Abstract
Proteins of the Drosophila behavior/human splicing (DBHS) family include mammalian SFPQ (PSF), NONO (p54nrb), PSPC1, and invertebrate NONA and Hrp65. DBHS proteins are predominately nuclear, and are involved in transcriptional and posttranscriptional gene regulatory functions as well as DNA repair. DBHS proteins influence a wide gamut of biological processes, including the regulation of circadian rhythm, carcinogenesis, and progression of cancer. Additionally, mammalian DBHS proteins associate with the architectural long noncoding RNA NEAT1 (Menε/β) to form paraspeckles, subnuclear bodies that alter gene expression via the nuclear retention of RNA. Here we describe the crystal structure of the heterodimer of the multidomain conserved region of the DBHS proteins, PSPC1 and NONO. These proteins form an extensively intertwined dimer, consistent with the observation that the different DBHS proteins are typically copurified from mammalian cells, and suggesting that they act as obligate heterodimers. The PSPC1/NONO heterodimer has a right-handed antiparallel coiled-coil that positions two of four RNA recognition motif domains in an unprecedented arrangement on either side of a 20-Å channel. This configuration is supported by a protein:protein interaction involving the NONA/paraspeckle domain, which is characteristic of the DBHS family. By examining various mutants and truncations in cell culture, we find that DBHS proteins require an additional antiparallel coiled-coil emanating from either end of the dimer for paraspeckle subnuclear body formation. These results suggest that paraspeckles may potentially form through self-association of DBHS dimers into higher-order structures.
Keywords: gene regulation, nuclear organization, protein structure, RNA-binding proteins
The Drosophila behavior/human splicing (DBHS) proteins have been described as multifunctional nuclear proteins, implicated in subnuclear body formation; transcription initiation; coactivation and corepression; constitutive and alternative splicing; transcriptional termination; and DNA repair (1) and contributing to circadian rhythm regulation (2, 3), tumor suppression (4), and Drosophila behavior (5). Mammalian genomes encode three paralogous DBHS proteins: NONO, SFPQ, and PSPC1, whereas invertebrates encode one or two. NONO and SFPQ are highly abundant, expressed in a variety of cell lines and tissues (1). In contrast, PSPC1 is more selectively expressed, found at low levels in HeLa cells, but expressed at equal levels with NONO and SFPQ in sertoli cells of the testis, acting as a coactivator of transcription (6).
Mammalian DBHS proteins are essential for the formation of subnuclear paraspeckles (7), where they interact with a long-noncoding RNA, NEAT1 (8–10), and influence its spatial arrangement (11). Paraspeckle formation is also dependent upon transcription of NEAT1, which acts as a structural scaffold, nucleating the bodies at least in part by recruiting DBHS proteins (12, 13). DBHS proteins functioning within paraspeckles bind to adenosine-to-inosine edited inverted-repeat hairpins in the 3′ UTR of specific mRNAs (13–15). This type of RNA is retained in the nucleus and sequestered in paraspeckles (16).
All DBHS proteins have a core conserved region of ∼300 aa. Their N- and C-terminal extensions differ giving total lengths of 400 (NONO) to >700 aa (SFPQ), with this extra polypeptide consisting primarily of polyproline/glutamine-rich low-complexity regions and functional sequences including nuclear localization signals. The DBHS core consists of clearly defined structural domains: two tandem RNA recognition motif (RRM) domains and a 100-aa segment of predicted coiled-coil (Fig. 1A and Fig. S1)—a putative protein:protein interaction motif. Between the second RRM and the coiled-coil lies a previously uncharacterized 52-aa conserved domain, NONA/paraspeckle (NOPS), which is definitive of the DBHS family. DBHS proteins form homo- and heterodimers via their core domains (17, 18) and have been shown to interact with nucleic acids through their RRM domains (1). Though RRM1 of DBHS proteins is canonical (containing four aromatic residues at conserved positions that are typically essential for RNA binding) (19), RRM2 is considered noncanonical: three of these conserved residues are substituted to Thr, Lys, and Ile, implying that either the RRM2s do not bind RNA, or that they bind it in an unexpected manner. Nevertheless, the residues at these positions within both RRMs are important for localizing DBHS proteins to paraspeckles (18).
Fig. 1.
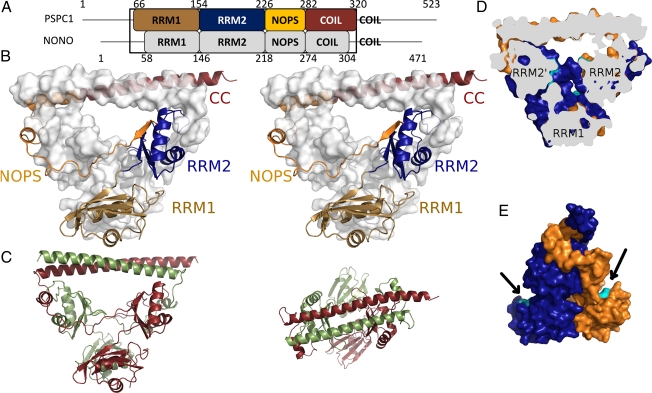
Structure of a PSPC1/NONO heterodimer. (A) Domain structure indicating RNA recognition motifs 1 and 2, NOPS domain, and coiled-coil. (B) Stereoview of PSPC1 (cartoon) and NONO (surface). (C) Side (Left) and top (Right) views of PSPC1 (red) and NONO (green). (D) Cutaway view revealing voids (putative RNA-binding residues, cyan). (E) Side view highlighting putative RNA-binding residues of RRM1 (cyan).
To shed light on how DBHS proteins mediate their specific functions in their many facets of gene regulation via protein:protein and protein:RNA interaction domains, we determined the structure of a heterodimer formed by the homologous conserved regions of PSPC1 and NONO. We have used this to guide further mutagenesis and fluorescence microscopy experiments to gain insight into the requirements for assembly of paraspeckles.
Results and Discussion
Pseudosymmetric Structure of a Truncated PSPC1/NONO Heterodimer.
The structure of the human PSPC1/NONO heterodimer encompasses four domains: RRM1, RRM2, NOPS, and coiled-coil. We previously showed that recombinant coexpression of these truncated proteins in Escherichia coli resulted in soluble heterodimeric complex (18), and full details of crystallization and X-ray data collection are provided elsewhere (20). Crystals prepared using a sample consisting of residues 61–320 of PSPC1 and 53–312 of NONO resulted in a structure containing one heterodimer per asymmetric unit (PDB ID code 3SDE; Fig. 1B and Movie S1). Small-angle X-ray scattering (SAXS) studies on an identical sample showed a close match between observed solution scattering and that calculated from our crystallographic model (Fig. S2).
The 70% sequence identity of the truncated proteins is reflected in striking twofold pseudosymmetry for the C-terminal part of the structure (Fig. 1C; rmsd 0.96 Å for 163 Cα pairs). In contrast, the two N-terminal RRM1 domains break the pseudosymmetry, but individually superimpose well on each other (rmsd 0.94 Å for 80 Cα pairs). Approximately 25% (4,500 Å2) of the solvent-accessible area of each monomer is buried as a result of heterodimer formation, and 43% of the residues of each monomer are directly involved in the interface as determined with PISA (21). The interaction interface is distributed in an extended manner throughout the whole protein, with all four domains contributing. The intimate, largely hydrophobic interaction suggests that these proteins are obligate dimers, although a gap volume index value of 2.25 (22) is typical of transient protein:protein interactions, suggesting dynamic exchange of dimer partners. Such exchange would be consistent with the evolution in vertebrates of multiple DBHS proteins, diversifying the functional repertoire of this family with various hetero- and homodimer combinations.
It has been noted that these proteins could not be functionally separated for assays such as paraspeckle targeting (18), a fact consistent with the intimate interface we observe. This observation is resoundingly significant for all experiments probing the function of specific DBHS proteins: it is essential to address the likelihood that each protein is dynamically involved in both homo- and heterodimers in cells. For example, the roles of NONO (2) and SFPQ (3) in regulating circadian rhythms were described separately, when it is likely that the NONO/SFPQ heterodimer is significantly involved in this process.
Observed Arrangement of Four Different RRM Domains Is Unprecedented.
The arrangement of four different RRM domains observed in the heterodimer is unique in known structures (Fig. 1B and Fig. S3). Of 30 structures containing tandem RRMs, only monomeric Raver (23) bears a superficial similarity in the arrangement of tandem RRMs with a monomer of PSPC1 or NONO. Structures with tandem RRM domains commonly act independently (as beads on a string), although the binding of nucleic acid can induce order (e.g., HuD:ARE complex) (24). Indeed, the only previous structures of oligomeric tandem RRM proteins [UP1:tDNA (25) and FUSE:FIR (26)] require the presence of nucleic acid to dimerize. The PSPC1/NONO dimer, however, forms its highly constrained structure in the absence of nucleic acid. Furthermore, homodimeric RRM structures [e.g., EPABP2 (27) and RBM12 (PDB ID 2EK6)] dimerize directly via their RRMs. Conversely, and intriguingly, the two pseudosymmetrically related noncanonical RRM2 domains of PSPC1/NONO form either side of a 20-Å solvent-filled channel at the center of the structure, and the two canonical RRM1 domains face outward (Fig. 1 D and E).
Predicting the RNA-binding mode of noncanonical RRMs is notoriously difficult (28), but we observe that the RRM2 arrangement is reminiscent of other oligomeric proteins that bind structured DNA: dimeric endonucleases recognize their double-stranded DNA substrates by placing pairs of nuclease domains at appropriate positions (29). Thus, we hypothesize that the highly unusual spatial arrangement of RRM domains reflects the complexity of unstructured and structured RNA substrates these proteins interact with. For example, the long noncoding RNA (lncRNA) NEAT1 contains regions of secondary structure, and inverted Alu repeats form hairpins depending on the extent of A-to-I editing. The twofold pseudosymmetric arrangement of RRMs in the heterodimer may be able to accommodate this duplex nucleic acid.
Definitive NOPS Domain Is a Unique Protein:Protein Interaction Motif.
The structure shows that the NOPS domain, characteristic of DBHS proteins, is unique in known protein:protein interaction domains and cannot be considered an independent structural entity (Fig. 2A). The role of the NOPS domain is to specifically bind to RRM2 of the partner DBHS protein (RRM2′, prime indicating the partner subunit), forming a substantial interaction surface (65% of the total dimer interface). This interaction also has a bearing on the structure of RRM2, which has atypical β-hairpins in loops 3 and 5 presenting hydrogen bonds to short β-strands in NOPS′ (Fig. 1C and Fig. S4). Loop 3 is variable in length in RRM domains, often forming an important extension to the RNA interaction interface as exemplified by the U1:U1A (30) and CUG repeat binding protein:RNA (31) complexes. In PSPC1/NONO, the NOPS domain interacts with the opposite side of the RRM's β-sheet to the canonical RNA-binding surface, leaving it free for further interactions.
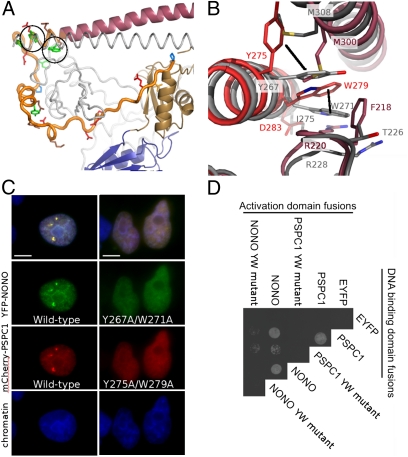
Fig. 2.
Conserved residues in the NOPS domain (NONO: Y267, W271; PSPC1: Y275, W279) are critical for dimerization and paraspeckle localization. (A) The NOPS domain of PSPC1 (yellow) with Y275 and W279 circled. (B) Conserved Tyr and Trp residues have radically different side-chain conformations in NONO and PSPC1. (C) Wild-type Tyr and Trp, and not Ala, are required for paraspeckle formation by full-length YFP-NONO and mCherry-PSPC1 fusion proteins in HeLa cells. (Scale bars: 5 μm.) (D) Wild-type Tyr and Trp, and not Ala, are necessary for hetero- and homodimer formation in yeast two-hybrid analyses of full-length PSPC1/NONO (EYFP as negative control).
Highly conserved residues at the C-terminal end of NOPS are involved in a complex cluster of hydrophobic interactions with both RRM2′ and the distal region of CC′ (Fig. 2 A and B). We therefore tested the effect of alanine mutations at two highly conserved positions (Tyr275 and Trp279; PSPC1 numbering) on full-length protein:protein interactions using the yeast two-hybrid system, and on protein localization in fluorescence microscopy paraspeckle-targeting assays (Fig. 2 C and D). We show that the mutants cannot interact with wild-type proteins or localize to paraspeckles, supporting our hypothesis that these residues are critical for functional DBHS dimerization. Interestingly, the observed structural conformations of Tyr275 and Trp279 in PSPC1 and NONO are unexpectedly different (Fig. 2B and Fig. S5), suggesting that structural plasticity associated with these residues may influence choice of homo- or heterodimerization partner in vivo.
Extended C-Terminal Coiled-Coil Motif Is Essential for Paraspeckle Localization.
The final 40–50 aa observed in each protein (270–320 in PSPC1) form an antiparallel coiled-coil, clamped at both ends by interactions with RRM2 and NOPS’ domains. A pronounced right-handed twist (Figs. 1B and 3A) is consistent with the observed hendecad (11mer) sequence repeat, rather than a heptad repeat observed in more common left-handed coiled coils (Fig. 3 A and B). To our knowledge, this right-handed antiparallel heterodimeric coiled-coil is unique. Although coiled coils play a variety of roles in protein structure and cellular function (32), only a limited number of proteins use the coiled-coil as a brace to place paired nucleic acid binding domains. A particularly striking example is the pseudosymmetric arrangement observed in a Mycoplasma type 1 restriction enzyme (PDB ID code 1YDX) (33). The structure described here is an example of such an arrangement being observed in an RRM-containing protein.
Fig. 3.
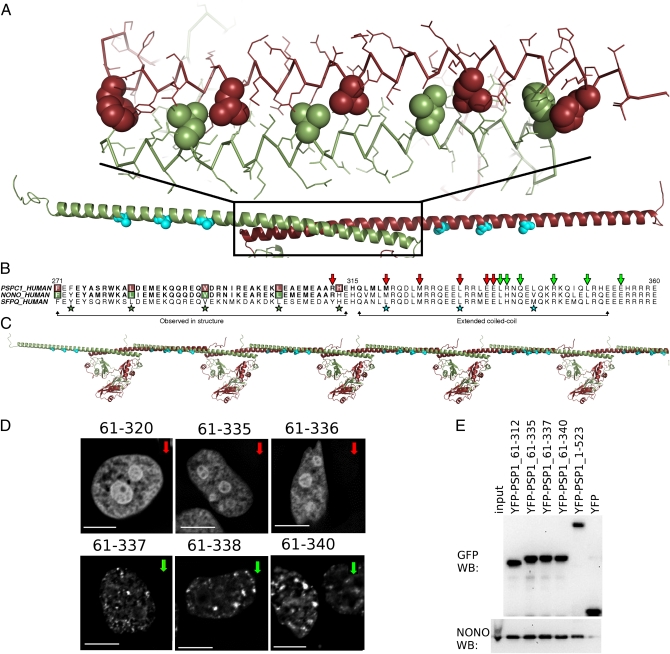
Coiled-coil formation is critical for paraspeckle targeting by DBHS proteins. (A) Observed coiled-coil (atomic model) and model of extended coiled-coil (cyan residues indicate hendecad repeat). (B) Sequence alignment of PSPC1, NONO, and SFPQ coiled coils; green arrows mark C termini of truncated PSPC1 proteins able to form paraspeckles; red arrows, those that cannot. (C) A schematic model for coiled-coil-mediated oligomerization of DBHS proteins. (D) YFP fusion proteins of PSPC1 proteins terminating after residue 337 colocalize with paraspeckles in HeLa cells; shorter proteins do not. (Scale bars: 5 μm.) (E) Wild-type endogenous NONO is coimmunoprecipitated by truncated YFP-PSPC1 proteins in an anti-GFP pull-down. Anti-GFP and anti-NONO Western blots.
What of the coiled-coil beyond that observed in our structure? It was necessary for us to test a number of C-terminal truncations of the proteins to obtain crystals (34), and we were aware that the coiled-coil was predicted to resume, after a five-residue stutter, for 40 residues beyond the crystallized C terminus (Fig. 3B). Furthermore, the first 42 aa of this additional region are highly conserved across the family (81% identity among human paralogues compared with ∼70% for the entire conserved region). Thus, we chose to investigate the significance of C-terminal truncation of PSPC1 to its localization to paraspeckles in HeLa cells with an established fluorescence microscopy assay (18) (Fig. 3D). Having previously shown that PSPC1 truncated at position 358 locates to paraspeckles (18), we investigated the same protein as crystallized (PSPC1 61–320) and found that it was not located in paraspeckles (Fig. 3D), suggesting a key role for the extended coiled-coil domain in paraspeckle targeting. A panel of PSPC1 truncation mutants, with increasingly long coiled-coil domains, revealed a sharp transition from diffuse nuclear localization to paraspeckle localization: PSPC1 61–337 is found in paraspeckles, whereas PSPC1 61–336 is not (Fig. 3D and Fig. S6). Nevertheless, coimmunoprecipitation experiments confirmed that all PSPC1 truncations, regardless of localization, interact with NONO (Fig. 3E), presumably as dimers.
Although a number of reasons may explain the sharp transition in localization, including disruption of a posttranslational modification, or a binding site for another protein or RNA, we hypothesize that a specific coiled-coil-mediated oligomerization is being interrupted by these truncations. Given that we previously showed that proteins including only the NOPS and coiled-coil of PSPC1 form nonparaspeckle nuclear fibrils (18), and that overexpressed protein samples incorporating this additional coiled-coil display poor solubility (34), it is possible that an interaction between these extended coiled coils allows DBHS dimers to form longer oligomers (Fig. 3C). Thus, paraspeckle assembly may be controlled in a manner consistent with other nuclear bodies, stress granules, and processing bodies for which protein self-association is important (35–37).
In conclusion, our structural and molecular biology characterization of the core conserved domains of PSPC1 and NONO expands the catalog of possible arrangements of RRM domains. The structure showcases the functional flexibility of this common module, needed to allow binding of unusual RNA substrates, such as the lncRNA NEAT1 or a retained, hyperedited RNA. The structure further provides a solid framework on which to interpret the activities of other important members of the DBHS family (SFPQ, NONA, and Hrp65). We also provide evidence that crucial interactions between NOPS and RRM2 domains dictate the formation of intimate DBHS protein heterodimers with significant consequences for their functional repertoire in vivo. Finally, by implicating an additional highly conserved coiled-coil region in localization of these proteins to subnuclear bodies, we provide insights into the complex molecular interactions that drive nuclear organization and function.
Methods
Expression, Purification, and Crystallization of PSPC1/NONO Heterodimer.
Recombinant PSPC1/NONO production, crystallization, and X-ray data collection at beamline MX2 of the Australian Synchrotron, of both native and selenomethionine forms, is described in detail elsewhere (20). See SI Methods for details.
X-Ray Structure Determination and Refinement.
Selenomethionine multiwavelength anomalous dispersion phasing and initial model building was performed with PHENIX (38) and resulted in modeling of 35% of the complete structure. The model was completed manually in COOT (39), cycled with refinement against the native data in REFMAC5 (40, 41). Water and ethylene glycol molecules were added before final rounds of refinement with BUSTER-TNT (42). Progress of refinement was monitored using R-free, the validation tools available in COOT, and MOLPROBITY (43). Refinement against native data to 1.9-Å resolution converged with residuals R = 0.183 and Rfree = 0.228. The asymmetric unit contains a single heterodimer, residues 66–320 of PSPC1 and residues 66–304 of NONO (494 aa), 470 waters, and 11 ethylene glycol molecules. The model has excellent geometry, with 98.6% of residues in most favored regions of the Ramachandran plot (Table S1 and Fig. S7).
Molecular graphical analysis was performed using PyMol (version 1.2r3pre; Schrödinger, LLC) Sequence alignments were produced with ALINE (44) and topology diagrams with TOPDRAW (45).
SAXS.
SAXS experiments were carried out at the SAXSWAXS beamline of the Australian Synchrotron on a concentration series (0.375–12 mg/mL) of protein sample. Data were analyzed with the ATSAS suite (46, 47). See SI Methods and Fig. S2 for details.
Plasmids for Fluorescent Protein Fusions.
Plasmids for fluorescent protein fusion experiments were generated using the pEYFP-C1 plasmid (Clontech) and standard molecular biology techniques. See SI Methods for details.
Localization Assays and Immunoprecipitation.
PSPC1 localization assays and immunoprecipitation experiments were carried out as previously described (18). See SI Methods for details.
Yeast Two-Hybrid Analyses.
Yeast two-hybrid experiments were carried out using the GAL4 DNA-binding domain fusions in pGBK-RC and GAL4 activation domain fusions in pGAD-RC, respectively, using the lithium acetate method according to Gietz and Woods (48). See SI Methods for details.
Supplementary Material
Acknowledgments
We thank Tom Caradoc-Davies, Nathan Cowieson, Christine Gee, Nigel Kirby, and Adrian Hawley. Aspects of this research were undertaken on the Macromolecular Crystallography and SAXSWAXS beamlines at the Australian Synchrotron (Victoria, Australia). Yeast two-hybrid plasmids and yeast strains were a kind gift from T. Ito (University of Tokyo). Support for this work was provided by National Health and Medical Research Council of Australia Project Grant 513880 (to C.S.B. and A.H.F.) and a fellowship (M.L.); a fellowship from the Western Australian Institute for Medical Research (A.H.F.); and a University of Western Australia scholarship (D.M.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3SDE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120792109/-/DCSupplemental.
References
- 1.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 2.Brown SA, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 3.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang G, Cui Y, Zhang G, Garen A, Song X. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc Natl Acad Sci USA. 2009;106:16794–16798. doi: 10.1073/pnas.0909022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanewsky R, Rendahl KG, Dill M, Saumweber H. Genetic and molecular analysis of the X chromosomal region 14B17-14C4 in Drosophila melanogaster: Loss of function in NONA, a nuclear protein common to many cell types, results in specific physiological and behavioral defects. Genetics. 1993;135:419–442. doi: 10.1093/genetics/135.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuwahara S, et al. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol Reprod. 2006;75:352–359. doi: 10.1095/biolreprod.106.051136. [DOI] [PubMed] [Google Scholar]
- 7.Fox AH, et al. Paraspeckles: A novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 8.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bond CS, Fox AH. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 17.Kiesler E, Miralles F, Ostlund Farrants AK, Visa N. The Hrp65 self-interaction is mediated by an evolutionarily conserved domain and is required for nuclear import of Hrp65 isoforms that lack a nuclear localization signal. J Cell Sci. 2003;116:3949–3956. doi: 10.1242/jcs.00690. [DOI] [PubMed] [Google Scholar]
- 18.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 20.Passon DM, Lee M, Fox AH, Bond CS. Crystallization of a paraspeckle protein PSPC1-NONO heterodimer. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1231–1234. doi: 10.1107/S1744309111026212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Nooren IM, Thornton JM. Diversity of protein–protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JH, Rangarajan ES, Yogesha SD, Izard T. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure. 2009;17:833–842. doi: 10.1016/j.str.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 25.Ding J, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crichlow GV, et al. Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition. EMBO J. 2008;27:277–289. doi: 10.1038/sj.emboj.7601936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J, McGivern JV, Nichols KW, Markley JL, Sheets MD. Structural basis for RNA recognition by a type II poly(A)-binding protein. Proc Natl Acad Sci USA. 2008;105:15317–15322. doi: 10.1073/pnas.0801274105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cléry A, Blatter M, Allain FH. RNA recognition motifs: Boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Middleton CL, Parker JL, Richard DJ, White MF, Bond CS. Substrate recognition and catalysis by the Holliday junction resolving enzyme Hje. Nucleic Acids Res. 2004;32:5442–5451. doi: 10.1093/nar/gkh869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katsamba PS, Bayramyan M, Haworth IS, Myszka DG, Laird-Offringa IA. Complex role of the beta 2-beta 3 loop in the interaction of U1A with U1 hairpin II RNA. J Biol Chem. 2002;277:33267–33274. doi: 10.1074/jbc.M200304200. [DOI] [PubMed] [Google Scholar]
- 31.Tsuda K, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 33.Calisto BM, et al. Crystal structure of a putative type I restriction-modification S subunit from Mycoplasma genitalium. J Mol Biol. 2005;351:749–762. doi: 10.1016/j.jmb.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 34.Lee M, Passon DM, Hennig S, Fox AH, Bond CS. Construct optimization for studying protein complexes: Obtaining diffraction-quality crystals of the pseudosymmetric PSPC1-NONO heterodimer. Acta Crystallogr D Biol Crystallogr. 2011;67:981–987. doi: 10.1107/S0907444911039606. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 36.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Z, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 43.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 45.Bond CS. TopDraw: A sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19:311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]
- 46.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 47.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 48.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.