Abstract
Mammalian sweet taste is primarily mediated by the type 1 taste receptor Tas1r2/Tas1r3, whereas Tas1r1/Tas1r3 act as the principal umami taste receptor. Bitter taste is mediated by a different group of G protein-coupled receptors, the Tas2rs, numbering 3 to ∼66, depending on the species. We showed previously that the behavioral indifference of cats toward sweet-tasting compounds can be explained by the pseudogenization of the Tas1r2 gene, which encodes the Tas1r2 receptor. To examine the generality of this finding, we sequenced the entire coding region of Tas1r2 from 12 species in the order Carnivora. Seven of these nonfeline species, all of which are exclusive meat eaters, also have independently pseudogenized Tas1r2 caused by ORF-disrupting mutations. Fittingly, the purifying selection pressure is markedly relaxed in these species with a pseudogenized Tas1r2. In behavioral tests, the Asian otter (defective Tas1r2) showed no preference for sweet compounds, but the spectacled bear (intact Tas1r2) did. In addition to the inactivation of Tas1r2, we found that sea lion Tas1r1 and Tas1r3 are also pseudogenized, consistent with their unique feeding behavior, which entails swallowing food whole without chewing. The extensive loss of Tas1r receptor function is not restricted to the sea lion: the bottlenose dolphin, which evolved independently from the sea lion but displays similar feeding behavior, also has all three Tas1rs inactivated, and may also lack functional bitter receptors. These data provide strong support for the view that loss of taste receptor function in mammals is widespread and directly related to feeding specializations.
Keywords: diet, sweetener
It is widely believed that most mammals perceive five basic taste qualities: sweet, umami, bitter, salty, and sour. The receptors for sweet, umami and bitter tastes are G protein-coupled receptors (GPCRs) (1). Sweet taste is mediated largely by a heteromer of two closely related Tas1r (type 1 taste receptor) family GPCRs: Tas1r2 and Tas1r3 (2–5). Tas1r1, another member of the Tas1r family, in combination with Tas1r3, forms an umami taste receptor (6). Tas1r receptors are class C GPCRs. Unlike sweet and umami tastes, bitter taste is mediated by Tas2r family GPCRs, which belong to class A GPCRs and are structurally unrelated to Tas1r family receptors (7, 8). The genes encoding Tas2r receptors, the Tas2r genes, differ substantially in gene number and primary sequences among species, most likely reflecting the likelihood that these genes are required for detecting toxic or harmful substances in a species’ ecological niche (9–11).
Direct evidence for a close correlation between taste function and feeding ecology comes from work on domestic and wild Felidae. Cats, obligate carnivores, are behaviorally insensitive to sweet-tasting compounds (12, 13). We proposed that this behavioral insensitivity was a consequence of the pseudogenization of Tas1r2 (14). Tas1r2 also is known to be pseudogenized in chicken, tongueless Western clawed frogs, and vampire bats (11, 15). The loss of the sweet taste receptor in chicken and vampire bats is consistent with their sweet insensitive behavior (16, 17). It is yet to be established how Western clawed frogs respond to sweeteners. In contrast with the feline, the giant panda lacks a functional umami taste receptor gene (Tas1r1) (18) and feeds primarily on bamboo. Nevertheless, the remainder of the taste system in both cats and giant pandas is similar to those of other mammals (12, 13, 18, 19).
Based on anatomical studies, it is likely that some aquatic mammals, such as sea lions (Carnivora) and dolphins (Cetacea)—species from two lineages that independently “returned” to the sea more than 35 and 50 million years ago, respectively (20)—have lost some taste function. Both animals exhibit an atrophied taste system, exemplified by few taste buds present in their lingual epithelium (21). Consistent with an atrophied taste system, both species exhibit a feeding behavior pattern that suggests that taste may not play a major role in food choice: they swallow their food whole, perhaps minimizing opportunities and needs for taste input (22, 23).
To further elaborate on the idea that taste behavior, taste receptor function, and feeding ecology are intimately interrelated, we have chosen a comparative approach. Specifically, we have tested two hypotheses. First, we hypothesized that mutations rendering sweet taste receptors dysfunctional should be observed in exclusively meat-eating species in addition to the cats. We selected for study a range of species from the order Carnivora to test this hypothesis. This group is particularly useful for testing this hypothesis because it includes species differing significantly in their dietary habits, ranging from obligate carnivores (e.g., domestic and wild cats) to relatively omnivorous species (e.g., bears) to rather strict herbivores (e.g., the giant panda) (24, 25). The second hypothesis tested here involved mammals that were both exclusive carnivores and were known anatomically to have an atrophied taste system. We hypothesized that not only sweet taste receptor function but receptors for other taste qualities, such as umami and bitter, would also be disrupted. To examine this prediction, we evaluated the molecular structure of the other Tas1rs in the sea lion and the Tas1rs and the Tas2rs in bottlenose dolphins from the order Cetacea.
We found that seven of the 12 species examined from the order Carnivora—only those that feed exclusively on meat—had pseudogenized Tas1r2 genes as predicted. Furthermore, we confirmed our hypothesis that, in addition to the loss of Tas1r2, both the sea lion and bottlenose dolphin lack Tas1r1 and Tas1r3 receptor genes, suggesting an absence of both sweet and umami taste-quality perception. Additionally, we failed to detect intact bitter receptor genes Tas2rs from the dolphin genome, suggesting that this modality may be lost, or its function greatly reduced, in dolphins. Thus, taste loss is much more widespread than previously thought, and such losses are consistent with altered feeding strategies.
Results
Molecular Cloning of Tas1r2 from Selected Species Within Carnivora.
To determine how widespread the pseudogenization of Tas1r2 is in the order Carnivora, and to evaluate how this relates to food habits of these species, we fully sequenced all six exons of Tas1r2 from 12 species within Carnivora using degenerate primers designed from conserved exon-intron boundary sequences among cat, dog, and giant panda Tas1r2s, three species with an assembled genome in the order Carnivora. Among these 12 species, we identified five that appear to have an intact Tas1r2: the aardwolf, Canadian otter, spectacled bear, raccoon, and red wolf. The intact Tas1r2 genes had entire complete coding sequences ranging from 2,511 to 2,517 bp.
Pseudogenized Tas1r2 genes in Pinnipedia.
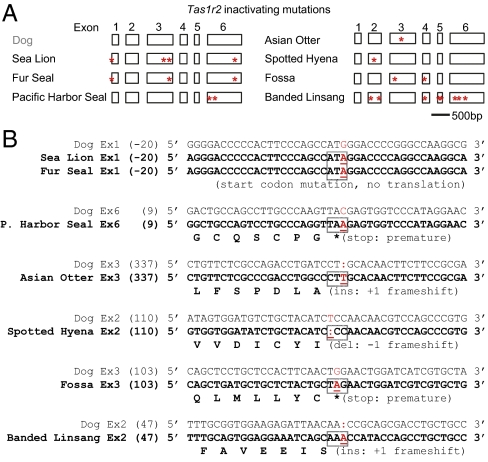
The sea lion, fur seal, and Pacific harbor seal belong to the Pinnipedia superfamily within the Caniformia suborder of Carnivora. In exon 1 of Tas1r2, both the sea lion and fur seal (Otariidae family) have an ATA instead of ATG in the start codon position. This mutation is predicted to prevent Tas1r2 from being translated (Fig. 1 A and B and SI Appendix, Fig. S1A). Additionally, we detected a 1-bp deletion between 579 and 580 bp in exon 3 of the sea lion Tas1r2 (Fig. 1A and SI Appendix, Fig. S1B); 2-bp deletions between 674 and 675 bp in exon 3 of the sea lion Tas1r2 and between 675 and 676 bp in exon 3 of the fur seal Tas1r2 (Fig. 1A and SI Appendix, Fig. S1C); and a 1-bp deletion between 802 and 803 bp in exon 6 of both species (Fig. 1A and SI Appendix, Fig. S1D). These defects in the coding sequence predict that the sea lion and fur seal Tas1r2 genes are pseudogenes.
Fig. 1.
Widespread pseudogenization of the sweet-taste receptor gene Tas1r2 in 7 species within Carnivora. (A) Schematic diagram shows the positions of ORF-disrupting mutations in Tas1r2 from selected species within Carnivora. The intact dog Tas1r2 gene structure is shown as a reference. The positions where ORF-disrupting mutations occurred are marked with a red asterisk (*). (B) A 42-bp–long nucleotide sequence containing the ORF-disrupting mutation that occurs closest to 5′ end of the gene is shown for each species. The aligned dog sequence is shown above it, and the amino acid sequence deduced from the nucleotide sequence up to the mutation site is shown underneath it. The codon that contains the ORF-disrupting mutation (marked in red and underlined) is indicated by a box.
Unlike in the sea lion and fur seal, Tas1r2 of the Pacific harbor seal (Phocidae family) has a normal ATG start codon; however, it has a nonsense mutation at 32 bp in exon 6 that leads to a premature stop codon (TAA; Fig. 1 A and B and SI Appendix, Fig. S2A). Additionally, it has a 2-bp deletion between 192 and 193 bp in the sixth exon that results in a frame shift and multiple stop codons thereafter (Fig. 1A and SI Appendix, Fig. S2B). These ORF-disrupting mutations would likely also render the Pacific harbor seal Tas1r2 gene defective.
Pseudogenized Tas1r2 in Asian small-clawed otter.
The Asian small-clawed otter, Canadian otter, and ferret all belong to the Mustelidae family within the Caniformia. In a previous study, we reported an intact Tas1r2 sequence in ferret, predicting a functional Tas1r2 receptor (24). Based on our current data, the Canadian otter appears to have an intact Tas1r2 gene as well. In contrast, we detected a T insertion at 360 bp in exon 3 of the Asian otter Tas1r2 based on the sequence alignment with the dog ortholog (Fig. 1 A and B and SI Appendix, Fig. S3), predicting a defective Tas1r2. The same insertion was found in DNA sample from a second Asian otter.
Pseudogenized Tas1r2 in spotted hyena.
The spotted hyena and aardwolf belong to the family Hyaenidae within the Feliformia. The aardwolf appears to have an intact Tas1r2 gene. In contrast, we detected a 1-bp ORF-disrupting deletion between 130 and 131 bp in exon 2 of Tas1r2 in the spotted hyena (Fig. 1 A and B and SI Appendix, Fig. S4). The same deletion was found in a second spotted hyena. Therefore, the spotted hyena Tas1r2 is a pseudogene.
Pseudogenized Tas1r2 in fossa.
Fossa is a species in the family Eupleridae within the Feliformia. Two ORF-disrupting mutations were found in exons of the fossa Tas1r2. Specifically, there is a nonsense mutation (A) at 125 bp in exon 3 that results in a stop codon (TAG; Fig. 1 A and B and SI Appendix, Fig. S5A). Moreover, in exon 4, we detected a T insertion at 58 bp that creates a stop codon immediately after (TGA, 58–60) (Fig. 1A and SI Appendix, Fig. S5B). The T insertion was found in three additional individuals, indicating that this mutation is fixed in the fossa Tas1r2 gene. In contrast, the nonsense mutation TAG (125 bp, exon 3) displayed polymorphism (TAG or TGG). Collectively, these ORF-disrupting mutations in the fossa Tas1r2 predict that the fossa Tas1r2 is a pseudogene.
Pseudogenized banded linsang Tas1r2.
The banded linsang belongs to the family Prionodontidae within the Feliformia. In exon 2, we detected an A insertion at 70 bp (Fig. 1 A and B and SI Appendix, Fig. S6A). Furthermore, we detected another 10-bp microdeletion between 274 and 275 bp in the second exon (Fig. 1A and SI Appendix, Fig. S6B), a 14-bp insertion between 78 and 91 bp in exon 4 (Fig. 1A and SI Appendix, Fig. S6C), a 20-bp microdeletion between 27 and 28 bp in exon 5 (Fig. 1A and SI Appendix, Fig. S6D), and another 2-bp deletion between 54 and 55 bp in exon 5 (Fig. 1A and SI Appendix, Fig. S6E). In exon 6, there is a 1-bp deletion between 210 and 211 bp, a 28-bp insertion between 235 and 262 bp, and a 1-bp deletion between 444 and 445 bp (Fig. 1A and SI Appendix, Fig. S6 F–H). With multiple ORF-disrupting mutations in the coding sequence, the banded linsang Tas1r2 is apparently defective.
In summary, we found that, in addition to cats, seven other species in the order Carnivora have been identified as possessing pseudogenized Tas1r2 genes. Moreover, the ORF-disrupting mutations that cause a pseudogenized Tas1r2 differ among species in different families and clades. Within a single family, the pseudogenization of Tas1r2 occurred in some species, but not others. A common feature shared by those species with pseudogenized Tas1r2 genes is that they are strict carnivores or piscivores (fish eaters) (25).
Evolutionary Analysis of Tas1r2 in Carnivora.
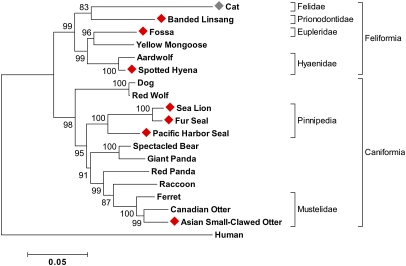
To gain insight into the evolution of the sweet taste receptor in the order Carnivora, we conducted a detailed evolutionary analysis of Tas1r2 from 18 species within Carnivora, including eight species (seven determined in this study and the domestic cat) with a pseudogenized Tas1r2 and 10 species (five determined in this study and five determined previously) with an intact Tas1r2. The human Tas1r2 was used as the outgroup for the analysis. We aligned the entire coding sequence of Tas1r2 from 18 carnivore species along with the human Tas1r2 sequence, then removed gaps, indels, and stop codons from the alignment, which results in 2,160-bp aligned nucleotides for phylogenetic analysis. A phylogenetic tree was built using the maximum-likelihood analysis method implemented in MEGA5 (Fig. 2) (26). The same tree was derived using the neighbor-joining method implemented in MEGA5 (SI Appendix, Fig. S7). We used our tree for further statistical analysis because our phylogenetic tree agrees well with trees proposed previously using other gene sequences or intron sequences (27, 28).
Fig. 2.
An evolutionary tree of Tas1r2 gene from 18 species within Carnivora. The evolutionary history is inferred by using the maximum-likelihood method based on the Tamura–Nei model (37) implemented in MEGA5 (26). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2,000 replicates) is shown next to the branches (38). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Species with a pseudogenized Tas1r2 are marked with a diamond (red and gray depict species characterized in this study or previously, respectively). The human Tas1r2 is used as the outgroup for the analysis.
To evaluate whether Tas1r2 is under strong purifying selection in the order Carnivora, we estimated the ratio (ω) of nonsynonymous to synonymous substitution rates by a likelihood method implemented in CODEML (29). The total nucleotides used for analysis were 2,160 bp after removing gaps, indels, and stop codons from the alignment. In model A, we analyzed this dataset of 18 species to evaluate the overall selective constraint on Tas1r2. With an assumption of a uniform ω, the average ω across the tree was estimated to be 0.1909, which is significantly <1 (model B), again indicating a strong overall negative selective pressure on Tas1r2 (P ∼ 0; Table 1).
Table 1.
Likelihood ratio tests of selective pressures on carnivore Tas1r2 gene
| Models | ω (dN/dS) | ln L* | np† | Models compared | 2Δ(ln L)‡ | P values |
| A. All branches have the same ω | ω = 0.1909 | −9,840.81 | 36 | |||
| B. All branches have the same ω= 1 | ω = 1 | −10,203.39 | 35 | B vs. A | 725.16 | ∼0 |
| C. The branches with a pseudogenized | ω1 = 0.13656 | |||||
| Tas1r2 have the ω2, other branches have ω1 | ω2 = 0.41974 | −9,806.38 | 37 | A vs. C | 68.86 | 1.1 × 10−16 |
| D. The branches with a pseudogenized | ω1 = 0.13253 | |||||
| Tas1r2 have ω2 = 1, other branches have ω1 | ω2 = 1 | −9,829.34 | 36 | D vs. C | 45.92 | 1.2 × 10−11 |
| E. Each branch has its own ω | Variable ω | −9,783.72 | 69 | C vs. E | 45.32 | 0.05955 |
Dataset: 18 species (all species, after removing gaps and nonsense mutations in pseudogenes).
*The natural logarithm of the likelihood value.
†No. of parameters.
‡Twice the difference in In L between the two models compared.
To examine whether the selective pressure is somewhat relaxed on species with a pseudogenized Tas1r2, we tested a two-ratio model (model C) with the assumption of a uniform ω for branches with a pseudogenized Tas1r2 (ω2) and for branches with an intact Tas1r2 (ω1), respectively. In this model, ω1 was estimated to be 0.13656, similar to that estimated in model A; ω2 was estimated to be 0.41974. This two-ratio model C fits significantly better than the above-mentioned one-ratio model A (P = 1.1 × 10−16; Table 1), indicative of a divergence in the selective pressure between the branches with a pseudogenized Tas1r2 and the branches with an intact Tas1r2. The selective purifying pressure is markedly relaxed in the branches with a pseudogenized Tas1r2.
To investigate whether the selective pressure is completely removed from the branches with a pseudogenized Tas1r2, we tested another two-ratio model (model D) that allows ω2 (pseudogenized) to be fixed to 1 and a uniform ω1 for the branches with an intact Tas1r2. This model D fits significantly less well than the above-mentioned two-ratio model C (P = 1.2 × 10−11; Table 1), suggesting that the selective pressure is partially but not completely relaxed. Finally, we tested an alternative model (E) in which ω is allowed to vary among branches, this model was found not to fit significantly better than a two-ratio model C (P = 0.05955).
Behavioral Taste-Testing of the Asian Small-Clawed Otter and Spectacled Bear.
To investigate the proposed relationship between Tas1r2 receptor structure and taste perception and diet, we carried out behavioral tests on two available carnivore species from among those we genotyped. Previously, we had tested sweet preferences in a number of species, including lesser panda, domestic ferret, haussa genet, meerkat, yellow mongoose, and Asiatic lion (24). With the exception of the Asiatic lion, all these species within Carnivora appear to have an intact sweet taste receptor and prefer sweet-tasting compounds. Thus, aside from cats, there have been no previous taste preference studies in carnivorans with a defective Tas1r2.
Here, we tested two Asian small-clawed otters, a species with defective Tas1r2, and four spectacled bears, which appear to have an intact sweet-taste receptor, for their preferences for a wide range of sweet-tasting compounds, including both sweet carbohydrates (natural sugars) and noncaloric sweeteners.
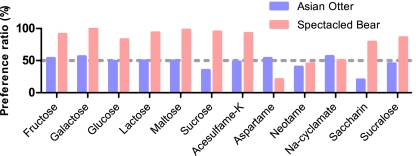
We used a two-bowl group test to assess preference for sweet-tasting compounds. When presented with a simultaneous choice between sugar-containing solution and water, the Asian otters showed no clear preference for sugars. The preference ratios range from 35.4% for sucrose (0.5 M) to 56.5% for galactose (0.8 M; Fig. 3). They showed indifference to, or avoidance of, a few nonnutritive sweeteners tested, including indifference to sucralose (45.5% at 5.03 mM) and avoidance to saccharin (20.6% at 6.2 mM; Fig. 3), indicating that the taste-test procedure is capable of demonstrating discrimination among tastants. The spectacled bears showed strong preferences for natural sugars, with preference ratios ranging from 83% for glucose (0.5 M) to 100% for galactose (0.8 M; Fig. 3). They also showed strong preferences for certain noncaloric sweeteners, including 5.0 mM sucralose (86.2%) and 6.2 mM acesulfame-K (92.9%) (Fig. 3). These results are congruent with the molecular data from both species.
Fig. 3.
Sweet-taste preferences of two genotyped species. Two Asian otter and four spectacled bears were tested behaviorally for their preferences for sweeteners using a two-bowl preference setup. One bowl contained sweetener solution and the other contained plain water. Dashed line indicates no preference (50%). Sweeteners were tested at the following concentrations: fructose (0.8 M), galactose (0.8 M), lactose (0.5 M), maltose (0.7 M), sucrose (0.5 M), acesulfame-K (6.0 mM), aspartame (10 mM), neotame (10.5 mM), saccharin (6.2 mM), and sucralose (5.0 mM).
Sea Lions and Dolphins Lack All Three Tas1r Receptors.
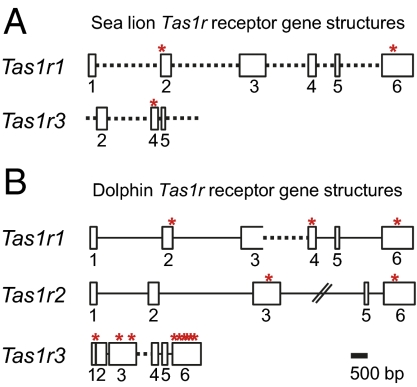
Given the atrophied taste system and unique feeding behavior in sea lions, we next investigated what happened to the other two Tas1rs in the sea lion. Using degenerate primers (SI Appendix, Table S1) and genomic DNA-based PCR, we sequenced the protein-coding regions of Tas1r exons. By aligning with the dog Tas1r1 sequence, we found that the sea lion Tas1r1 has a 1-bp deletion in exon 2 between 53 and 54 bp (Fig. 4A and SI Appendix, Fig. S8A), and an 11-bp deletion in exon 6 between 302 and 303 bp (Fig. 4A and SI Appendix, Fig. S8B). Tas1r3 has a 1-bp deletion between 72 and 73 bp in exon 4 (Fig. 4A and SI Appendix, Fig. S8C). Only exons 2, 4, and 5 have been sequenced for sea lion Tas1r3. As was the case for Tas1r2, sea lion Tas1r1 and Tas1r3 are also pseudogenes.
Fig. 4.
The sea lion and dolphin Tas1r receptor genes are inactivated by pseudogenization. (A) Sea lion Tas1r1 (Upper) and Tas1r3 (Lower) gene structures with boxes representing exons, and lines representing introns. Regions where ORF-disrupting mutations were found are marked with an asterisk. Regions with no sequence coverage are labeled with dashed lines. Only three exons have been sequenced for the sea lion Tas1r3. (B) Schematic drawings of the dolphin Tas1r1 (Top), Tas1r2 (Middle), and Tas1r3 (Bottom) gene structures. The symbols are the same as in A. The straight line with slashes indicates the intron is not proportionally scaled.
Dolphin is an aquatic mammal that evolved independently from the sea lion but displays similar feeding behavior and dramatic loss-of-taste system (23). We searched the draft dolphin genome (2.59X) for Tas1r genes using the dog Tas1r amino acid sequences (TBlastN). We found several contigs with sequence similarity to dog Tas1rs (∼70–80% amino acid identity). ABRN01270722 contains the first two exons and part of the third exon of dolphin Tas1r1, and ABRN01270723 contains exons 4–6 of Tas1r1. Dolphin intron 2 has a mutation in the donor splice site at the 5′ end (AT instead of GT; Fig. 4B) that would interfere with splicing of the Tas1r1 mRNA, a 5-bp deletion in exon 4 that disrupts the ORF of dolphin Tas1r1 (Fig. 4B and SI Appendix, Fig. S9A), and a 2-bp deletion in exon 6 (Fig. 4B). These ORF-disrupting mutations render dolphin Tas1r1 nonfunctional.
Using the dog Tas1r2 amino acid sequence, we retrieved only contig ABRN01341268 and determined potential coding sequences of dolphin Tas1r2 exons 1–3 and 5–6. Exon 4 appears to have been lost entirely based on using dog, cow, mouse, or human sequences, because queries and in exons 3 and 6 ORF-disrupting mutations were found (Fig. 4B). Specifically, there is a 20-bp ORF-disrupting insertion between 488 and 507 bp in exon 3 (Fig. 4B and SI Appendix, Fig. S9B), and there is a 1-bp ORF-disrupting deletion in exon 6 between 378 and 379 bp (Fig. 4B). Thus, dolphin Tas1r2 also is a pseudogene.
Next, we retrieved two contigs ABRN01316859 (exons 1–3) and ABRN01316858 (exons 4–6), covering the entire coding sequences of the dolphin Tas1r3 gene. Multiple ORF-disrupting mutations were found throughout the coding region (Fig. 4B). A 7-bp deletion between 99 and 100 bp in exon 6 is shown in SI Appendix, Fig. S9C. These data indicate that dolphin Tas1r3 also is a pseudogene. Like the sea lion, the bottlenose dolphin lacks all three Tas1r receptors.
Pseudogenization of Dolphin Tas2r Receptor Genes.
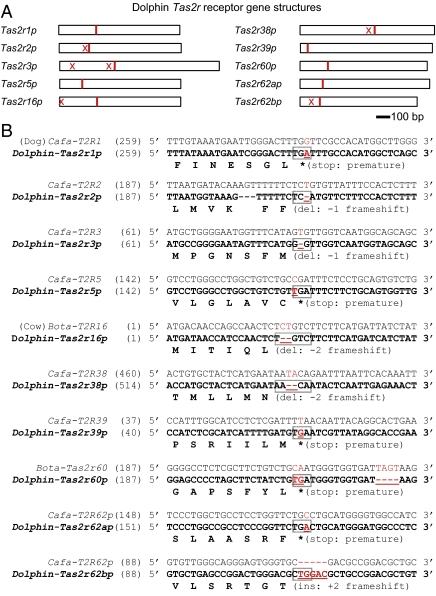
Because all three Tas1r receptor genes are pseudogenized in dolphins, what happened to genes mediating another major taste quality: bitter? To address this question, TBlastN searches were performed on the dolphin genome using 15 dog and 16 cow intact Tas2r genes as queries. Because Tas2rs are intronless genes, it was relatively easy to identify the dolphin Tas2r genes. The query results revealed only 10 pseudogenes, and no intact dolphin Tas2r receptor gene was found. The genes are named after their dog or cow orthologs (Tas2r1p, Tas2r2p, Tas2r3p, Tas2r5p, Tas2r16p, Tas2r38p, Tas2r39p, Tas2r60p, Tas2r62ap, and Tas2r62bp). Pseudogenization was caused by ORF-disrupting mutations (including nonsense mutations and frameshift mutations; Fig. 5 A and B and SI Appendix, Document 1). Sequence similarities (nucleotide) between dolphin Tas2rs and their orthologs in dog and cow are between 75% and 86%. A neighbor-joining tree was constructed by using aligned dolphin, dog, and cow Tas2r sequences (SI Appendix, Fig. S10) to show the evolutionary relationship among dog, cow, and dolphin bitter receptors. Despite the relatively low coverage of the dolphin genome assembly, the fact that none of the 10 identified Tas2r receptors is intact hints that this modality may also be lost, or its function greatly reduced.
Fig. 5.
Dolphin Tas2rs are pseudogenes. (A) Schematic diagram shows the premature stop codons (either nonsense mutations or stop codons resulting from prior frameshift mutations, depicted with a red line) and only the frame-shift mutations (X) before the premature stop codons in dolphin Tas2rs. Tas2r receptor genes are shown in scale. No start codon is detected for Tas2r62bp. (B) A 42-bp–long nucleotide sequence containing the ORF-disrupting mutation that occurs closest to 5′ end of the gene is shown for each of the 10 identified dolphin Tas2r receptor genes; the aligned dog sequence is shown above it, and the amino acid sequence deduced from the nucleotide sequence up to the mutation site is shown underneath it. The codon that contains the ORF-disrupting mutation (marked in red and underlined) is indicated by a box.
Discussion
Widespread Pseudogenization of Tas1r2 in the Order Carnivora.
In the present study, by examining Tas1r2 receptor genes in additional carnivore species, both closely and distantly related to Felidae, we found that functional loss of Tas1r2 is widespread and independent in Carnivora. ORF-disrupting mutations (e.g., stop codons, insertion or deletion of nontriplet nucleotides) were detected in seven nonfeline carnivore species: the sea lion, fur seal, Pacific harbor seal, Asian small-clawed otter, spotted hyena, fossa, and banded linsang, as well as in one cetacean species: the bottlenose dolphin. Looking specifically at the obvious mutations, with the exception of the sea lion and fur seal (sister species in the family Otariidae), none of the mutations disrupting the ORF of Tas1r2 were shared between any two species of these seven.
The widespread loss of Tas1r2 in species of the order Carnivora, the variance in their lineages, and the independent pseudogenizing mutations among species in distinct families strongly support the hypothesis that the loss of Tas1r2 occurred independently many times during the evolution of the order Carnivora. It seems most likely that changes in dietary behavior in the process of becoming obligate carnivores may favor the loss of a functional Tas1r2, arguing for convergent evolution of pseudogenized Tas1r2. Another case of convergent evolution of taste was found in the Tas2r38 receptor gene. Mutations in the Tas2r38 gene have resulted in independent loss of phenylthiocarbamide sensitivity in some humans, chimpanzees, and macaques (30, 31). Notably, the mutations in chimpanzees and macaques are start codon mutations, resembling the case with sea lion and fur seal Tas1r2s (30, 31).
Our evolutionary analysis has provided further evidence to support the view of convergent evolution of pseudogenized Tas1r2. Similar to many other vertebrate genes, we found that Tas1r2 is under an overall strong purifying selection. During evolution, several species at separate times may have changed their diet to a more carnivorous one. As the animal moved closer toward obligate carnivore it became increasingly likely that loss of the Tas1r2 gene would be well tolerated, reflected by the fact that the relaxation of functional constraint occurred in branches with a pseudogenized Tas1r2, although not completely.
Sweet Taste Receptor Genes and Taste Behavior.
Cats with a defective Tas1r2 gene do not prefer sweet carbohydrates and noncaloric sweeteners and probably cannot detect them. Is this also true for others? We addressed this question in the current study by behavioral taste-testing of the Asian small-clawed otter. Similar to the cat family Felidae, the two Asian otters we tested did not show a preference for sucrose at a concentration up to 800 mM, or for other sugars or artificial sweeteners. These data are limited in the number of animals tested and the number and kinds of behavioral tests conducted and, consequently, they warrant caution in interpretation. However, a preference for sweet sugars is a powerful and clear phenotype in most mammals studied, so we believe its absence in the Asian otter is striking. Thus, we tentatively conclude that the Asian otter is not capable of detecting compounds that taste sweet to humans, consistent with their lack of a functional sweet taste receptor Tas1r2/Tas1r3 and their carnivorous diet. In contrast, the spectacled bear, with an intact Tas1r2 gene, shows strong preferences for sugars and some nonnutritive sweeteners, consistent with the presence of a functional sweet taste receptor and with their omnivorous diet.
Major Taste Loss in Two Lineages of Aquatic Mammals.
In the present study, we found that the sweet, umami, and perhaps bitter taste receptor are all pseudogenized in dolphin, and the sweet and umami taste receptors are pseudogenized in sea lions (we have no data on bitter receptor structure in this species). Consistent with the genetic data, anatomical studies of the dolphin taste system revealed that only few taste bud-like structures are present in small pits in the root region of dolphin tongues (32). No buds are found in the canonical taste structures, including fungiform, foliate, and circumvallate papillae (32). Taste bud numbers are greatly reduced in sea lions as well (21). Only a few studies from two groups were conducted to examine the abilities of dolphins and sea lions to detect sour, bitter, salty, and sweet taste stimuli; umami was not examined in these studies (33, 34). Dolphins could not detect sucrose in one study (33) and showed reduced sensitivity in another study (34). Furthermore, dolphins detected only elevated levels of quinine (bitter), and a California sea lion showed no response to quinine sulfate but did respond to elevated levels of quinine monohydrochloride dihydate (0.40 parts per trillion; from 3 to ∼4 orders above human detection threshold) (33, 34). However, these data need to be interpreted cautiously because only a limited number of animals were examined. Nevertheless, these behavioral data agree largely with our molecular data. For instance, no sugar detection (or reduced sensitivity to sugar) correlates well with the pseudogenization of Tas1r2 and Tas1r3 receptor genes. The remaining response (if any) to sugar could be due to another property of sugar solutions, such as osmolarity or viscosity of sugars (13). Dolphins appeared to maintain some response to quinine at high concentration, but we failed to detect intact Tas2r receptor genes from the dolphin genome database. One explanation for this finding is that due to the low coverage, and therefore incompleteness, of the dolphin genome (2.59X), there may exist intact bitter receptor genes in the genome eluding initial genome sequence coverage. An alternative explanation is that quinine may become irritating at such high concentrations, and therefore elicit irritation responses (35). Nevertheless, with the reported greatly elevated threshold to quinine and the abundance of pseudogenized Tas2r receptor genes in the available genome database (10 of 10 genes), we predict that bitter taste function of dolphins is also greatly reduced, if not completely absent.
Recently, sweet, umami, and bitter taste receptors have been implicated in several extraoral functions (36). Pseudogenization of Tas1r receptor genes in dolphins and sea lions and Tas2r receptor genes in dolphin indicates that these receptors cannot be involved in extraoral (e.g., gut, pancreas) chemosensation (36) in these species. Thus, to the extent that these extraoral taste receptors are functionally significant in rodents and humans, these functions must have been assumed by other mechanisms in the species we have identified here with pseudogenized receptors. What these other mechanisms are remains to be determined, and further assessment of the relationships among taste receptor structure, dietary choice, and the associated metabolic pathways will lead to a better understanding of the evolution of diet and food choice as well as their mechanisms.
Materials and Methods
Details about collection of DNA samples, sequencing Tas1r2 from selected species within Carnivora, sequencing sea lion Tas1r1 and Tas1r3, data mining of the dolphin whole-genome shotgun assembly, evolutionary analysis, and taste testing as well as associated references are described in SI Appendix, Materials and Methods. Primers for amplification of taste receptors are listed in Table S1. Dataset for evolutionary analysis is provided in Dataset S1.
Supplementary Material
Acknowledgments
We thank Dr. Oliver Ryder and Leona Chemnick from the Zoological Society of San Diego/Conservation and Research for Endangered Species for providing the DNA samples of carnivores, the Naples Zoo at Caribbean Gardens for providing a fossa DNA sample, and Dr. Stephen O'Brien (National Cancer Institute) for providing DNA samples of a southern fur seal and a raccoon; Dr. Robert Zingg (Zoological Garden of Zurich) for allowing access to certain individual carnivores and for assistance with behavioral taste testing; and Dr. Joshua Plotkin for reading the manuscript. This work was made possible by National Institutes of Health Grant DC010842 (to P.J.) and institutional funds from the Monell Chemical Senses Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.T.D. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. JN130349–JN130360 (sea lion, fur seal, Pacific Harbor seal, Asian small-clawed otter, spotted hyena, fossa, banded linsang, aardwolf, Canadian otter, spectacled bear, raccoon, and red wolf Tas1r2 sequences, respectively); JN413105 (sea lion Tas1r1); JN413106 (sea lion Tas1r3); JN622015 (dolphin Tas1r1); JN622016 (dolphin Tas1r2); JN622017 (dolphin Tas1r3); and JN622018–JN622027 (dolphin Tas2rs)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118360109/-/DCSupplemental.
References
- 1.Bachmanov AA, Beauchamp GK. Taste receptor genes. Annu Rev Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmanov AA, et al. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses. 2001;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Max M, et al. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 4.Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- 5.Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 6.Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 7.Chandrashekar J, et al. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 8.Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 9.Go Y, Satta Y, Takenaka O, Takahata N. Lineage-specific loss of function of bitter taste receptor genes in humans and nonhuman primates. Genetics. 2005;170:313–326. doi: 10.1534/genetics.104.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liman ER. Use it or lose it: Molecular evolution of sensory signaling in primates. Pflugers Arch. 2006;453:125–131. doi: 10.1007/s00424-006-0120-3. [DOI] [PubMed] [Google Scholar]
- 11.Shi P, Zhang J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol. 2006;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
- 12.Bartoshuk LM, Jacobs HL, Nichols TL, Hoff LA, Ryckman JJ. Taste rejection of nonnutritive sweeteners in cats. J Comp Physiol Psychol. 1975;89:971–975. doi: 10.1037/h0077172. [DOI] [PubMed] [Google Scholar]
- 13.Beauchamp GK, Maller O, Rogers JG. Flavor preferences in cats (Felis catus and Panthera sp.) J Comp Physiol Psychol. 1977;91:1118–1127. [Google Scholar]
- 14.Li X, et al. Pseudogenization of a sweet-receptor gene accounts for cats’ indifference toward sugar. PLoS Genet. 2005;1:27–35. doi: 10.1371/journal.pgen.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, et al. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol Biol Evol. 2010;27:2642–2650. doi: 10.1093/molbev/msq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganchrow JR, Steiner JE, Bartana A. Behavioral reactions to gustatory stimuli in young chicks (Gallus gallus domesticus) Dev Psychobiol. 1990;23:103–117. doi: 10.1002/dev.420230202. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Elias DJ, Shumake SA, Gaddis SE. Taste preferences of the common vampire bat (Desmodus-Rotundus) J Chem Ecol. 1982;8:715–721. doi: 10.1007/BF00988313. [DOI] [PubMed] [Google Scholar]
- 18.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pontius JU, et al. Agencourt Sequencing Team; NISC Comparative Sequencing Program Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhen MD. Evolution of marine mammals: Back to the sea after 300 million years. Anat Rec (Hoboken) 2007;290:514–522. doi: 10.1002/ar.20545. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura K, Shindoh J, Kobayashi K. Scanning electron microscopy study of the tongue and lingual papillae of the California sea lion (Zalophus californianus californianus) Anat Rec. 2002;267:146–153. doi: 10.1002/ar.10093. [DOI] [PubMed] [Google Scholar]
- 22.Miller D. Seals and Sea Lions. St. Paul, MN: Voyageur; 1998. p. 72. [Google Scholar]
- 23.Werth AJ. Adaptations of the cetacean hyolingual apparatus for aquatic feeding and thermoregulation. Anat Rec (Hoboken) 2007;290:546–568. doi: 10.1002/ar.20538. [DOI] [PubMed] [Google Scholar]
- 24.Li X, et al. Analyses of sweet receptor gene (Tas1r2) and preference for sweet stimuli in species of Carnivora. J Hered. 2009;100(Suppl 1):S90–S100. doi: 10.1093/jhered/esp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowak RM. Walker's Carnivores of the World. Baltimore: Johns Hopkins Univ Press; 2005. [Google Scholar]
- 26.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flynn JJ, Finarelli JA, Zehr S, Hsu J, Nedbal MA. Molecular phylogeny of the carnivora (mammalia): Assessing the impact of increased sampling on resolving enigmatic relationships. Syst Biol. 2005;54:317–337. doi: 10.1080/10635150590923326. [DOI] [PubMed] [Google Scholar]
- 28.Yu L, et al. Phylogenetic utility of nuclear introns in interfamilial relationships of Caniformia (order Carnivora) Syst Biol. 2011;60:175–187. doi: 10.1093/sysbio/syq090. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki N, et al. Identification of non-taster Japanese macaques for a specific bitter taste. Primates. 2010;51:285–289. doi: 10.1007/s10329-010-0209-3. [DOI] [PubMed] [Google Scholar]
- 31.Wooding S, et al. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimura K, Kobayashi K. A comparative morphological study on the tongue and the lingual papillae of some marine mammals-particularly of four specis of Odontoceti and Zalophus. Odontology. 1997;85:385–507. [Google Scholar]
- 33.Kuznetsov VB. A method of studying chemoreception in the Black Sea bottlenose dolphin (Tursiops truncatus) In: Sokolov VY, editor. Morfologiya, Fiziologiya I Akustika Morskikh Mlekopitayushchikh. Moscow: Nauka; 1974. [Google Scholar]
- 34.Friedl WA, et al. In: Taste Reception in the Pacific Bottlenose Dolphin (Tursiops Truncatus Gilli) and the California Sea Lion (Zalophus Californianus). Sensory Abilities of Cetaceans: Laboratory and Field Evidence, NATO ASI Series. Thomas JA, Kastelein RA, editors. New York: Plenum; 1990. pp. 447–454. [Google Scholar]
- 35.Liu L, Simon SA. Responses of cultured rat trigeminal ganglion neurons to bitter tastants. Chem Senses. 1998;23:125–130. doi: 10.1093/chemse/23.2.125. [DOI] [PubMed] [Google Scholar]
- 36.Behrens M, Meyerhof W. Gustatory and extragustatory functions of mammalian taste receptors. Physiol Behav. 2011;105:4–13. doi: 10.1016/j.physbeh.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.