Abstract
In honey bees (Apis mellifera), the development of a larva into either a queen or worker depends on differential feeding with royal jelly and involves epigenomic modifications by DNA methyltransferases. To understand the role of DNA methylation in this process we sequenced the larval methylomes in both queens and workers. We show that the number of differentially methylated genes (DMGs) in larval head is significantly increased relative to adult brain (2,399 vs. 560) with more than 80% of DMGs up-methylated in worker larvae. Several highly conserved metabolic and signaling pathways are enriched in methylated genes, underscoring the connection between dietary intake and metabolic flux. This includes genes related to juvenile hormone and insulin, two hormones shown previously to regulate caste determination. We also tie methylation data to expressional profiling and describe a distinct role for one of the DMGs encoding anaplastic lymphoma kinase (ALK), an important regulator of metabolism. We show that alk is not only differentially methylated and alternatively spliced in Apis, but also seems to be regulated by a cis-acting, anti-sense non–protein-coding transcript. The unusually complex regulation of ALK in Apis suggests that this protein could represent a previously unknown node in a process that activates downstream signaling according to a nutritional context. The correlation between methylation and alternative splicing of alk is consistent with the recently described mechanism involving RNA polymerase II pausing. Our study offers insights into diet-controlled development in Apis.
Keywords: polyphenism, ubiquitin, spliceosome
As in other eusocial insects, the defining feature of honey bee (Apis mellifera) biology is reproductive division of labor between female members of the colony (1). This remarkable evolutionary invention involves postembryonic epigenetic reprogramming of global gene expression by dietary cues in growing larvae to generate two distinct female “castes” from a single genome (2–5). Nurse bees use differential feeding with royal jelly to determine the developmental trajectory of a newly hatched female so that it develops into either a long-lived and highly reproductive queen or a short-lived functionally sterile worker bee (6, 7).
Larval differentiation is an incremental process that proceeds over a period of 6 d by using discrete switches that activate both hormonal signaling and the expression of many genes belonging to several distinct functional categories (4). Initially, the growing larva retains a certain degree of plasticity, but it is widely believed that after 3 d of growth larval commitment to a given trajectory is irreversible (6), suggesting that caste determination in honey bee larvae is a multistep, threshold-based process. Although the capacity of the honey bee genome to respond to an external stimulus by a developmental reaction is ultimately under genetic control, the mechanism by which a growing larva makes a choice between two alternative developmental pathways is not well understood.
Recently some of us reported that the queen phenotype can also be induced without the nutritional signals from royal jelly by interfering with postembryonic de novo methylation of DNA in growing larvae (8). Silencing DNA methyl-transferase 3 in newly hatched embryos results in the majority of the treated individuals emerging from the pupal stage as queens, indicating that differential methylation is essential to epigenetic control of caste determination. Our goal in the present study was to characterize the dynamics of context-dependent methylation patterns during caste determination in Apis by contrasting methylomes from larval queen and worker head with previously published results from adult queen and worker brain methylomes and relating them to coordinated changes in metabolism and gene expression.
We report that 2,399 of 6,086 methylated genes show differential patterns of methylation (DMG) in queen and worker larvae, compared with only 560 in adult brain. We characterize in detail one of the DMGs coding for anaplastic lymphoma kinase (ALK) and propose that this important regulator of metabolism is a key player in linking the nutritional environment with downstream signaling in Apis by modulating nutrient-independent activation of the phosphatidylinositol 3-kinases (PI3K) cascade. Our results tie genome-wide methylation signatures to gene expression and hormonal regulation and suggest how dietary changes might affect the modulation of metabolic fluxes and signal transduction via epigenetic machinery. In a broader context, our study highlights the context-dependent role of DNA methylation in the dynamic process of orchestrating the complex interplay between environment and genome to determine phenotypic outcomes.
Results
Characterization and Comparative Analyses of Larval Methylomes.
We used DNA extracted from 96 ± 1-h-old larval heads to map at single-base resolution the genome-wide methylation patterns in queens and workers. This developmental stage represents the critical time point at which caste-specific differentiation is essentially irrevocable (6). The sequencing of bisulfite-converted larval DNA yielded a dataset of 145,193,761 and 119,560,692 raw reads (from 36 to 76 bp) for queens and workers, respectively, with 90,114,079 and 81,417,785 of these reads mapped to unique genomic regions. After filtering, the median number of reads covering each CpG was 26 and 25.
To determine whether larval methylomes are unique or similar to previously characterized adult honey bee methylomes (9), we compared our results with previously published adult brain methylation patterns in both queens and workers (Table 1). We found that the total number of methylated genes is virtually the same: 6,086 methylated genes in larval head and 5,854 in adult brain. The small difference in the number of methylated genes might reflect the higher sequencing coverage of the larval methylome, allowing for more confidence in making methylation calls. We also found that the total number of methylated cytosines (mCs) is higher in larval head than in adult brain (Table 1). This difference is due to additional worker-specific sites: in larvae there were 9,143 mCs unique to queen larvae, 25,560 mCs unique to worker larvae, and 66,325 mCs found in both castes (Table 1). As in the brain, mCs were located predominantly in exons.
Table 1.
Comparison of adult brain and larval methylomes in queens and workers
| % upmethylated DMGs |
|||||||||
| No. of methylated genes |
No. of mCs |
No. of DMGs |
Adult brain |
Larvae head |
|||||
| Adult brain | Larvae head | Adult brain | Larvae head | Adult brain | Larvae head | Q | W | Q | W |
| 5,854* | 6,086* | 70,000 | 100,000 | 561 | 2,399 | 44 | 56 | 18 | 82 |
Q, queen; W, worker.
*A small number of Apis genes seem to be methylated only in one tissue (420 in brain and 510 in larvae). However, it was not possible to determine whether these are genuine cases of genes methylated in a tissue-specific manner or artifacts caused by uneven coverage and/or filtration issues.
The key difference between larval and adult methylomes was the number of DMGs; we detected 2,399 in larvae (Table S1), compared with only 561 in adult brain (Table 1). This striking difference is consistent with the fundamental difference between adult brain and larval tissues. Brain cells are postmitotic (nondividing), whereas development of larval tissues is associated with high growth rate and protein turnover. Another possible explanation is that queen and worker adult brains differ less in functionality relative to queen and worker larvae.
The vast majority of larval DMGs (1,967 or 82%) were up-methylated in workers. We expected that queens would have lower levels of methylation because silencing de novo DNA methylation in larvae results in the queen phenotype. In accord with Lyko et al. (9), all methylated genes showed higher levels of evolutionary conservation than nonmethylated genes (Table S1). Another characteristic feature of methylated genes was the type of functions associated with conserved domains found in their products. The most common were DNA-binding domains such as helicases, zinc fingers, RNA recognition motifs, and certain protein-binding motifs, including the WD β-transducin repeat implicated in coordinating multiprotein assemblies In total, 31 evolutionarily conserved domains were over-represented in methylated genes (Table S2).
Critical Metabolic Networks in Apis Are Enriched in Methylated Genes.
Given the central role of nutritional cues in epigenetic control of larval development, we hypothesized that key metabolic pathways in Apis should be enriched in methylated genes. To test this idea we mapped the Apis proteins to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and refined these annotations using orthology with Drosophila and human genes. The combined mapping results were further refined using National Center for Biotechnology Information annotations. Using this approach we found enrichment in methylated genes in a few functionally connected groups of genes belonging to well-characterized pathways, namely the tricarboxylic acid (TCA) cycle, the ubiquitin–proteasome pathway, the inositol phosphate/TOR (target of rapamycin) pathway, the spliceosome, and the cell division and cytoskeletal network (Table 2). The enrichment of these essential gene networks in methylated genes is consistent with previous findings that methylated genes in Apis are not only highly conserved evolutionarily but also provide essential “housekeeping” functions required by virtually every type of cell (10, 11). In addition, many genes encoding hormone receptors, and all genes implicated in responses to juvenile hormone (JH), were found to be methylated or differentially methylated (Table S1).
Table 2.
Central metabolic pathways in Apis are enriched in methylated genes
| Pathway | Estimated no. of genes | No. of methylated genes | No. of DMGs |
| TCA and related reactions | 43 | 32 (75) | 13 (30) |
| TOR/PI3/insulin | 87 | 66 (76) | 38 (44) |
| Ubiquitin–proteasome | 108 | 96 (89) | 46 (43) |
Values in parentheses are percentages. More details in SI Materials and Methods.
TCA cycle.
The majority of Apis genes in the TCA network (75%) were methylated (Table 2 and Table S3), and in most cases the nonmethylated genes were paralogs of a methylated gene that provides a core activity in a given complex. Fig. S1 shows our annotation of the TCA cycle in Apis and highlights all methylated and differentially methylated genes.
In the context of epigenetic regulation, the differential methylation of the gene coding for ATP citrate lyase (ATPCL) in larvae is particularly interesting. This important enzyme, which converts citrate released from mitochondria to cytosol to acetyl-CoA, has recently been identified as a key protein linking cellular metabolism to epigenetic regulation via histone acetylation. It has been shown that ATPCL activity is required to link growth factor-induced increases in nutrient metabolism to the regulation of histone acetylation and gene expression (12). This finding adds more weight to the notion that components of royal jelly with histone deacetylase inhibitory activities (13) might play a role in larval development by controlling gene expression via chromatin modifications.
In Apis the TCA cycle has also been implicated in generating different metabolic fluxes related to insulin signaling in brains of adult worker honey bees engaged in different activities [i.e., feeding brood (“nurses”) and foraging for nectar and pollen]. The TCA and insulin/TOR pathways share a common element, phosphorenolpyruvate carboxykinase, which in many organisms is encoded by two genes producing one mitochondrial and one cytoplasmic enzyme. Apis has only the cytoplasmic insulin-regulated enzyme that is described as a major player in the gluconeogenic pathway (Fig. S1). Pathway analyses of microarray data revealed that nurses and foragers differ in adult brain energy metabolism gene expression and insulin signaling (14).
Inositol phosphate/TOR pathway.
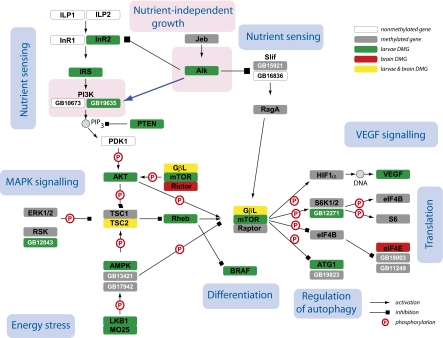
Our analyses revealed that the entire TOR/PI3K/insulin gene network was highly enriched in methylated genes (Fig. 1 and Table S3). The genes found to be differentially methylated, such as mTOR, GβL, raptor, or TSC2, represent the central nodes of this network and are known to be critical components of these highly interconnected pathways (Fig. 1). The TOR/PI3 pathway represents the main network of multigenic interactions linking nutrient sensing with the regulation of cell growth and metabolism. Insect fat body cells have the capacity to sense nutrients and activate the TOR/PI3K network, leading in turn to the activation of a systemic growth signal. The signaling through this pathway is linked to other metabolic and/or regulatory gene networks, including the TCA cycle. For example, the important TCA cycle enzyme ATPCL (differentially methylated in Apis) that links cellular metabolism to epigenetic regulation via histone acetylation is up-regulated by insulin in mammalian liver (15). Several differentially expressed and differentially methylated genes belonging to the TOR/PI3/insulin signaling cascade have been reported in previous studies in the context of queen development (4, 8, 16). TOR (up-regulated in 72- to 96-h-old queen larvae) is also a negative regulator of autophagic cell death and therefore plays an integral role in an apoptotic process of tissue disintegration by which the number of ovarioles in workers is reduced to only a few relative to nearly 200 in queen ovaries (4). One particularly interesting finding revealed by our examination of this pathway is the methylation status of ALK and its putative ligand Jellybelly (JEB). Our analyses of ALK and their implications for phenotypic plasticity are presented below.
Fig. 1.
Annotation of the insulin/TOR network in Apis showing methylated and differentially methylated genes. In those cases in which more than one paralog has been found only one is referred to by the consensus protein name, with the others designated by the GB numbers from the official honey bee gene list (www.beebase.org). The nutrient-independent activation of the PI3K pathway by ALK is highlighted in pink. Gene annotation is shown in Table S3.
Ubiquitin–proteasome pathway.
As shown in Fig. S2 and Table 2, 96 of 108 genes (89%) belonging to the highly conserved ubiquitin–proteasome pathway were methylated. A high enrichment of both methylated genes and DMGs was found at each step E1–E3 of this pathway and in all multienzyme complexes/classes (Table S3). DMGs are found predominantly in larvae, but some are also differentially methylated in the adult brain (ARF-BP1 and Apc4; Fig. S2). The ubiquitin–proteasome pathway is the key mechanism for proteolysis that has been implicated in protein folding, cell cycle progression, and apoptosis (17). This complex and tightly controlled pathway has the capacity to precisely regulate the timing of protein degradation during the cell cycle or after exposure to specific external stimuli and is implicated in epigenetic regulation (18), including histone methylation (19).
Spliceosome, splicing factors, and DNA methylation.
Recently we reported that methylated CpGs in Apis show a strong tendency to be localized in the vicinity of splicing sites and in the genomic regions encoding alternatively spliced cassette exons (9, 11). Here we show that in Apis the spliceosomal complex and splicing factors are also enriched in methylated genes (Table S4). The connection between DNA methylation and alternative splicing is further strengthened here by additional analyses of the larval methylation profiles showing that alternative splicing is more common in methylated spliced genes than in nonmethylated spliced genes. We found that methylated and spliced genes were significantly enriched in alternatively spliced genes (1,119 of 3,313) compared with nonmethylated spliced genes (679 of 2,860; Fisher exact test: P = 4.4E-18).
The correlation of differentially methylated regions with the level of alternatively spliced exons in Apis suggests that methyl tags could play a role in selecting which exons are included in mature transcripts. Splicing occurs cotranscriptionally and involves an interplay of protein complexes with the kinetics of RNA polymerase II elongation and RNA topology. Evidence in model organisms shows how the rate of RNA synthesis affects its secondary structure, which in turn affects splicing (19–21). Drugs that inhibit elongation favor inclusion of exons that would not be used efficiently at higher synthesis rates. Similar results were obtained using inhibitors of histone deacetylation that increase elongation rate by making chromatin more open (19). At this point we do not have a clear understanding of the involvement of DNA methylation in RNA processing in Apis. One possibility is that mCs interfere with the rate of RNA synthesis by affecting DNA-binding proteins mediating RNA polymerase II pausing and leading to alterations in the topology of nascent RNA molecules, which in turn could affect the commitment of spliceosomes to a given catalytic event. As in other species, various components of the epigenetic control systems in Apis must be interconnected to operate in a combinatorial fashion. Together these results add more weight to the notion that methylation is a critical component of the gene splicing machinery.
Differentially Methylated and Differentially Expressed Genes in Queen and Worker Larvae.
Previous efforts to understand how differential feeding of honey bee larvae regulates caste determination focused on gene expression and hormonal regulation (2–4, 22). It has been shown that young larvae destined to become queens predominantly up-regulate physio-metabolic genes, whereas worker larvae up-regulate more developmental genes, including those involved in the development of organs that are unique to workers (e.g., pollen collecting baskets). These differences underpin two kinds of major alterations in the original developmental plan during female caste differentiation. One type affects general growth and specific organs (ovaries) and is best described as “incremental alterations.” The other type, referred to as “character state alterations,” is manifested by the lack or presence of entire organs such as wax glands, or leg baskets in one caste. We compared a battery of several hundred genes differentially expressed during larval growth stages L3–L5 (48–96 h after hatching) (4) with our list of methylated genes. We found that all genes reported to be differentially expressed in queen and worker larvae at stages L3, L4, and L5 of larval growth were methylated, and many were differentially methylated. Several examples of differentially expressed genes that also were differentially methylated in queen and worker larvae are shown in Table S4. In agreement with this study, these genes belong to central metabolic and regulatory pathways. In contrast, genes expressed in a tissue- or life stage-specific pattern, such as olfactory receptors, odorant binding proteins, or cuticular proteins, were generally not methylated (Table S5).
The queen-inducing properties of JH (23) have implicated this important messenger molecule in cell signaling underlying larval developmental plasticity. Barchuk et al. (4) have identified more than 52 genes whose expression is strongly affected by treating worker larvae with JH. We found that not only were all JH-responsive genes methylated, but several were also differentially methylated (Table S6). However, it is not possible from these results to infer whether the increase in JH circulating titers that occurs during L3 in queen-destined larvae triggers differential methylation, or vice versa. This is an important topic for future study.
ALK Is an Organizer of Context-Dependent Metabolism in Apis.
We used our list of DMGs to find the conserved Apis genes previously identified in other species as organizers of metabolism and cellular signaling in a context-dependent manner. To this end, we found that one of the DMGs codes for ALK, an important protein with a critical capacity to enable growth in a nutrient-independent fashion by directly activating PI3K and bypassing InR (insulin receptor) and TOR.
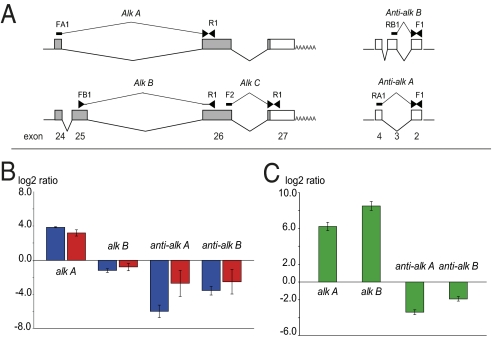
In Apis, alk (Amalk) is a relatively large gene (21 kb) spanning 27 exons transcribed and alternatively spliced into numerous isoforms (Fig. 2). The gene is differentially methylated, with the majority of methylated CpGs localized in the intron between exons 25 and 26 and one methylated CpG in exon 9.
Fig. 2.
Simplified gene model of Amalk and its pattern of methylation in different phenotypes. (A) alk gene model and the positions of methylated CpGs. (B) CpG percentage methylation levels in alk intron 26 (highlighted in blue) in the adult brain and larval heads of both queens and workers.
Although intronic methylation in Apis is rare, the position of this region is consistent with the idea that DNA methylation is involved in controlling alternative splicing of the upstream cassette exon 25. According to a recently described mechanism, DNA methylation inhibits a conserved DNA-binding protein, CTCF, that promotes inclusion of weak/conditional upstream exons by mediating RNA polymerase II pausing (24). Our data provide some support for this model. The intronic region is differentially methylated not only between queens and workers but also within the same caste between tissues with different metabolic fluxes, such as adult brain and larval head (Fig. 2B).
We also found that low methylation correlated with exon 25 inclusion at high frequency. In the adult brain, where the intronic region shows much lower methylation relative to larvae (Fig. 2B), the majority of the available ALK ESTs (81%) contained exon 25 (SI Materials and Methods). Thus, differential methylation of this intronic sequence seems to affect the dynamics of exon 25 inclusion and generates proteins with different amino-terminals in the intracellular domain that could interact with distinct partners. Furthermore, the existence of multiple transcript variants produced by this gene (Figs. 2A and 3A) suggests that Amalk is expressed in a highly contextual combinatorial manner. We illustrate this complexity in Fig. 3B by showing the alternative use of exon 25 in adult brain relative to larval head. In both queens and workers alk-A (no exon 25) was up-regulated in the brain, whereas alk-B containing exon 25 was slightly down-regulated.
Fig. 3.
Relative expression of alk and anti-alk in selected tissues. (A) Section of the alk gene model showing the transcript variants produced by alternative splicing of alk and anti-alk and the positions of PCR primers used for variant-specific amplifications. (B) Expression of alternative transcripts alk-A, alk-B, and anti-alk-A, anti-alk-B in adult brain relative to larval head. Quantitative data for alk-C representing total expression of the alk gene were used for normalization. alk-A represents the short transcript with no exon 25, whereas alk-B contains exon 25. anti-alk-B contains an alternatively spliced exon 3 of anti-alk, which is missing in anti-alk-A. Blue bars, workers; red bars, queens. (C) Expression of the same transcripts in the adult queen brain relative to ovary. The reference gene was ugt. (experimental details in SI Materials and Methods). Two biological replicates were used in each experiment. Error bars represent SD.
Another noteworthy feature of Amalk is the presence of an antisense transcript that fully overlaps with the 6.4 kb of its 3′ region. Anti-Amalk is an example of a cis-acting natural antisense transcript that is polyadenyaled and alternatively spliced but does not code for a protein (Figs. 2A and 3A). Our analyses suggest that it might have a complex role in controlling the expression of Amalk, especially in a tissue-specific manner. For example, two alternatively spliced variants, anti-alk-A and anti-alk-B, are both down-regulated in adult brain relative to larval head, with the lowest expression found in worker brains (Fig. 3C).
Although these results are correlative, the remarkably complex regulation of ALK in Apis suggests that this gene has been recruited to link different nutritional environments with the cellular responses. In mammals and Drosophila ALK protects neural progenitor growth against reductions in amino acids and insulin-like peptides during nutrient restriction via two mechanisms (25). First, ALK suppresses the growth requirement for amino acid sensing via Slimfast/Rheb/TOR complex 1 and second, ALK, rather than insulin-like receptor, primarily activates the PI3–kinase pathway. We also found that in Apis the expression of ALK is higher in the adult brain relative to some other tissues, such as ovaries (Fig. 3B). Taken together, our data suggest that in Apis ALK might modulate PI3–kinase signaling according to nutritional input in both adult and larval honey bees. The high adaptability of this critical regulatory node is provided by flexible methylation marks, including methylated ALK and its ligand JEB (Fig. 1), allowing for rapid rewiring of the whole network, depending on the tissue type and on inputs from other signaling pathways.
Discussion
Caste determination in honey bees always has been a classic example of nutritionally mediated phenotypic plasticity, and our findings contribute to understanding this phenomenon in epigenetic terms. In a growing larva dynamic methylation marking across the genome is a means through which confirmation of environmental experience, including nutritional status, is received and recorded by the genetic hardware, resulting in differential gene expression. Thus, by connecting environmental exposure with gene networks, the dynamic epigenome enables plasticity of phenotype in a fixed genotype (26–28). The developmental trajectory of a female bee embryo is determined by environmental signals that enable durable alterations in gene expression potential of a single genetic makeup. It is a continuing process driven by metabolic flux and incremental discrete switches involving hundreds of genes whose combined and continuous effects contribute to this process. The presence of compounds with histone deacetylase inhibitor activities in royal jelly (13) suggests that the queen/worker developmental dichotomy is driven by a combinatorial action of DNA methylation, histone modification, and possibly other elements that respond to changes in the in vivo microenvironment brought about by fluctuations of metabolite levels. Some of the small metabolites, such as methyl or acetyl groups, may play a direct role in connecting metabolism with signal transduction by posttranslational modification of specific residues (12, 29).
The epigenome is a multifactorial and highly dynamic molecular system including cytosine methylation, histone modifications, various noncoding RNAs, and chromatin complexes, whose role is to ensure that gene expression is not only faithful to a specific developmental program but also flexible to ensure adaptable responses to environmental influences (5, 11, 30). Such environmentally induced gene expression patterns are also durable and can influence adult phenotype. An emerging view in this field is that epigenetic control systems act as facilitators rather than enforcers of cellular and/or organismal determination (28). One possibility is that some of the alternatively spliced transcript variants that correlate with variable methylation levels in queens and workers are examples of transcriptional events that are mechanistically less probable and can only occur in a specific environmental context (28). These intermittent transcripts could play an important role in resetting critical network nodes as part of a “decision-making” process by which the nutrient status of an organism is evaluated to generate a balanced cellular response.
The intricate interactions between genotypic and environmental influences on phenotypic plasticity in honey bees illustrate the limits to predictions of phenotypes from knowledge of genotypes and raise a question as to how generalizable are the specific results presented in this article. Social insects provide excellent material for comparative analysis because reproductive division of labor between workers and queens has arisen independently in evolution at least 12 times (1). Such studies will greatly accelerate progress in understanding the roles of DNA methylation in organisms with different levels of complexity by allowing scientists to determine how epigenetic networks have evolved in insects and what consequences they have had for morphological and behavioral change.
Materials and Methods
Detailed protocols are provided in SI Materials and Methods.
Bisulfite Sequencing (BS-Seq) of Larval DNAs.
Genomic DNAs extracted from honey bee larvae (details in SI Materials and Methods) were used to generate BS-Seq libraries using a previously published protocol (31). Data processing, mapping, and methylation assessments are described in ref. 32. Postmapping steps were carried out using the tools included in libngs (http://github.com/sylvainforet/libngs). We assessed the methylation status of each CpG dinucleotide and differential methylation between queen and worker larvae with our previously published methods (9, 32). P values were corrected for multiple testing using the Benjamini and Hochberg method (33). These tests were carried out using the R statistical environment (http://www.r-project.org).
Other Bioinformatics Analyses.
Gene ontology (GO) annotations of the honey bee predicted genes were produced using blast2go software (34). Pathway annotations were retrieved from the KEGG website (35). The annotations of the pathways presented in this article were manually refined using homology to the fly and human genes. Enrichment in GO terms and KEGG pathways was assessed using the GOstat module of the Bioconductor platform (www.bioconductor.org).
To produce spliced alignments of all of the honey bee transcriptome sequences available from GenBank, we used SibSim4 (http://sibsim4.sourceforge.net/). False positives caused by sequencing errors or spurious alignments were avoided by only using canonical splice sites, falling into annotated gene models, and supported by at least five reads.
Supplementary Material
Acknowledgments
We thank Paul Helliwell and Joanna Maleszka for providing the biological material used in this study. Work in the R.M. laboratory was supported by Australian Research Council Grant DP1092706 and National Health and Medical Research Council Grant 585442. Work in the S.E.J. laboratory is supported by National Institutes of Health (NIH) Grant GM60398 and the Howard Hughes Medical Institute. S.E.J. is an Investigator of the Howard Hughes Medical Institute. S. Feng is a Special Fellow of the Leukemia and Lymphoma Society. G.E.R.'s contributions were supported by NIH Director's Pioneer Award 1DP1OD006416.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing data reported in this paper have been deposited in the GenBank database (accession no. SRA047112.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202392109/-/DCSupplemental.
References
- 1.Wilson EO. The Insect Societies. Cambridge, MA: Harvard Univ Press; 1971. [Google Scholar]
- 2.Hartfelder K, Engels W. Social insect polymorphism: Hormonal regulation of plasticity in development and reproduction in the honeybee. Curr Top Dev Biol. 1998;40:45–77. doi: 10.1016/s0070-2153(08)60364-6. [DOI] [PubMed] [Google Scholar]
- 3.Evans JD, Wheeler DE. Gene expression and the evolution of insect polyphenisms. Bioessays. 2001;23:62–68. doi: 10.1002/1521-1878(200101)23:1<62::AID-BIES1008>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Barchuk AR, et al. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol. 2007;7:70. doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maleszka R. Epigenetic integration of environmental and genomic signals in honey bees: The critical interplay of nutritional, brain and reproductive networks. Epigenetics. 2008;3:188–192. doi: 10.4161/epi.3.4.6697. [DOI] [PubMed] [Google Scholar]
- 6.Weaver N. Physiology of caste determination. Annu Rev Entomol. 1966;11:79–102. doi: 10.1146/annurev.en.11.010166.000455. [DOI] [PubMed] [Google Scholar]
- 7.Drapeau MD, Albert S, Kucharski R, Prusko C, Maleszka R. Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 2006;16:1385–1394. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honey bees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 9.Lyko F, et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foret S, Kucharski R, Pittelkow Y, Lockett GA, Maleszka R. Epigenetic regulation of the honey bee transcriptome: Unravelling the nature of methylated genes. BMC Genomics. 2009;10:472. doi: 10.1186/1471-2164-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet. 2011;27:127–131. doi: 10.1016/j.tig.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spannhoff A, et al. Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Rep. 2011;12:238–243. doi: 10.1038/embor.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ament SA, Corona M, Pollock HS, Robinson GE. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc Natl Acad Sci USA. 2008;105:4226–4231. doi: 10.1073/pnas.0800630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda H, Noguchi T, Iritani N. Transcriptional regulation of fatty acid synthase gene and ATP citrate-lyase gene by Sp1 and Sp3 in rat hepatocytes(1) FEBS Lett. 1999;464:113–117. doi: 10.1016/s0014-5793(99)01700-7. [DOI] [PubMed] [Google Scholar]
- 16.Patel A, et al. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS ONE. 2007;2:e509. doi: 10.1371/journal.pone.0000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 18.Mishra SK, et al. Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature. 2011;474:173–178. doi: 10.1038/nature10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 20.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc Natl Acad Sci USA. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wirtz P, Beetsma J. Induction of caste differentiation in the honey bee Apis mellifera L. by juvenile hormone. Entomol Exp Appl. 1972;15:517–520. [Google Scholar]
- 24.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng LY, et al. Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell. 2011;146:435–447. doi: 10.1016/j.cell.2011.06.040. [DOI] [PubMed] [Google Scholar]
- 26.Mathers JC. Session 2: Personalised nutrition. Epigenomics: A basis for understanding individual differences? Proc Nutr Soc. 2008;67:390–394. doi: 10.1017/S0029665108008744. [DOI] [PubMed] [Google Scholar]
- 27.Feinberg AP. Epigenomics reveals a functional genome anatomy and a new approach to common disease. Nat Biotechnol. 2010;28:1049–1052. doi: 10.1038/nbt1010-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Combes AN, Whitelaw E. Epigenetic reprogramming: Enforcer or enabler of developmental fate? Dev Growth Differ. 2010;52:483–491. doi: 10.1111/j.1440-169X.2010.01185.x. [DOI] [PubMed] [Google Scholar]
- 29.Metallo CM, Vander Heiden MG. Metabolism strikes back: Metabolic flux regulates cell signaling. Genes Dev. 2010;24:2717–2722. doi: 10.1101/gad.2010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabor Miklos GL, Maleszka R. Epigenomic communication systems in humans and honey bees: From molecules to behavior. Horm Behav. 2011;59:399–406. doi: 10.1016/j.yhbeh.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 31.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen PY, Cokus SJ, Pellegrini M. BS Seeker: Precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 34.Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.