Abstract
Noncanonical amino acids have proved extremely useful for modifying the properties of proteins. Among them, extensively fluorinated (fluorous) amino acids seem particularly effective in increasing protein stability; however, in the absence of structural data, the basis of this stabilizing effect remains poorly understood. To address this problem, we solved X-ray structures for three small proteins with hydrophobic cores that are packed with either fluorocarbon or hydrocarbon side chains and compared their stabilities. Although larger, the fluorinated residues are accommodated within the protein with minimal structural perturbation, because they closely match the shape of the hydrocarbon side chains that they replace. Thus, stability increases seem to be better explained by increases in buried hydrophobic surface area that accompany fluorination than by specific fluorous interactions between fluorinated side chains. This finding is illustrated by the design of a highly fluorinated protein that, by compensating for the larger volume and surface area of the fluorinated side chains, exhibits similar stability to its nonfluorinated counterpart. These structure-based observations should inform efforts to rationally modulate protein function using noncanonical amino acids.
Keywords: de novo-designed proteins, protein structure, coiled-coil proteins, unnatural amino acids, hexafluoroleucine
The development of methods that allow noncanonical amino acids to be either genetically encoded or incorporated in a residue-specific manner into proteins together with native ligation strategies to produce semisynthetic proteins have allowed a wide range of noncanonical side chains to be introduced into proteins (1–3). The introduction of fluorinated amino acids into proteins has attracted particular interest, because although essentially absent from biology, fluorine has proved a remarkably useful element to probe the workings of biological molecules. For example, fluorinated substrates have been extensively used to investigate enzyme mechanisms, and 19F NMR has proved a valuable tool for studying structure, dynamics, and interactions of fluorine-labeled proteins, peptides, lipids, and nucleic acids (4–9). Fluorinated molecules also have important medical applications, exemplified by 20% of all pharmaceuticals containing fluorine, which improves pharmacokinetic properties (10).
Additional interest in creating highly fluorinated proteins stems from the fact that perfluorinated small molecules possess unique physicochemical properties that are not found in nature. These properties underlie their exceptional chemical and thermal stability and their unusual tendency to undergo phase separation—the so-called fluorous effect (11–14). This inertness underpins many of the important uses for perfluorocarbons (for example, as nonstick polymers such as polytetrafluoroethylene, as fire retardants, and in medical applications, including its use as blood substitutes and anesthetics) (10, 15–17). The phase-segregating properties of perfluorocarbons have been effectively exploited in fluorous organic synthesis strategies that allow rapid purification of perfluorocarbon-tagged molecules into fluorinated solvents from complex reaction mixtures (12, 14).
Inspired by the novel properties of fluorocarbons, there have been numerous studies aimed at using extensively fluorinated (or fluorous) amino acids to modulate the properties of proteins and particularly, increase their thermal stability (11, 13). Thus, fluorous analogs of hydrophobic amino acids such as leucine, valine, and phenylalanine have been incorporated into both natural and de novo-designed proteins either biosynthetically or by chemical synthesis (18–21). Proteins with sequences containing up to ∼25% fluorous residues have been synthesized without gross structural perturbation. In almost all cases, fluorination significantly enhances stability to thermal unfolding, chemical denaturation, and proteolytic degradation, with minimal impact on the biological activity of the protein or peptide (18, 21–24).
A particularly intriguing property of perfluorocarbons is their unusual self-segregating properties. In principle, the self-segregating properties of fluorocarbons could be applied to direct the specific association of proteins through fluorous interactions—a property that could be extremely useful. However, evidence for such fluorous interactions in proteins seems mixed. Studies on two parallel coiled-coil systems, one soluble and one membrane-bound, found that incorporation of hexafluoroleucine (hFLeu) at the a and d positions of the canonical coiled-coil heptad repeat led to self-segregation of the fluorinated and nonfluorinated peptides (18, 22, 25). In contrast, studies in our laboratory using an antiparallel four-helix bundle system failed to find convincing evidence for fluorous self-segregation when hFLeu was, similarly, introduced at a and d positions (26, 27).
Despite the numerous studies on extensively fluorinated proteins and peptides, the origin of their enhanced stability, whether through favorable fluorocarbon–fluorocarbon interactions or simple differences in hydrophobicity, remains a matter of debate. To date, no structures of highly fluorinated proteins have been reported, which severely hinders our understanding of how interactions between fluorocarbon side chains within the core of the protein contribute to the dramatic changes in observed stability.
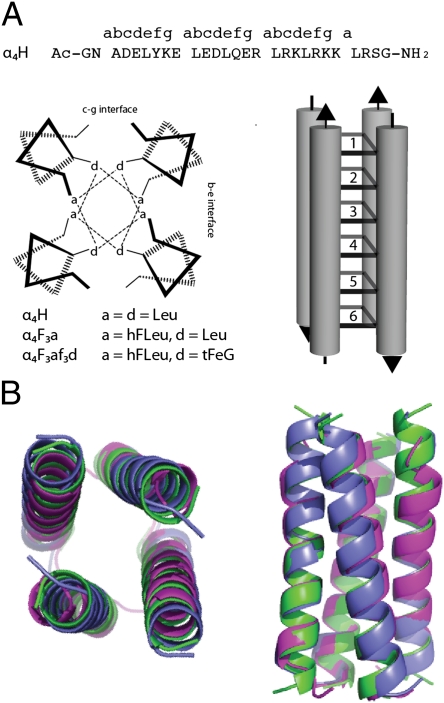
In this report, we present high-resolution X-ray structures for three de novo-designed proteins: α4H (20), α4F3a (26), and α4F3af3d. These proteins are designed to form antiparallel four α-helix bundles in which the hydrophobic core is packed in six layers by residues at the canonical a and d positions of the helical repeat, which is illustrated in Fig. 1. In α4H, the hydrophobic core contains leucine (Leu) at each a and d position, whereas in α4F3a, the Leu residues at the three a positions are substituted for hFLeu; therefore, 50% of the core is now fluorocarbon. Both proteins fold in a cooperative two-state transition; for α4H, ΔG°fold = −18.0 ± 0.2 kcal/mol (1), whereas the incorporation of a total of 24 trifluoromethyl groups in the core of α4F3a leads to a significant increase in stability with ΔG°fold = −27.6 ± 0.1 kcal/mol (26).
Fig. 1.
Design of α4-proteins. (A) The sequences and helical wheel diagram for the α4-proteins illustrating positions of the hydrophobic a and d residues in the antiparallel four-helix bundle topology. The hydrophobic core of these proteins comprises six layers formed by a and d residues as illustrated in the diagram. (B) End and side views of the overlay of backbone atom traces determined from the crystal structures of α4H (green), α4F3a (blue), and α4F3af3d (purple).
A careful comparison of the structures of α4H and α4F3a allowed us to design and structurally characterize the third protein, α4F3af3d, to test the hypothesis that changes in buried hydrophobic surface area, rather than favorable interactions between fluorinated residues, are responsible for the increased stability imparted by fluorination. α4F3af3d contains smaller trifluoroethylglycine residues (tFeG) at the d positions, which compensate for the larger hFLeu residues at a positions. We were able to obtain the crystal structure of this protein and show that, despite containing 36 trifluoromethyl groups in the core, it is actually slightly less stable than α4H.
Results
Structure of α4H.
As a reference structure against which to compare the effects of fluorination, we first determined the structure of α4H. The protein crystallized in space group I41, and we used standard molecular replacement methods to solve its structure at a resolution of 1.36 Å; statistical data for the structure are given in Table S1. The asymmetric unit comprises an antiparallel dimer of two peptides (A and B chains), with the electron density being well-defined for all but the last two residues of chain A and the first residue and last two residues of chain B. The antiparallel four-helix bundle structure, shown in Fig. 1, was generated from the dimer of crystallographically nonequivalent peptide chains by the appropriate symmetry operation.
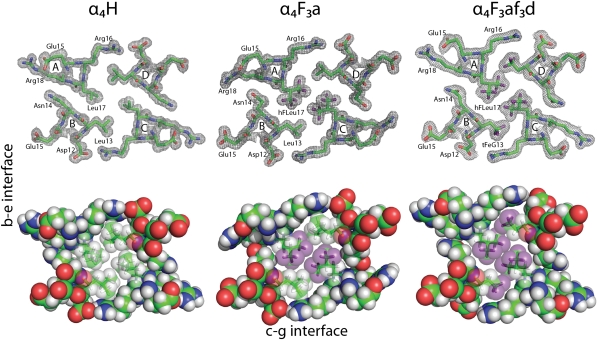
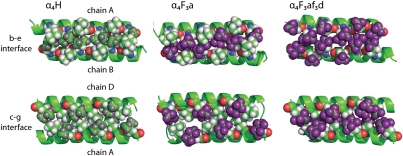
The modeled electron density for a cross-section of α4H is shown in Fig. 2 with stereoviews shown in Fig. S1. In accord with the intended design, the antiparallel orientation of the α-helices in α4H is enforced by complementary electrostatic interactions between residues in the c and g positions (c–g interface) and residues in the b and e positions (b–e interface). The two interfaces are nonequivalent, and in the case of α4H, this inequality results in a larger spacing between helices of the c–g interface, which is formed by knobs into holes packing of the Leu residues at d positions (Fig. 3), than the b–e interface, which is formed by knobs into holes packing of the Leu residues at a positions (Fig. 3). The program SOCKET (28) was used to further analyze the structure of α4H: the protein adopts a left-handed coiled coil with interhelix angles of 152.36° (b–e interface) and 169.15° (c–g interface). The program also verified the knobs into holes packing arrangement of the Leu residues; additional details are in SI Methods and Fig. S2.
Fig. 2.
Cross-sections through the hydrophobic cores of α4H, α4F3a, and α4F3af3d. (Upper) Representative electron density (2Fo − Fc) maps for each protein, with residues contoured at 1.0 σ. (Lower) Space-filling representations of the hydrophobic core illustrating how fluorination conserves the tight packing of side chains. Fluorine atoms are colored purple.
Fig. 3.
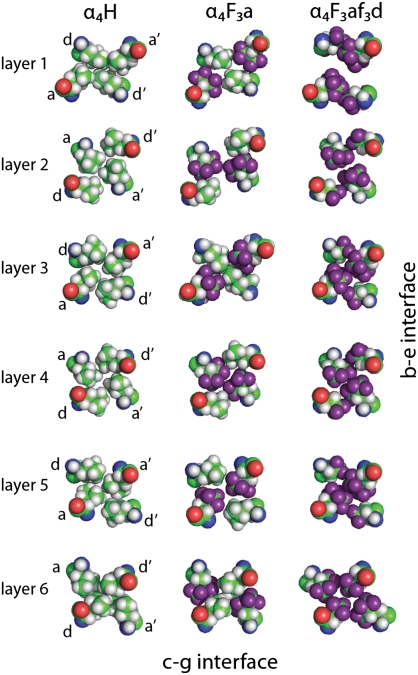
Comparison of the packing of the b–e and c–g interfaces by fluorinated and nonfluorinated resides in the structures of α4H, α4F3a, and α4F3af3d. Layer 1 of the core is shown to the left; layer 6 is to the right. In α4H, Leu residues at a positions are colored dark gray to distinguish them from Leu residues at d positions. In α4F3a and α4F3af3d, fluorine atoms are colored purple.
Structure of α4F3a.
Of the various fluorinated versions of α4H that we have synthesized, the structure of α4F3a was of particular interest, because it is one of the most stable fluorinated proteins on a per-residue basis (ΔΔGfold = −0.8 kcal/mol per hFLeu). α4F3a crystallized under similar conditions to α4H and in the same space group I41, minimizing the possibility that altered crystal contacts may be responsible for any changes to the protein structure. We were able to determine its structure at 1.54 Å (Table S1) and resolve all but the last two residues of the A chain and last residue of the B chain. In particular, the electron density for all of the hFLeu residues in α4F3a is well-defined and clearly indicates the shape and orientation of the trifluoromethyl moieties (Fig. 2). The trifluoromethyl groups have full occupancy and do not seem to undergo rapid rotation, at least at the cryogenic temperatures of data acquisition. In each residue, the two trifluoromethyl groups adopt a staggered configuration that minimizes steric repulsion between the trifluoromethyl groups and the β-carbon of hFLeu.
Overall, the incorporation of 72 fluorine atoms into α4F3a is remarkably nonperturbing to the structure of the protein: the helices move slightly farther apart, displacing the backbone atoms of α4F3a by an rmsd of only 0.95 Å from the coordinates of α4H (Fig. 1). Interactions between hFLeu residues play an important role in forming the b–e interface of the four-helix bundle. Knobs into holes packing of hFLeu in adjacent layers of the core results in a tightly packed fluorinated stripe that runs along the entire b–e interface, which is illustrated in Fig. 3. The c–g interface, in contrast, is formed by knobs into holes packing of the Leu residues (Fig. 3).
Comparison of Core Packing Between α4H and α4F3a.
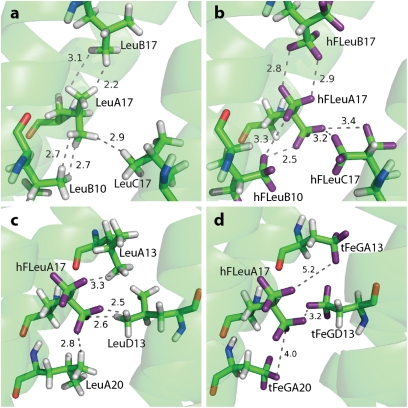
We were particularly interested in how fluorination might alter interactions between residues in the hydrophobic cores of α4H and α4F3a. Fig. 4 compares, in detail, the interaction of one buried residue, LeuA17, in α4H with the corresponding residue, hFLeuA17, in α4F3a. The distances between the fluorine atoms of hFLeuA17 and adjacent hFleu residues range between 2.5 and 3.2 Å, whereas the distances between the hydrogen atoms of LeuA17 (which were modeled into the structure to facilitate comparison) and adjacent Leu residues vary between 2.2 and 3.1 Å. These differences are consistent with the shorter van der Waals radius for hydrogen of 1.2 Å compared with fluorine (1.35 Å). The trifluoromethyl groups of hFLeuA17 also form extensive contacts with the methyl groups of adjacent Leu residues in α4F3a (Fig. 4), with fluorine–hydrogen distances of 2.5–3.3 Å. Overall, there is no evidence from the structure that the hFLeu residues adopt conformations that would either maximize fluorine–fluorine contacts or minimize fluorine–hydrocarbon contacts, which would be predicted if favorable fluorous interactions between residues were important. We also found no evidence for dipolar interactions between trifluoromethyl groups and polar groups in the protein (as judged by proximity and alignment of the groups), which have been observed for some fluorinated compounds bound to proteins (10).
Fig. 4.
Analysis of contacts between hydrocarbon and fluorocarbon side chains. In A–D, the residue at position A17 is oriented similarly to facilitate comparison. (A) Distances between LeuA17 and adjacent Leu residues in α4H. (B) The equivalent distance measurements between hFLeuA17 and adjacent hFLeu residues in α4F3a. (C) Distances between hFLeuA17 and adjacent Leu residues in α4F3a. (D) Distances between hFLeuA17 and adjacent tFeG residues in α4F3af3d; note that the tFeG residues adopt conformations that position the trifluoromethyl group farther away from hFLeu.
We also carefully compared the packing arrangement of Leu and hFLeu residues in α4H and α4F3a for each of the six layers of the hydrophobic core, which is shown in Fig. 5. In α4H, the central four layers of the core are packed, and therefore, the Leu residues at a positions extend to the center of the protein core and make van der Waals contacts with Leu residues at the corresponding a′ positions. The Leu residues at the d positions are less deeply buried and oriented to the c–g interface. In the outer layers (layers 1 and 6), the situation is reversed. Thus, Leu residues in d positions extend to the center of the helical bundle, making contact with their counterparts at d′, whereas those residues at a and a′ positions are oriented out to the b–e interface. The reason for this change in packing arrangement is not obvious.
Fig. 5.
Comparison of core packing (a and d positions) for each layer of α4H, α4F3a, and α4F3af3d. The pattern of hydrophobic contacts is generally unchanged by fluorination, despite the hFLeu side chain being significantly larger; exceptions are layer 3 of α4F3a, where Leu inserts between hFLeu residues, and layer 1 of α4F3af3d, where fraying of the core results in a cleft opening across the b–e interface.
In α4F3a, the larger hFLeu residues pack the core in an arrangement very similar to the arrangement seen for Leu in α4H (Fig. 5). Thus, in layers 1 and 6 of the core, the hFLeu residues at the a and a′ positions point to the b–e interface, allowing the Leu residues at the d and d′ positions to extend across the core and make van der Waals contacts with each other. In the central layers, the a position hFLeu residues extend into the center to make van der Waals contacts with hFLeu residues at a′. The only significant difference between the core packing of α4F3a and α4H involves the packing of layer 3. Here, Leu13 in chains A and C of α4F3a extends into the center of the core and disrupts the packing of hFLeu17 in chains B and D. This layer seems to be somewhat mobile, because Leu13 in the A and C chains can be modeled in two conformations, each with ∼50% occupancy (Fig. S3). This change in the packing arrangement is not seen in the chemically equivalent but crystallographically nonequivalent layer 4 of α4F3a. This finding may be explained if both central layers of the core have some inherent mobility in solution, but crystal packing effects freeze out each layer in a different conformation in the crystal.
Structural Basis for Enhanced Stability of α4F3a.
The high-resolution structures of α4H and α4F3a provide an opportunity to rationalize the enhanced stability imparted by fluorination. We first considered whether the fluorinated residues are able to pack more efficiently into the hydrophobic core, where packing efficiency is defined as the volume occupied by the peptide chains divided by the total volume of the core. The formation of cavities within proteins is known to be destabilizing (29, 30), and therefore, a more efficiently packed core should be associated with increased stability. To calculate the packing efficiency of the core, we used truncated structures of α4H and α4F3a in which the surface-exposed side chains in the b, c, e, f, and g positions were mutated in silico to alanine. This mutation was done to prevent small changes in the conformation of the solvent-exposed side chains from affecting the calculation. From these structures, we calculated the total van der Waals volume of the core and the sum of the van der Waals volumes of the individual peptide chains.
We calculated the tetrameric core of α4H to have a total volume of ∼8,730 Å3, of which the peptide chains occupy ∼7,820 Å3, which results in a packing efficiency of ∼90%. In α4F3a, substitution of hFLeu for Leu increases the volume of each peptide chain by an average of 96 Å3 or an increase of 5% over the peptide chain volume of α4H. This finding represents an increase of 32 Å3/hFLeu residue, and this figure is in good agreement with previous calculations on the volume of hFLeu. The total volume of the α4F3a tetramer core expands to ∼9,220 Å3, an increase of ∼6%; therefore, the packing efficiency of α4F3a is essentially unchanged at ∼89%. Thus, the additional stability imparted by fluorination does not result from more efficient packing of the protein core; however, the density of the core is slightly increased, because fluorine is 19 times heavier than hydrogen.
We next consider whether fluorous interactions (i.e., favorable van der Waals type interactions between fluorocarbon residues) may account for the stability of α4F3a, because such interactions have often been hypothesized to account for the high stability of highly fluorinated proteins (18, 20, 21, 25). The structure of the mixed hydrocarbon–fluorocarbon core of α4F3a provides a unique opportunity to test this hypothesis. If such fluorous interactions were important in α4F3a, fluorocarbon–fluorocarbon contacts should be maximized at the expense of fluorocarbon–hydrocarbon contacts. However, as discussed above, there is no evidence from the structure that this change is the case. Fluorous contacts could be increased by repacking layers 1 and 6, and therefore, the hFLeu residues form contacts across the C2 axis of the helical bundle. Instead, Leu residues at the d positions interpose between hFLeu residues—the same type of packing seen in α4H. (Fig. 5). Moreover, in layer 3 of α4F3a, which is the only layer that differs in its packing from α4H, contacts between the two hFLeu residues are disrupted by the leucines in the d position, which would lead to a loss of putative fluorous interactions.
Finally, we considered whether the increased stability of fluorinated proteins could be explained simply by the increase in hydrophobicity of the fluorinated residues. It is well-established that changes in the stability of natural proteins correlate with changes in buried hydrophobic surface area or hydrophobic volume associated with protein folding (31). Additionally, although fluorocarbons are often described as being intrinsically more hydrophobic hydrocarbons, the larger volume and surface area of fluorocarbons are often overlooked in such comparisons; when these factors are counted, fluorocarbons and hydrocarbons exhibit similar hydrophobicities (32). The structure of α4F3a allowed the increase in buried hydrophobic surface area associated with the introduction of hFLeu to be experimentally measured as ∼20 Å2/residue. Using the generally accepted value of ∼30 cal/mol per Å2 for the hydrophobic effect in proteins (31), α4F3a would be expected to be ∼7.2 kcal/mol more stable than α4H. The experimentally determined stabilization ΔΔGofold = 9.6 kcal/mol is somewhat greater, but it may be considered to be in reasonable agreement given the approximate nature of the calculation.
Design of a Highly Fluorinated α4-Protein Lacking Enhanced Stability.
With the insights gained from the structure of α4F3a, we designed a protein to test whether fluorous interactions or conventional hydrophobic volume and surface area changes associated with fluorination contribute more to protein stability. This peptide, α4F3af3d, incorporates smaller tFeG residues at d positions and hFLeu at a positions, and therefore, the entire core is now packed with fluorocarbon side chains. The smaller volume and surface area of tFeG with respect to Leu almost exactly compensate for the larger hFLeu side chain, with the result that α4F3af3d has essentially the same volume and surface area as α4H while containing 50% more fluorine than α4F3a. Therefore, if fluorous interactions contribute significantly to stability, α4F3af3d should be more stable than α4F3a; however, if conventional hydrophobic effects dominate, α4F3af3d should have a similar stability to α4H.
We determined the free energy of folding for α4F3af3d using guanidinium hydrochloride as the denaturant. The protein unfolds at low concentrations of guanidinium hydrochloride in a cooperative two-state transition; fits to the unfolding curve yielded ΔG°fold = −17.8 ± 1.0 kcal/mol (Fig. S4). Therefore, α4F3af3d exhibits very similar stability to α4H (ΔG°fold = −18.0 ± 0.2 kcal/mol), which is consistent with our prediction that stability is primarily affected by changes in hydrophobic surface area and volume.
α4F3af3d crystallized under similar conditions to the other proteins in space group P21212, and its structure was determined at 1.72 Å resolution (Table S1). The modeled electron density for a cross-section of α4F3af3d is shown in Fig. 2. The backbone atoms overlay those atoms of α4H, with an rmsd of 1.02 Å. (Fig. 1); however, no electron density was visible for the first four residues of the A and C chains, indicating that these residues are unstructured. This lack of structure disrupts the first layer of the core, and therefore, the two hFLeu residues are oriented to the c–g interface and are separated by 4.6 Å, opening up a narrow cleft in this layer (Fig. 3). Fig. 4D shows details of the contacts made by one residue, hFLeuA17, with adjacent tFeG side chains in the core, and it may be compared with the equivalent residues in α4H and α4F3a (Fig. 4 A–C). Notably, the adjacent tFeGA13 residue points away from the hFLeu residue, resulting in longer fluorine–fluorine distances between neighboring residues than seen in the structure of α4F3a.
Examination of the hydrophobic core packing (Fig. 5) reveals that the remaining layers adopt an arrangement very similar to α4H. In layers 2–5, hFLeu residues in the a positions extend into the center of the core to make contact with their counterparts at a′ positions. The tFeG residues in the d positions are oriented to the c–g interface, where they pack in a knobs into holes fashion with tFeG residues from the adjacent peptide chain. In layer 6 the tFeG residues point into the center, and the hFLeu residues abut them on either side. Using a similar analysis to the analysis described above, we calculated the packing efficiency for the core of α4F3af3d. Fraying of the helices reduces the buried hydrophobic surface area slightly, and therefore, the total core volume is 8,360 Å3; the volume occupied by the protein chains is 7,390 Å3, giving a packing efficiency of ∼88%, which is very similar to α4H and α4F3a.
Discussion
The field of protein design is now advancing to embrace amino acids beyond the 20 canonical residues (1–3). However, to successfully exploit the potential of new amino acids to augment the functions of natural proteins, it is important to understand both how the novel residues modulate the structures of the proteins that they are incorporated into and how structural changes, in turn, give rise to changes in the physical and biological properties of the protein. In the case of fluorinated amino acids, numerous studies have shown their use in stabilizing proteins against thermal unfolding and chemical denaturation; however, the explanation for this enhanced stability has remained a matter of debate. The studies reported here provide detailed structural information on how highly fluorinated amino acids are accommodated within a protein and provide insights into the origin of the stabilizing effect.
The X-ray structures of α4F3a and α4F3af3d reveal that large numbers of fluorine atoms (72 and 108 atoms, respectively) can be incorporated into proteins with only minimal perturbation of their structure, although the hFLeu side chain is some 32 Å3 (∼30%) larger than Leu. It has been conjectured that the unusual phase-segregating properties of per-fluorinated molecules, ingeniously exploited in organic synthesis (12, 14), could be used to direct protein–protein interactions in a manner orthogonal to the conventional hydrophobic effect (20–22, 25); however, the structures of α4F3a and α4F3af3d reveal no evidence for preferential fluorous interactions between fluorinated residues.
Instead, the increased thermodynamic stability of α4F3a can be adequately explained by the increases in buried hydrophobic surface area and volume that accompany fluorination. In other words, the same principles that underpin the stability of natural proteins (efficient packing of side chains and conventional hydrophobic effects) seem to be responsible for the enhanced stability of fluorinated proteins. Furthermore, although almost all studies have reported increases in protein stability on incorporation of fluorinated residues, the design of α4F3af3d, which has a highly fluorinated core, shows that fluorination per se does not stabilize proteins. Thus, if changes in residue size are controlled for, extensively fluorinated proteins can be designed that have very similar structures and stabilities to their natural counterparts.
The above discussion raises the question of why fluorination has proved such a generally successful strategy for increasing protein stability. We suggest that this success may be because of the fact that fluorination closely preserves the shape of side chains, which is important for the correct packing of side chains within the hydrophobic core, while increasing size and hydrophobicity. This preservation allows the fluorinated residue to be introduced with minimal adjustment of the surrounding structure, which was shown by the structure of α4F3a. The alternative approach to increasing residue hydrophobicity would be to add extra carbon atoms to the side chain (e.g., by changing an alanine to a valine). However, such modifications will also change the side chain's shape, possibly giving a poor fit that can compromise stability and/or biological activity of the protein.
Another question is whether fluorous interactions can really be used to facilitate specific interactions between proteins. To address this question, it is necessary to consider the nature of the fluorous effect in more detail. Although the phase separation of fluorocarbon:hydrocarbon solvent mixtures is often ascribed to fluorophilic or fluorous interactions between fluorocarbon molecules, this classification is not strictly correct. The phenomenon arises because the cohesive dispersion forces between two hydrocarbon molecules are greater than between two fluorocarbon molecules or between a fluorocarbon and a hydrocarbon molecule (because hydrocarbons are more polarizable than fluorocarbons). Thus, fluorocarbons are excluded from the hydrocarbons. More generally, the mutual solubility (or immiscibility) of a mixture of two nonpolar solvents is related to the difference in the solubility parameter δ (Eq. 1),
where ΔEV is the energy of vaporization and V is the molal volume of the pure liquid at a given temperature (33, 34). As the difference in δ between the two solvents increases, the heat of mixing becomes more unfavorable until they are no longer miscible. As discussed in the work by Scott (33), fluorocarbons have low δ-values, because they have both low boiling points and larger molal volumes than hydrocarbons.
Clearly, there are many differences when considering the hydrophobic interface between two proteins and the immiscibility of two liquids, such that the principles discussed above that underlie the segregating tendency of small fluorocarbon molecules cannot be simply applied to protein–protein interactions. Notably, protein interfaces are highly structured and formed by specific interactions between side chains, whereas solvent–solute interactions are transient, nonspecific, and dynamic. We contend that steric effects play a far more important role in specifying hydrophobic interfaces between proteins than the potential differences in dispersion forces between fluorocarbon and hydrocarbon residues. Moreover, although fluorinated residues are similar in shape, they are not completely isosteric with their hydrocarbon counterparts, and therefore, the influence of steric effects can never be entirely ignored.
The peptides described in the literature (18, 22) that seem to exhibit fluorous segregation were designed to form parallel coiled coils, whereas our studies use peptides that form antiparallel coiled coils. As discussed previously by others (35, 36), the oligomerization state of parallel coiled coils is very sensitive to changes in the volume of the hydrophobic core, whereas the antiparallel arrangement is far more robust. Significantly, fluorination also induced a change in the oligomerization state of the self-segregating peptides from a dimeric to a tetrameric coiled coil (18), which is consistent with the larger volume of the fluorinated side chains introduced into the core. Thus, the self-segregating behavior of these peptides might be better ascribed to steric effects rather than a manifestation of fluorous segregation per se.
In conclusion, fluorination represents a unique tool for stabilizing proteins by providing the ability to increase hydrophobicity while closely preserving the shape of the side chain. In most cases, the perturbation is likely to be small enough not to significantly alter the structure and function of the protein. We hope that the insights gained from this study will aid future efforts to modulate protein stability and protein–ligand interactions using this versatile class of noncanonical amino acids.
Methods
A detailed description of the synthesis, crystallization, X-ray structure determination, and determination of ΔGfold for the peptides described in this study is provided in SI Methods (1). Note that we initially reported a value of ΔG°fold for α4H of −20.3 kcal/mol (20). The value of ΔG°fold = −18.0 ± 0.2 kcal/mol is a more recent measurement (26) obtained using an autotitrator under the same conditions as the other peptides discussed here. We consider this measurement to be more accurate.
Supplementary Material
Acknowledgments
We thank Dr. David Smith of LS-CAT for help with remote data collection. Use of the Advanced Photon Source was supported by US Department of Energy, Office of Basic Energy Sciences, Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3TWE, 3TWF, and 3TWG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120112109/-/DCSupplemental.
References
- 1.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 2.Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Valiyaveetil FI, Sekedat M, Muir TW, MacKinnon R. Semisynthesis of a functional K+ channel. Angew Chem Int Ed Engl. 2004;43:2504–2507. doi: 10.1002/anie.200453849. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki Y, Buer BC, Al-Hashimi HM, Marsh ENG. Using fluorine nuclear magnetic resonance to probe changes in the structure and dynamics of membrane-active peptides interacting with lipid bilayers. Biochemistry. 2011;50:5979–5987. doi: 10.1021/bi200639c. [DOI] [PubMed] [Google Scholar]
- 5.Danielson MA, Falke JJ. Use of 19F NMR to probe protein structure and conformational changes. Annu Rev Biophys Biomol Struct. 1996;25:163–195. doi: 10.1146/annurev.bb.25.060196.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerig JT. Fluorine NMR of proteins. Prog Nucl Magn Reson Spectrosc. 1994;26:293–370. [Google Scholar]
- 7.Yu J-X, Kodibagkar VD, Cui W, Mason RP. 19F: A versatile reporter for non-invasive physiology and pharmacology using magnetic resonance. Curr Med Chem. 2005;12:819–848. doi: 10.2174/0929867053507342. [DOI] [PubMed] [Google Scholar]
- 8.Buer BC, Chugh J, Al-Hashimi HM, Marsh ENG. Using fluorine nuclear magnetic resonance to probe the interaction of membrane-active peptides with the lipid bilayer. Biochemistry. 2010;49:5760–5765. doi: 10.1021/bi100605e. [DOI] [PubMed] [Google Scholar]
- 9.Evanics F, Kitevski JL, Bezsonova I, Forman-Kay J, Prosser RS. 19F NMR studies of solvent exposure and peptide binding to an SH3 domain. Biochim Biophys Acta. 2007;1770:221–230. doi: 10.1016/j.bbagen.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Müller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: Looking beyond intuition. Science. 2007;317:1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 11.Neil E, Marsh G. Towards the nonstick egg: Designing fluorous proteins. Chem Biol. 2000;7:R153–R157. doi: 10.1016/s1074-5521(00)00139-3. [DOI] [PubMed] [Google Scholar]
- 12.Horváth IT, Rábai J. Facile catalyst separation without water: Fluorous biphase hydroformylation of olefins. Science. 1994;266:72–75. doi: 10.1126/science.266.5182.72. [DOI] [PubMed] [Google Scholar]
- 13.Yoder NC, Yüksel D, Dafik L, Kumar K. Bioorthogonal noncovalent chemistry: Fluorous phases in chemical biology. Curr Opin Chem Biol. 2006;10:576–583. doi: 10.1016/j.cbpa.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Luo Z, Zhang Q, Oderaotoshi Y, Curran DP. Fluorous mixture synthesis: A fluorous-tagging strategy for the synthesis and separation of mixtures of organic compounds. Science. 2001;291:1766–1769. doi: 10.1126/science.1057567. [DOI] [PubMed] [Google Scholar]
- 15.Böhm H-J, et al. Fluorine in medicinal chemistry. ChemBioChem. 2004;5:637–643. doi: 10.1002/cbic.200301023. [DOI] [PubMed] [Google Scholar]
- 16.Hagmann WK. The many roles for fluorine in medicinal chemistry. J Med Chem. 2008;51:4359–4369. doi: 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]
- 17.Lowe KC. Engineering blood: Synthetic substitutes from fluorinated compounds. Tissue Eng. 2003;9:389–399. doi: 10.1089/107632703322066570. [DOI] [PubMed] [Google Scholar]
- 18.Bilgiçer B, Xing X, Kumar K. Programmed self-sorting of coiled coils with leucine and hexafluoroleucine cores. J Am Chem Soc. 2001;123:11815–11816. doi: 10.1021/ja016767o. [DOI] [PubMed] [Google Scholar]
- 19.Chiu H-P, et al. Helix propensity of highly fluorinated amino acids. J Am Chem Soc. 2006;128:15556–15557. doi: 10.1021/ja0640445. [DOI] [PubMed] [Google Scholar]
- 20.Lee K-H, Lee H-Y, Slutsky MM, Anderson JT, Marsh ENG. Fluorous effect in proteins: de novo design and characterization of a four-α-helix bundle protein containing hexafluoroleucine. Biochemistry. 2004;43:16277–16284. doi: 10.1021/bi049086p. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, et al. Fluorinated coiled-coil proteins prepared in vivo display enhanced thermal and chemical stability. Angew Chem Int Ed Engl. 2001;40:1494–1496. doi: 10.1002/1521-3773(20010417)40:8<1494::AID-ANIE1494>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 22.Bilgiçer B, Kumar K. De novo design of defined helical bundles in membrane environments. Proc Natl Acad Sci USA. 2004;101:15324–15329. doi: 10.1073/pnas.0403314101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H-Y, Lee K-H, Al-Hashimi HM, Marsh ENG. Modulating protein structure with fluorous amino acids: Increased stability and native-like structure conferred on a 4-helix bundle protein by hexafluoroleucine. J Am Chem Soc. 2006;128:337–343. doi: 10.1021/ja0563410. [DOI] [PubMed] [Google Scholar]
- 24.Gottler LM, Lee H-Y, Shelburne CE, Ramamoorthy A, Marsh ENG. Using fluorous amino acids to modulate the biological activity of an antimicrobial peptide. ChemBioChem. 2008;9:370–373. doi: 10.1002/cbic.200700643. [DOI] [PubMed] [Google Scholar]
- 25.Bilgiçer B, Kumar K. Synthesis and thermodynamic characterization of self-sorting coiled coils. Tetrahedron. 2002;58:4105–4112. [Google Scholar]
- 26.Buer BC, de la Salud-Bea R, Al Hashimi HM, Marsh ENG. Engineering protein stability and specificity using fluorous amino acids: The importance of packing effects. Biochemistry. 2009;48:10810–10817. doi: 10.1021/bi901481k. [DOI] [PubMed] [Google Scholar]
- 27.Gottler LM, de la Salud-Bea R, Marsh ENG. The fluorous effect in proteins: Properties of α4F6, a 4-α-helix bundle protein with a fluorocarbon core. Biochemistry. 2008;47:4484–4490. doi: 10.1021/bi702476f. [DOI] [PubMed] [Google Scholar]
- 28.Walshaw J, Woolfson DN. Socket: A program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 29.Eriksson AE, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 30.Serrano L, Kellis JT, Jr, Cann P, Matouschek A, Fersht AR. The folding of an enzyme. II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 31.Sharp KA, Nicholls A, Fine RF, Honig B. Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science. 1991;252:106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- 32.Mecinović J, et al. Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the “hydrophobic wall” of carbonic anhydrase. J Am Chem Soc. 2011;133:14017–14026. doi: 10.1021/ja2045293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott RL. The solubility of fluorocarbons. J Am Chem Soc. 1948;70:4090–4093. doi: 10.1021/ja01192a036. [DOI] [PubMed] [Google Scholar]
- 34.Hildebrand JH, Cochran DRF. Liquid-liquid solubility of perfluoromethylcyclohexane with benzene, carbon tetrachloride, chlorobenzene, chloroform and toluene. J Am Chem Soc. 1949;71:22–25. [Google Scholar]
- 35.Betz SF, DeGrado WF. Controlling topology and native-like behavior of de novo-designed peptides: Design and characterization of antiparallel four-stranded coiled coils. Biochemistry. 1996;35:6955–6962. doi: 10.1021/bi960095a. [DOI] [PubMed] [Google Scholar]
- 36.Harbury PB, Tidor B, Kim PS. Repacking protein cores with backbone freedom: Structure prediction for coiled coils. Proc Natl Acad Sci USA. 1995;92:8408–8412. doi: 10.1073/pnas.92.18.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.