (Bornaviridae) (15) have shown how NPs assemble with RNA to form RNPs. Interestingly, recent studies by two research groups on the structure of the Lassa fever virus (LASV) (Arenaviridae) NP have added to our understanding of the functions of virally encoded NPs beyond their role in the packaging of viral genomic RNA (16–18). Both groups concur that the LASV C-terminal domain possesses 3′–5′ RNA-specific exoribonuclease activity, whereas they hold different opinions on the function of the N-terminal domain. Qi et al. describe a LASV NP(1-569)-dTTP complex structure and propose that the LASV NP is responsible for the RNA cap-snatching mechanism that initiates its transcription (18). In contrast, Hastie and colleagues report the structure of a LASV(1-340)-ssRNA complex showing that the N-terminal domain possesses an RNA-binding crevice for the viral genome (17). Bearing in mind the potential relationship between the two negative single-strand RNA viruses CCHFV and LASV, and our lack of knowledge of the biological role and precise mechanisms of NPs in the bunyaviruses, we conducted a structural and biochemical analysis of CCHFV NP.
Results
CCHFV NP Exists as a Monomer in Vitro.
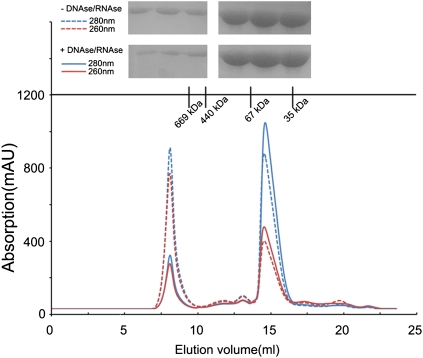
The full-length CCHFV NP protein (strain YL04057) was expressed and purified as a recombinant protein in Escherichia coli. As oligomerization of NPs has been observed in several (−)ssRNA viruses (9, 11, 12, 19), we first determined the oligomeric state of CCHFV NP. Gel filtration and SDS−PAGE showed that CCHFV NP mainly exists in a monomeric form with DNase/RNase treatment (Fig. 1), which differs from other reported (−)ssRNA viruses except for Rift Valley fever virus (RFV) NP in its compact conformation (10).
Fig. 1.
Size exclusion chromatography (SEC) of CCHFV NP. CCHFV NP samples with or without DNase/RNase treatment (5 mg/mL) were injected, respectively, onto a Superdex 200 10/300 GL column. Retention volume was about 15 mL for the monomeric form of CCHFV NP. (Upper) Retention volumes are shown for the molecular weight standards and SDS/PAGE analysis of the SEC elution fractions corresponding to the two peaks.
Overall Structure of CCHFV NP.
The crystal structure of monomeric CCHFV NP (residues 1–482) was determined using the single-wavelength anomalous dispersion (SAD) method and refined to 2.3 Å resolution with a final Rwork value of 22.4% (Rfree = 25.7%) (Table 1). The final model contains the full-length polypeptide, except for residues L181-S194 and S367-N371, which could not be built due to a lack of interpretable electron density, indicating their high structural flexibility.
Table 1.
Data collection and refinement statistics
| Parameters | Native | Selenomethionine peak |
| Data collection statistics | ||
| Cell parameters | ||
| a | 58.3 | 57.1 |
| b | 67.9 | 68.2 |
| c | 131.5 | 131.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Space group | P212121 | P212121 |
| Wavelength used, Å | 1.0000 | 0.9798 |
| Resolution, Å | 50.00–2.30 (2.34–2.30)‡ | 50.00–3.10 (3.15–3.10) |
| No. of all reflections | 155,036 (8,610) | 89,319 (6,840) |
| No. of unique reflections | 22,556 (1,196) | 12,908 (980) |
| Completeness (%) | 99.1 (99.7) | 99.9 (100.0) |
| Average I/σ(I) | 13.5 (4.9) | 9.0 (6.1) |
| Rmerge (%)* | 6.1 (36.2) | 10.2 (41.9) |
| Refinement statistics | ||
| No. of reflections used (σ(F) > 0) | 21,212 | |
| Rwork (%)† | 22.4 | |
| Rfree (%)† | 25.7 | |
| rmsd bond distance, Å | 0.025 | |
| rmsd bond angle, ° | 2.461 | |
| Average B value, Å2 | 37.4 | |
| Ramachandran plot | ||
| Res. in favored regions, % | 93.0 | |
| Res. in generously allowed regions, % | 3.1 | |
| Res. in disallowed regions, % | 2.9 | |
*Rmerge = ΣhΣl | Iih−<Ih> |/ΣhΣI <Ih>, where <Ih> is the mean of the observations Iih of reflection h.
†Rwork = Σ(||Fp(obs)|−|Fp(calc)||)/ Σ|Fp(obs)|; Rfree is an R factor for a preselected subset (5%) of reflections that was not included in refinement.
‡Numbers in parentheses are corresponding values for the highest resolution shell.
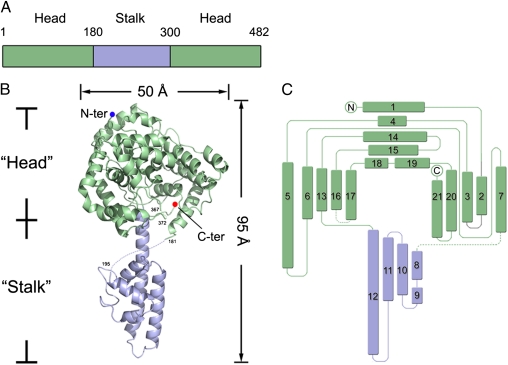
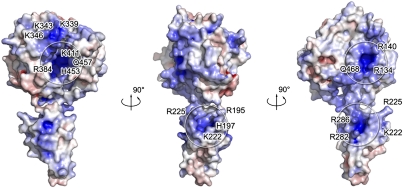
CCHFV NP possesses a racket-shaped overall structure with dimensions of 40 × 50 × 95 Å, and features two major parts: a “head” domain (M1-I180 and A300-I482) and a “stalk” domain (R181-A299) (Fig. 2). Both head and stalk domains are predominantly composed of α-helices. There is a large positively charged cavity located at the center of the head domain, and a positively charged region in the stalk domain adjacent to the head domain (Fig. 3). Although CCHFV NP has no primary sequence homologs, a comparison of the structure of the head domain with reported structures in the Protein Data Bank (PDB) using the DALI structure comparison service (20) revealed a high structural similarity with the N-terminal domain of LASV NP (18) (PDB code: 3MX5, Z score = 15.5). Alignment of these two structures gives an overall root-mean-square deviation (rmsd) of 3.2 Å for all Cα atoms of the 259 aligned residues (Fig. S1).
Fig. 2.
Structure of CCHFV NP. (A) Schematic diagram of the domain organization in the primary sequence of CCHFV NP. The stalk and head domains are colored as light blue and green, respectively. (B) Overall structure in cartoon representation. Missing residues are linked by dotted lines. (C) Topology diagram. Head and stalk domains are colored in green and light blue, respectively.
Fig. 3.
Potential RNA-binding region of CCHFV. The electrostatic surface potential of CCHFV NP was calculated using adaptive Poisson-Boltzmann solver (APBS) tools. The positive surface is colored blue, the negative surface, red, with limits ±10 kbT/ec. Positive residues are labeled on CCHFV NP, suggesting the presence of several positively charged grooves that may be involved in RNA binding.
LASV belongs to the Arenaviridae family and is a single-stranded ambisense RNA virus with two genomic RNA segments encoding four genes, and its NP is responsible for encapsulating the viral genomic RNA into ribonucleoprotein (21). Qi and colleagues presented the first full-length LASV NP structure and proposed that the full-length LASV NP contains an RNA-specific 3′–5′ exonuclease activity (18). This exonuclease activity was confirmed by an independent group who located this function to the C-terminal domain (16). However, Qi et al. also suggested that the N-terminal domain of LASV NP contains an RNA cap-binding function, whereas a newly reported LASV(1-340)-ssRNA complex structure shows that the N-terminal domain actually possesses RNA-binding activity (17). The CCHFV NP head domain shows high structural similarity with the LASV NP N-terminal domain, despite poor primary sequence similarity (Fig. S1).
Cap-Binding Ability of CCHFV NP.
Because the CCHFV NP head domain shows high structural similarity to the LASV NP N-terminal domain, we first investigated whether they have similar functions. We first examined the cap-binding ability of the CCHFV NP head domain. Results of isothermal titration calorimetry showed, unexpectedly, that the monomeric CCHFV NP binds cap analogs, i.e., m7G, m7Gp, and m7Gppp, with extremely low or no binding affinity in vitro (Fig. S2), compared with the cap-binding affinities reported for bona fide virally encoded cap-binding proteins (22). Attempts to cocrystallize or soak crystals of CCHFV NP with cap analogs were also carried out, but no extra electron density was observed in the potential cap-binding site suggested by structural comparisons with LASV NP. We therefore conclude that CCHFV NP, at least in its monomeric form, is unlikely to bind with the cap in vitro.
RNA-Binding Affinity of CCHFV NP.
We subsequently examined whether CCHFV NP binds RNA at a range of different concentrations using electrophoretic mobility shift assays (EMSAs) (Fig. S3) and found that CCHFV NP's binding affinity with a 24-nt ssRNA probe was weak, because free RNA could still be observed even at a NP:RNA molar ratio of 16:1. Surface plasmon resonance spectroscopy analysis also revealed poor binding affinity with a poly(U) oligonucleotide (Fig. S4). These results are consistent with the weak nucleic acid-binding affinity observed during purification (Fig. 1). When treated with DNase and RNase (1 μg/mL), CCHFV NP could easily be separated from E. coli nucleic acids, suggesting that the binding affinity of CCHFV NP for nucleic acids is weak.
CCHFV NP and Host Defense Mechanism.
Both CCHFV and LASV antagonize the host IFN response to infection by interfering with the activation pathway of IRF-3 (23). Further studies have suggested that, in LASV, the exoribonuclease activity encoded by the C-terminal domain of LASV NP is responsible for this suppression of innate immunity (16, 18). However, results from IFN induction reporter assays driven by INF-β or IFN-stimulated response element (ISRE) promoters indicated that CCHFV NP did not induce a significant type I IFN response (Fig. S5), suggesting that CCHFV NP may not be responsible for interruptions of the IRF-3 pathway.
Moreover, previous research has demonstrated host cell caspase-3–dependent proteolysis of CCHFV NP into two fragments when caspase activity is induced during infection and this has been proposed as a host defense mechanism against CCHFV infection (24). We confirmed this observation when purified caspase-3 was added to CCHFV NP protein in vitro (Fig. S6B). The cleavage site of caspase-3 on CCHFV NP was identified to be 266DEVD269 and is located in a loop region connecting two long central helices in the stalk domain (Fig. S6A). We therefore propose that host cell defenses against CCHFV infection may recognize the stalk domain of NP and thus antagonize the biological function of NP.
CCHFV Nucleoprotein Has Endonuclease Activity.
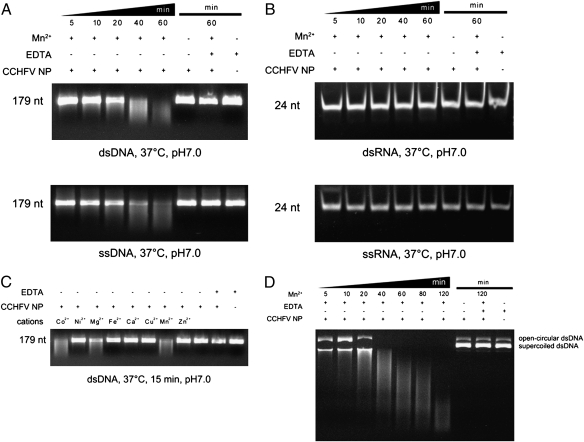
The inconsistency between the biological functions of CCHFV NP and its structural homolog, LASV NP, led us to further investigate the biological function of CCHFV NP. Results showed that CCHFV NP has a divalent cation-dependent endonuclease activity in vitro (Fig. 4). We assayed the nuclease activity of the CCHFV NP using ss/dsDNA (Fig. 4A) and ss/dsRNA substrates (Fig. 4B) with or without secondary structure. Using 179-nt dsDNA and ssDNA, we found that CCHFV NP has an intrinsic nuclease activity on both single- and double-stranded DNA that is stimulated to different extents by divalent cations such as Mn2+, Co2+, and Mg2+. Other divalent cations had little or no effect on CCHFV NP nuclease activity (Fig. 4C). In contrast, purified NP did not hydrolyze ssRNA or dsRNA in the presence or absence of Mn2+, suggesting that the nuclease activity of CCHFV NP is DNA specific (Fig. 4B). CCHFV NP also completely degraded circular dsDNA and highly structured dsDNA (λ-DNA), showing that it is a sequence nonspecific endonuclease (Fig. 4D and Fig. S7A).
Fig. 4.
DNA-specific endonuclease activity of CCHFV NP. (A) Time series of in vitro dsDNA (Upper) and ssDNA (Lower) and (B) dsRNA (Upper) and ssRNA (Lower) degradation assay products. Reaction products of 0.3 μM purified CCHFV NP with 100 ng/μL substrates at 37 °C in a final volume of 10 μL are shown after 5, 10, 20, 40, and 60 min. Reaction products were loaded onto a 20% (wt/vol) polyacrylamide gel and stained with ethidium bromide. (C) Effect of divalent cations on CCHFV NP nuclease activity. Purified CCHFV NP (0.3 μM) and dsDNA substrate were incubated with CoCl2, NiCl2, MgCl2, FeSO4, CaCl2, CuCl2, MnCl2, and ZnCl2 at a concentration of 1 mM. CCHFV NP alone, CCHFV NP and EDTA, or CCHFV NP and EDTA with Mn2+ were used as controls. All reactions were stopped by the addition of 10 mM EDTA. (D) Effect of CCHFV NP endonuclease activity on circular double-stranded plasmids. CCHFV NP was incubated with circular double-stranded plasmids for different lengths of time. All reactions were stopped by the addition of 10 mM EDTA.
Because the head domain shows little structural similarity with other endonucleases, we carried out further experiments to confirm the above findings. We selected several residues, i.e., Y374, R384, E387, K411, H453, and Q457, which appear to be potentially involved in the putative active site, for mutational analysis (Fig. 5B). Results of in vitro endonuclease assays with site-specific mutants R384A, E387A, K411A, H453A, and Q457A revealed that they had significant, but different degrees of impact, whereas Y374A had only marginal effects (Fig. 5C). Not surprisingly, these positions are absolutely conserved in related NP sequences (Fig. S8). Because the only pocket within the whole protein large enough for substrate binding is located in the head domain, we wondered whether the stalk domain plays any role in this endonuclease activity. We generated a construct (NPΔ180–300) of the head domain in which stalk domain residues were replaced by a flexible linker consisting of glycine residues. The head domain alone showed equivalent endonuclease activity to the wild-type protein (Fig. S9), indicating that the head domain is responsible for the endonuclease activity of CCHFV NP.
Fig. 5.
CCHFV NP head domain has endonuclease activity. (A) Structure comparison between the CCHFV NP head domain (from residue K59) and the N-terminal domain of LASV NP. Proteins are shown as cartoon representations in different colors. (B) Potential DNase active site of CCHFV NP head domain. Residues whose hydrophilic side chain is near the potential site for DNA binding are shown as colored sticks. (C) Nuclease activity of relevant mutants at the potential active site of CCHFV NP.
In addition, we measured the specific activity of CCHFV NP. CCHFV NP showed DNA endonuclease activity as high as 1.3 × 105 units/mg (Fig. S7B). Considering the molecular weight difference between CCHFV NP (56 kDa) and DNase I (31 kDa), this specific activity is comparable with that of a typical DNA nuclease. Taken together, all these results reveal that CCHFV NP possesses a unique endonuclease activity.
Discussion
Our work reveals that bunyavirus nucleoprotein has an endonuclease function not present in other negative single-stranded RNA viruses. Interestingly, the paper by Qi and colleagues also reported that both trimeric and hexameric forms of LASV NP can degrade dsDNA, whereas they did not comment on which part of the LASV NP is responsible for this DNase activity (18). The similar enzymatic activity of the CCHFV and LASV nucleoproteins raises the possibility that the function of (−)ssRNA virus nucleoproteins is more complex than previously anticipated. Further work is needed to determine the precise role of CCHFV NP during the life cycle of CCHFV.
There are two positively charged regions in the structure of the CCHFV NP head domain. One is a large positively charged crevice, constituted by a number of residues (K339, K343, K346, R384, K411, H453, and Q457) and the other region is constituted by R134, R140, and Q468. The stalk domain also contains a positively charged region consisting of residues R195, H197, K222, R225, R282, and R286 (Fig. 3). The presence of these three positively charged surfaces suggests they may play a role in RNA binding.
However, because the recombinant CCHFV NP protein (1-482) expressed in E. coli mainly exists as a monomer and shows weak binding affinity with cellular nucleic acids, we infer that recombinant CCHFV NP does not bind strongly to DNA or RNA. This is quite different from the situation during the purification of LASV NP (1-340) which, like some other reported (−)ssRNA viruses (10, 17), remained strongly and stably bound to host cell nucleic acids.
Most of the above positively charged residues are well conserved in the NP primary sequences of the Nairovirus family (Fig. S7). When we compared our structure with that of the LASV NP N-terminal domain bound with RNA (Fig. S10), we found that the region of CCHFV equivalent to the RNA-binding LASV NP α6 helix is probably located at residues L181-S194, residues which are missing from our final structure due to their intrinsic structural flexibility. Currently, several structures of (−)ssRNA virus nucleoproteins have been reported, including rabies virus (13), VSV (14), and BDV (15). These three NP structures demonstrate that RNA binds nonspecifically in a deep positively charged groove. In contrast, similar deep positively charged RNA-binding grooves are not obvious in the NPs of CCHFV and LASV, indicating that there is variation in the RNA-binding mechanism within the (−)ssRNA viruses (Fig. S11).
Although the encapsulation of genomic RNA depends on the formation of a capsid assembled from nucleoproteins, some virally encoded nucleoproteins show weak binding affinity with RNA in vitro (9). One reason for this is that, although the positively charged residues of negative-strand RNA virus nucleoproteins actually interact with the phosphate groups of the RNA backbone, they are not the major driving force for viral genomic RNA encapsulation. Instead, virus RNA plays a pivotal role in the formation of RNP, the so-called “RNA-stabilized oligomerization” process. According to the experiments reported above, monomeric CCHFV NP shows very weak binding affinity with RNA in vitro, and the head domain may mainly function as an endonuclease under our experimental conditions. Further work is required to determine whether it participates in RNA binding and if so, how the virus coordinates these two different functions.
Conclusion
The correct assembly of viral genomic RNA by nucleocapsid proteins is crucial for the assembly and maturation of single-stranded viruses and is a significant process in protecting viral RNA from host cell degradation. Recent structural work on LASV NP has extended our knowledge of virally encoded NP proteins beyond the packaging of viral genomic RNA (16–18), but the exact function of the N-terminal domain is still under discussion. Here we found that, despite the structural similarity between the head domain of CCHFV NP and the N-terminal domain of LASV NP, they may not have equivalent functions. CCHFV NP shows weak binding with RNA and does not show any cap-snatching affinity in vitro. However, our work shows that CCHFV has a new endonuclease function, which has not previously been described in any of the members of this virus family.
The crystal structure of CCHFV NP reported in this work extends our knowledge of bunyavirus nucleoproteins. CCHFV NP has a racket-shaped overall structure, composed of a head domain that resembles the topology found in LASV NP but has a nuclease function in place of LASV's cap-binding function. The electrostatic potential surface of CCHFV NP displays several positive regions, whereas CCHFV NP unexpectedly shows very weak binding activity with RNA in vitro. These data provide a new insight into the biological role of NPs in bunyavirus replication and provide valuable information for the development of intervention strategies designed to lessen the pathogenic burden of this important group of viral infections.
Materials and Methods
Protein Production.
Full-length CCHFV NP (strain YL04057) (3) was cloned into the pGEX-6p-1 (GE Healthcare) vector with BamHI and XhoI restriction sites using the cloning primers 5′-CGGGATCCATGGAAAACAAAATCGA-3′ (forward) and 5′-CCGCTCGAGTTAGATGATGTTGGCGC-3′ (reverse). The accuracy of the inserts was verified by sequencing.
The recombinant plasmid was transformed into E. coli strain BL21 (DE3) and overexpressed as a GST fusion protein. The cells were cultured at 37 °C in 800 mL LB media containing 100 μg/mL ampicillin. Once OD600 reached ∼0.6, the culture was transferred to 16 °C, and protein was induced by incubating with 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for an additional 16 h. Harvested cells were resuspended in lysis buffer containing 20 mM Hepes (pH 6.8), 500 mM NaCl, 1 μg/mL DNase I, and 1 μg/mL RNase and homogenized with a low-temperature ultra-high pressure cell disrupter (JNBIO). The lysate was centrifuged at 25,000 × g for 30 min at 4 °C to remove cell debris. The supernatant was then loaded twice onto a GST column preequilibrated with lysis buffer, and the GST tag was removed by digestion with PreScission protease (GE Healthcare) overnight at 4 °C. Eluted CCHFV NP protein was further purified by Superdex-200 gel filtration chromatography (GE Healthcare). SDS-PAGE analysis revealed over 95% purity of the final purified recombinant protein. The purified protein was then concentrated to 10 mg/mL in a buffer containing 20 mM Hepes (pH 6.8), 200 mM NaCl.
A CCHFV NPΔ180–300 truncation was generated by overlap-extension PCR using two sets of cloning primers: 5′-CGGGATCCATGGAAAACAAAATCG-3′ (forward1), 5′-CCGCTCGAGTTAGATGATGTTGGC-3′ (reverse1) and 5′-AGAAGGAACTTGGGAGGAGGAGGAGGAGGTGCACAGATT-3′ (forward2), 5′-CAATCTGTGCACCTCCTCCTCCTCCTCCCAAGTTCCTTC-3′ (reverse2). We removed the stalk domain (I180-Q300) using standard molecular cloning techniques and replaced it with five glycines. This truncation product was then inserted into a pGEX-6p-1(GE Healthcare) vector using the BamH1 and XhoI restriction sites. The protein production and purification process was the same as that for the full-length CCHFV NP protein described above.
Crystallization.
Initial crystallization conditions were screened by the hanging drop vapor-diffusion method using Hampton Research crystal screening kits at 16 °C. Crystals were obtained by mixing 1 μL of the protein solution with an equal volume of a reservoir solution and equilibrating the mixed drop against 300 μL of reservoir solution.
Small crystals first appeared after 2 d in 200 mM KCl and 20% (wt/vol) PEG3350. Further optimization with additive and detergent screens (Hampton Research) was performed, and the final optimized crystals were grown in a buffer containing 20 mM KCl, 10 mM MnCl2, and 12% (wt/vol) PEG3350. Rod-like crystals grew to a final size of 20 × 40 × 150 μm within 5 d. Selenomethionine substituted crystals appeared in the same condition, reaching their final size within 10 d. Crystals were harvested and cryoprotected in the well solution containing an additional 20% (vol/vol) glycerol and ash cooled in a dry nitrogen stream at 100 K for X-ray data collection.
X-Ray Data Collection, Processing, and Structure Determination.
Diffraction data for the native crystal of CCHFV NP was first collected to 2.3 Å at 100 K using a MAResearch M165 CCD detector on beamline 1W2A at the Beijing Synchrotron Radiation Facility. Anomalous diffraction data for selenomethionine derivatives were collected to 3.1 Å at 100 K using an ADSC Q315 CCD detector on beamline BL17 at the Photon Factory (KEK, Japan). All datasets were indexed, integrated, and scaled using the HKL2000 package (25). The orthorhombic crystal form belongs to space group P212121 with cell parameters a = 58.3 Å, b = 67.9 Å, and c = 131.5 Å.
Heavy atom searching, initial phase calculations, and density modifications were performed with PHENIX (26). The resulting electron density map was displayed with COOT (27) and an initial model was built manually. Several rounds of simulated annealing, restrained individual atomic displacement parameter refinement, energy minimization, and individual B-factor refinement were carried out with PHENIX (26). Solvent molecules were located from stereochemically reasonable peaks in the σA-weighted 2Fo−Fc difference Fourier electron density map (1.2 σ). Model geometry was verified using PROCHECK (28). All structure figures were drawn with PyMOL (29). Coordinates and structure factors have been deposited with the RCSB under accession code: 3U3I.
Endonuclease Activity Assays.
The DNA cleavage assay was performed by incubating 0.3 μM CCHFV NP with 100 ng/μL dsDNA or ssDNA at 37 °C in a final volume of 10 μL. The reaction buffer was 20 mM Hepes pH 7.0, 200 mM NaCl, and 1 mM metal salts. Reactions were stopped by the addition of EDTA to a final concentration of 10 mM, and reaction products were loaded on a 1.5% agarose gel and stained with ethidium bromide. The effect of divalent cations on the DNase activity of CCHFV NP was tested by incubating DNA substrates with protein in the presence of different metal salts: CoCl2, NiCl2, MgCl2, FeSO4, CaCl2, CuCl2, MnCl2, and ZnCl2. The circular double-stranded plasmid pGEX-6p-1 (GE Healthcare) and digested λ-DNA were also tested as substrates and the reaction products were loaded on a 1.5% agarose gel and stained with ethidium bromide. The reaction buffer and conditions for 24-nt RNA cleavage were the same as those for the DNA cleavage assay, and the reaction products were loaded onto 20% polyacrylamide gels and stained with ethidium bromide.
Isothermal Titration Calorimetry.
The binding affinities of cap analogs of wild-type CCHFV NP, i.e., m7G, m7Gpp, and m7Gppp, were measured using isothermal titration calorimetry (ITC). A 0.1-mM CCHFV NP protein solution was titrated against 0.3 mM cap-analog solutions (Sigma-Aldrich) using a VP-ITC microcalorimeter (MicroCal). ITC data were collected at 25 °C and analyzed using ORIGIN (MicroCal Software).
Supplementary Material
Acknowledgments
We thank the staff at the Beijing Synchrotron Radiation Facility and Photon Factory for their help with data collection, and Prof. Hong-bing Shu from Wuhan University who supplied the plasmids for INF induction reporter assays. This work was supported by The National Basic Research Program of China program (973 program) (no. 2010CB530100, 2010CB833602), the National Natural Science Foundation of China (no. 31000332, 31170678), and the Special Program for Key Basic Research of the Ministry of Science and Technology (no. 2007FY210700).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinate and structure factor have been deposited in the Protein Data Bank (www.rcsb.org) with the accession code 3U3I.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200808109/-/DCSupplemental.
References
- 1.Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol. 2006;80:8834–8842. doi: 10.1128/JVI.00752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flick R, Whitehouse CA. Crimean-Congo hemorrhagic fever virus. Curr Mol Med. 2005;5:753–760. doi: 10.2174/156652405774962335. [DOI] [PubMed] [Google Scholar]
- 3.Wei PF, et al. Serial expression of the truncated fragments of the nucleocapsid protein of CCHFV and identification of the epitope region. Virol Sin. 2010;25:45–51. doi: 10.1007/s12250-010-3067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogg MMPJ, Patterson JL. RNA binding domain of Jamestown Canyon virus S segment RNAs. J Virol. 2007;81:13754–13760. doi: 10.1128/JVI.01492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborne JCER, Elliott RM. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J Virol. 2000;74:9946–9952. doi: 10.1128/jvi.74.21.9946-9952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mir MABB, Brown B, Hjelle B, Duran WA, Panganiban AT. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J Virol. 2006;80:11283–11292. doi: 10.1128/JVI.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohl BPBJ, Barr JN. Investigating the specificity and stoichiometry of RNA binding by the nucleocapsid protein of Bunyamwera virus. RNA. 2009;15:391–399. doi: 10.1261/rna.1367209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gött P, Sr, Stohwasser R, Schnitzler P, Darai G, Bautz EK. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology. 1993;194:332–337. doi: 10.1006/viro.1993.1263. [DOI] [PubMed] [Google Scholar]
- 9.Ferron F, et al. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7:e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raymond DD, Piper ME, Gerrard SR, Smith JL. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci USA. 2010;107:11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng AK, et al. Structure of the influenza virus A H5N1 nucleoprotein: Implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008;22:3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 13.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 14.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 15.Rudolph MG, et al. Crystal structure of the borna disease virus nucleoprotein. Structure. 2003;11:1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA. 2011;108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hastie KM, et al. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci USA. 2011;108:19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi X, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu YM, Zeng DD, Huang Q, Fan CH. A scaffolded DNA origami algorithm for polygon network structures and hollow three-dimensional structures. Acta Biophys Sin. 2010;26:750–758. [Google Scholar]
- 20.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchmeier MJ, Peters CJ, de la Torre JC. Arenaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1791–1827. [Google Scholar]
- 22.Guilligay D, et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol. 2008;15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Sobrido L, Giannakas P, Cubitt B, García-Sastre A, de la Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlberg H, Tan YJ, Mirazimi A. Induction of caspase activation and cleavage of the viral nucleocapsid protein in different cell types during Crimean-Congo hemorrhagic fever virus infection. J Biol Chem. 2011;286:3227–3234. doi: 10.1074/jbc.M110.149369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. In: Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Part A, Methods in Enzymology. Carter CW, Sweet RM, editors. Vol 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 29.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.