Abstract
Defining the function of the genes that, like RUNX1, are deregulated in blood cell malignancies represents an important challenge. Myeloid leukemia factors (MLFs) constitute a poorly characterized family of conserved proteins whose founding member, MLF1, has been associated with acute myeloid leukemia in humans. To gain insight into the functions of this family, we investigated the role of the Drosophila MLF homolog during blood cell development. Here we report that mlf controls the homeostasis of the Drosophila hematopoietic system. Notably, mlf participates in a positive feedback loop to fine tune the activity of the RUNX transcription factor Lozenge (LZ) during development of the crystal cells, one of the two main blood cell lineages in Drosophila. At the molecular level, our data in cell cultures and in vivo strongly suggest that MLF controls the number of crystal cells by protecting LZ from degradation. Remarkably, it appears that the human MLF1 protein can substitute for MLF in the crystal cell lineage. In addition, MLF stabilizes the human oncogenic fusion protein RUNX1-ETO and is required for RUNX1-ETO–induced blood cell disorders in a Drosophila model of leukemia. Finally, using the human leukemic blood cell line Kasumi-1, we show that MLF1 depletion impairs RUNX1-ETO accumulation and reduces RUNX1-ETO–dependent proliferation. Thus, we propose that the regulation of RUNX protein levels is a conserved feature of MLF family members that could be critical for normal and pathological blood cell development.
Keywords: fruit fly, cancer, proteasome
Development of the hematopoietic system relies on the activity of several signaling pathways and transcription factors that have been conserved from mammals to fly (1). Drosophila has emerged as a model organism to gain insight into the gene regulatory networks controlling hematopoiesis. Deregulation of these networks lies at the origin of several diseases in humans, including leukemia and lymphoma. Interestingly, these hematologic malignancies are frequently associated with recurring chromosomal abnormalities (2). Along with their clinical prognostic value, these rearrangements have led to the identification of many genes involved in the etiology of cancer; thus, characterizing the function of these genes in normal and pathological blood cell development is of particular interest.
One notorious gene identified by cloning translocation breakpoints is RUNX1/AML1, which is affected by the t(8;21)(q22;q22) translocation found in ∼15% of all cases of acute myeloid leukemia (AML) (3). RUNX1 belongs to the RUNX transcription factor family, which is characterized by the presence of a highly conserved DNA binding domain (4). RUNX proteins activate or repress transcription in a context-dependent manner, thereby regulating cell proliferation and differentiation in a variety of metazoans. In particular, RUNX genes have been shown to regulate hematopoiesis in both vertebrates and Drosophila. For instance, RUNX1 plays a prominent role in definitive hematopoietic stem cell emergence, as well as in megakaryocyte and lymphocyte differentiation in mammals, and mutations affecting RUNX1 are among the most frequent genetic abnormalities associated with blood cell malignancies in humans. These alterations promote leukemia by altering RUNX1 dosage or by producing mutant proteins that act as dominant negative and/or display neomorphic activities, as for RUNX1-ETO, the product of the t(8;21)(q22;q22) translocation, which comprises the RUNX1 DNA-binding domain fused to the transcriptional corepressor ETO (5). In Drosophila, the RUNX factor Lozenge (LZ) is specifically expressed in and required for development of one of the two main blood cell types: the crystal cells, a megakaryocyte-like lineage that participates in clotting (6). In fact, LZ interacts and cooperates with the pan-hematopoietic GATA transcription factor Serpent (SRP) to activate the crystal cell differentiation program (7–9). This cooperation is conserved in mammals, where it controls megakaryopoiesis (10–12) and hematopoietic stem cell development (13). Moreover, reminiscent of the situation in humans, RUNX1-ETO expression in the Drosophila LZ+ blood cell lineage induces a preleukemic phenotype characterized by a switch of cell fate from differentiation to self-renewal (14). Thus, Drosophila provides a valuable model for studying the normal and oncogenic functions of RUNX factors during hematopoiesis.
The myeloid leukemia factor (MLF) family comprises a small group of evolutionarily conserved genes whose founding member was first identified as the target of the t(3;5)(q25;q35) translocation associated with myelodysplastic syndrome (MDS) and AML (15). This translocation generates a fusion protein between the N-terminal region of nucleophosmin (NPM; a multifunctional nucleolar protein) and most of MLF1. Whereas NPM, which is involved in other translocations, seems to act by providing a dimerization domain and a nucleolar targeting sequence (16), little is known about MLF1 activity. Significantly, however, increased MLF1 expression correlates with poor prognosis in AML and with malignant progression in MDS (17), and MLF1 ectopic expression affects the myeloerythroid lineage switch and cell cycle progression in cell cultures (18–20). Nonetheless, MLF1 function in hematopoiesis remains poorly defined. Only one MLF gene is present in Drosophila, whereas there are two MLF paralogs in vertebrates (21). As in mammals, MLF encodes for a nucleocytoplasmic shuttling protein with no recognizable structural domain apart from a 14-3-3 binding motif (22). However, its in vivo function remains unclear, given that mlf mutant flies are subviable and do not exhibit any obvious developmental defect (23).
Given the conservation between MLF proteins and the parallels between mammalian and fly blood cell development, we assessed whether MLF controls hematopoiesis in Drosophila. In particular, we found that mlf expression is activated in the crystal cells by SRP/LZ, and that mlf regulates the expansion of this lineage. At the molecular level, our data indicate that MLF controls the number of crystal cells by protecting LZ from proteasome-mediated degradation. Interestingly, human MLF1 is able to substitute for mlf function in the crystal cell lineage. Furthermore, mlf is required for the activity and stable expression of the human leukemogenic protein RUNX1-ETO in Drosophila, whereas human MLF1 depletion causes a decrease in RUNX1-ETO protein and impairs RUNX1-ETO–dependent proliferation in human leukemic blood cells. Thus, we propose that the control of RUNX levels is a conserved function of MLF proteins that play an important role in the control of blood cell homeostasis.
Results
mlf Is Expressed in the Crystal Cell Lineage in Response to SRP/LZ.
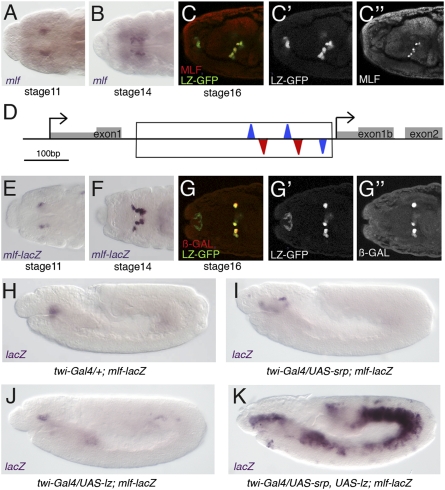
We first explored mlf expression in the Drosophila hematopoietic system. As in vertebrates, in Drosophila hematopoiesis occurs in different waves (1). In the embryo, prohemocytes emerge from the head mesoderm and differentiate into plasmatocytes and crystal cells. In the larva, blood cell progenitors, plasmatocytes, and crystal cells are present in the body cavity and in a specialized organ, the lymph gland, that will release its content at pupariation. Previous work suggested that mlf is expressed in embryonic crystal cells (23). In line with these results, in situ hybridization and immunostaining showed that mlf is expressed from stage 11 in a bilateral cluster of cells that correspond to the crystal cells, as demonstrated by costaining with lz-driven expression of GFP (LZ-GFP) (Fig. 1 A–C). Moreover, high levels of nuclear MLF were detected in larval crystal cells both in the lymph gland and in circulation (Fig. S1). In addition, low levels of MLF were consistently detected throughout the lymph gland (Fig. S1). Thus, mlf may control crystal cell development and larval lymph gland homeostasis.
Fig. 1.
mlf expression is activated by SRP/LZ in the crystal cell lineage. (A–C) mlf is expressed in crystal cells. A and B, in situ hybridization; C, coimmunostaining with GFP (expressed under the control of lz-Gal4). Embryonic stages are indicated. (D) Schematic representation of mlf regulatory sequences. The two alternative promoters and first exons are shown. Conserved GATA (WGATAR) and RUNX (TGYGGTY) consensus binding sites are indicated by blue and red triangles, respectively. The genomic region used to generate the transgenic lines is boxed. (E–G) lacZ expression driven by mlf enhancer is detected in crystal cells. E and F, in situ hybridization; G, coimmunostaining with GFP (expressed under the control of lz-Gal4). Embryonic stages are indicated. (H–K) Twist-driven coexpression of SRP and LZ activates mlf-lacZ expression throughout the mesoderm. lacZ in situ hybridization on stage 11 embryos of the indicated genotypes.
Our analysis of mlf cis-regulatory regions revealed that its first intron contains closely linked GATA and RUNX consensus binding sites that are conserved in other Drosophila species (Fig. 1D), a hallmark of SRP/LZ direct target genes (9). To test whether this cis element is sufficient to recapitulate mlf expression in crystal cells, we generated transgenic lines expressing lacZ under its control. Interestingly, mlf-lacZ expression was detected in embryonic (Fig. 1 E–G) and larval crystal cells (Fig. S1). In addition, lacZ and mlf transcription was activated throughout the mesoderm in response to the ectopic expression of both SRP and LZ, but not in response to SRP or LZ alone (Fig. 1 H–K and Fig. S2). These results suggest that this cis regulatory module is a target of the SRP/LZ complex, and that SRP/LZ activates mlf expression in the crystal cell lineage.
mlf Controls Crystal Cell Development.
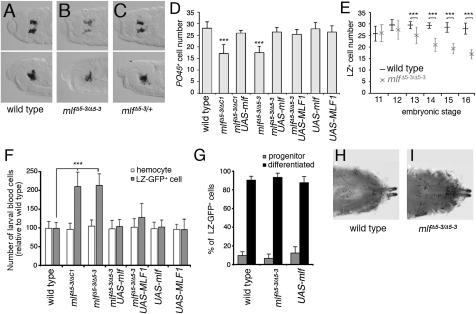
We then asked whether mlf controls hematopoiesis. In situ hybridizations against the plasmatocyte differentiation marker peroxidasin (pxn) and the crystal cell differentiation marker PO45 were used to monitor hemocyte differentiation in embryos carrying a homozygous null mutation in mlf (mlfΔ5–3/Δ5–3) (23). Consistent with the mlf expression pattern, mlf loss did not impair plasmatocyte differentiation (Fig. S3). In contrast, mlf loss affected crystal cell development (Fig. 2). PO45 levels were lower in mlf embryos compared with WT or heterozygous siblings (Fig. 2 A–C), and the number of PO45-expressing cells was reduced (Fig. 2D). Importantly, reexpressing mlf solely in the crystal cells using the lz-Gal4 driver restored normal crystal cell numbers in mlfΔ5–3 homozygous embryos (Fig. 2D), and similar observations were seen using transheterozygous null mutations in mlf (mlfΔ5–3/ΔC1). Furthermore, counting LZ-expressing cells from embryonic stages 11–16 showed that their number was initially WT but decreased progressively in the absence of mlf (Fig. 2E). Therefore, MLF is necessary for crystal cell maintenance in the embryo, and it acts cell-autonomously after the onset of lz expression.
Fig. 2.
mlf controls embryonic and larval crystal cell homeostasis. (A–C) Decreased expression of the crystal cell differentiation marker PO45 is observed in the absence of mlf. Dorsal (Upper) and lateral (Lower) views showing PO45 expression in stage 15 embryos of the indicated genotypes. (D) Stage 15 mlf embryos have fewer PO45-expressing cells. Expression of mlf or its human homolog MLF1 under the control of lz-Gal4 in mlf mutant backgrounds restores WT crystal cell number. (E) Immunostaining against LZ reveals a progressively decreasing number of LZ-expressing hemocytes in mlf embryos. (F and G) lz-Gal4, UAS-GFP third instar wandering larvae of the indicated genotypes were bled to determine the relative number of circulating hemocytes or LZ-GFP+ cells (F), as well as the proportion of differentiated (PO45+) and progenitor (PO45−) LZ-GFP+ cells (G). (F) Loss of mlf induces an increase in the number of circulating larval LZ-GFP+ cells that is suppressed by expression of mlf or human MLF1 in this lineage. (G) mlf does not affect the ratio of progenitor to differentiated LZ-GFP+ cells. (H and I) Posterior segments of third instar larvae showing that mlf loss induces an increase in the number of crystal cells, as demonstrated by their blackening after heat treatment. ***P < 0.0001 compared with WT, Student t test.
We also assessed the function of mlf in the larval lymph gland. In third instar larvae, the lymph gland consists of a pair of primary lobes with prohemocytes in their medullary zone and differentiated cells in their cortical zone, and several smaller secondary lobes containing only prohemocytes (1). In mlf mutant larvae, the lymph glands were hypertrophied and displayed aberrant precocious differentiation (Fig. S4); the expression of the prohemocyte marker tepIV was strongly reduced, whereas both crystal cells and plasmatocytes were present in the secondary lobes. Thus, mlf is required to maintain lymph gland homeostasis, and it prevents premature blood cell differentiation in this organ.
Remarkably, by monitoring circulating larval blood cells, we observed that mlf loss induced an increase in the number of LZ-GFP+ cells, whereas the total hemocyte population was not significantly changed (Fig. 2F). Consequently, the proportion of circulating LZ-GFP+ cells rose from 4.8% in WT to 10.3% in mlf larvae. As in the embryo, LZ-GFP+ cell count was restored to WT when mlf was specifically reexpressed in this lineage (Fig. 2F). However, the proportion of LZ-GFP+ cells expressing PO45 was similar in WT and mlf mutant larvae (Fig. 2G), suggesting that mlf loss does not prevent LZ-GFP+ cell differentiation in crystal cells. Consistent with these results, heat treatment, which activates the melanization cascade in mature crystal cells, revealed the presence of more crystal cells in mlf mutants than in WT controls (Fig. 2 H and I). Thus, although mlf loss results in fewer crystal cells in the embryo, it promotes crystal cell expansion in the larvae. Of note, MLF overexpression did not seem to affect crystal cell number or differentiation in either the embryo or the larva (Fig. 2 D, F, and G), suggesting that MLF is not a limiting factor.
We then asked whether human MLF1 could replace mlf function in the crystal cell lineage. Accordingly, we expressed MLF1 under the control of the lz-Gal4 driver, and determined the number of crystal cells present in the embryo or in circulation in the larvae. Although MLF1 expression did not alter the number of crystal cells in a WT background, it did rescue mlf-associated hematopoietic defects (Fig. 2 D and F). This suggests that although MLF1 exhibits only 23% of identity with MLF (21), the function of MLF proteins has been conserved from Drosophila to human.
mlf Promotes SRP/LZ-Induced Transactivation by Interfering with LZ Degradation.
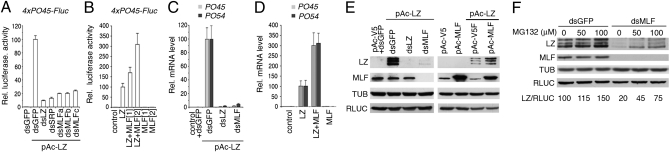
As mentioned above, SRP/LZ-induced gene transcription is pivotal for crystal cell differentiation. Interestingly, in a genome-wide RNAi screen, we recently identified mlf as a potential regulator of SRP/LZ in the Drosophila Kc blood cell line (24). We thus used this system to decipher MLF's mechanism of action. As reported previously (24), transfection of an expression plasmid for LZ (pAc-LZ) activated a reporter gene under the control of the PO45 crystal cell-specific enhancer (4xPO45-Fluc), and knockdown endogenous srp expression by dsRNA impaired this transactivation, indicating that 4xPO45-Fluc activity reflects SRP/LZ-induced transcription (Fig. 3A). Using three different dsRNAs targeting mlf, we showed that mlf depletion strongly reduced 4xPO45-Fluc activation by SRP/LZ. Conversely, cotransfection of increasing amounts of pAc-MLF with pAc-LZ resulted in a dose–response enhancement of the reporter gene activity (Fig. 3B). To extend these observations, we asked whether mlf also controls the induction of SRP/LZ target genes within their genomic context. Real-time quantitative PCR (qPCR) experiments demonstrated that the transactivation of PO45 and Bc/PO54 (another crystal cell-specific differentiation marker) observed on LZ expression was also impaired in the presence of dsRNA against mlf and was enhanced when mlf expression plasmid was cotransfected (Fig. 3 C and D). Therefore, MLF behaves as a coactivator for SRP/LZ.
Fig. 3.
MLF is required for LZ transactivation and stability in Kc cells. (A and B) Luciferase reporter assays. (A) dsRNA against mlf impairs LZ-induced transactivation of the p45PO45-Fluc plasmid. Three different dsRNA probes targeting mlf were tested. (B) Increasing amounts of pAc-MLF expression plasmid enhance pAc-LZ–induced transactivation of p45PO45-Fluc. (C and D) Real-time qPCR assays. (C) dsRNAs against mlf inhibit LZ-induced activation of PO45 and PO54 transcription. (D) Increasing levels of pAc-MLF expression plasmid promote pAc-LZ–induced PO45 and PO54 expression. (E) Western blot analysis showing that LZ levels decrease on depletion of MLF by dsRNA (Left) and increase on cotransfection of pAc-MLF with pAc-LZ (Right). Endogenous tubulin (TUB) and pAc-expressed Renilla luciferase (RLUC) were used as loading and transfection controls. (F) Inhibition of the proteasome by MG132 increases LZ levels in MLF-depleted Kc cells. The relative levels of LZ (normalized to RLUC) are shown in the lower part of the panel.
Strikingly, Western blot analysis revealed that mlf dsRNA not only efficiently knocked down MLF expression, but also strongly decreased pAc-driven expression of LZ (Fig. 3E). In contrast, pAc-driven expression of Renilla luciferase was not affected. Conversely, overexpression of MLF increased LZ levels. Thus, it appears that MLF regulates LZ expression posttranscriptionally. Furthermore, treatment with the proteasome inhibitor MG132 resulted in increased LZ levels and partially restored LZ accumulation in MLF-depleted cells (Fig. 3F). These data raise the possibility that MLF promotes SRP/LZ-induced transactivation by protecting LZ from degradation by the proteasome.
mlf Controls Crystal Cell Development by Impinging on LZ Levels in Vivo.
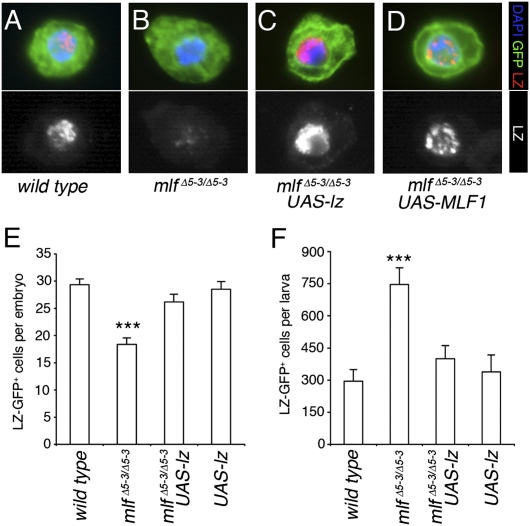
The foregoing results suggested that MLF might regulate crystal cell development by controlling LZ levels. This hypothesis is consistent with the drop in LZ+ cell number in mlf embryos but seems at odds with its increase in mlf larvae. However, in line with our observation in Kc cells, immunofluorescent labeling revealed significantly reduced LZ expression in LZ-GFP+ circulating cells of mlf mutant larvae compared with control larvae (Fig. 4 A and B). Of note, GFP expression, which is driven under the control of lz, did not seem markedly affected by the loss of mlf, indicating that mlf is not required for lz transcription but is critical for LZ protein accumulation. Moreover, in the eye disk, where mlf is ubiquitously expressed but not up-regulated in the LZ+ cells, mlf loss also decreased LZ levels, but only modestly (Fig. S5), suggesting that LZ stability is differentially regulated in the hematopoietic and visual systems. Importantly, expressing human MLF1 in a mlf mutant background not only rescued crystal cell number (Fig. 2 D and F), but also restored LZ expression (Fig. 4D and Fig. S6A), providing further evidence that MLF proteins regulate LZ levels. We thus reasoned that increasing LZ expression might rescue the crystal cell defects caused by mlf loss. Consequently, we generated mlf mutants carrying the lz-Gal4 driver together with a UAS-GFP transgene and a UAS-lz transgene. In these conditions, we observed strong LZ expression in the crystal cells of mlf mutant larvae (Fig. 4C and Fig. S6A). Furthermore, the expression of the UAS-lz transgene suppressed not only the drop in crystal cell number in mlf embryos, but also the rise in crystal cell number in mlf larvae (Fig. 4 E and F). Therefore, these two apparently opposed phenotypes likely stem from a common cause—a decrease in LZ levels.
Fig. 4.
Decreased LZ levels are responsible for MLF-associated crystal cell phenotypes. (A–D) MLF stabilizes LZ in vivo. Fluorescent immunostaining against LZ in circulating larval blood cells from lz-Gal4, UAS-GFP third instar larva of the indicated genotype is shown. Nuclei were stained with DAPI. The lower panels show LZ immunostaining only. (E and F) Reexpression of LZ under the control of the lz-Gal4 restores WT LZ-GFP+ cell number in the embryo (E) and in third instar larvae circulation (F). GFP expression was used to score the number of LZ-GFP+ cells in stage 14–15 embryos (E) or in third instar wandering larvae (F) of the indicated genotypes.
Human Leukemic Fusion Protein RUNX1-ETO Is Regulated by MLF Proteins.
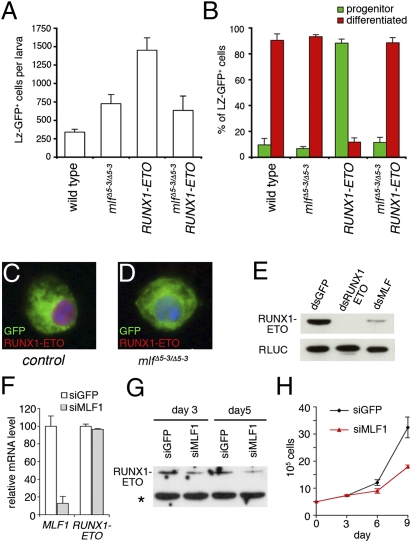
We recently showed that expression of the human RUNX oncogenic fusion protein RUNX1-ETO in the LZ+ blood cell lineage induces a preleukemic switch in flies (14). Indeed, it inhibits crystal cell differentiation and promotes LZ-GFP+ progenitor amplification in the larvae (14) (Fig. 5 A and B). In light of the foregoing results and of the link between MLF and leukemia in humans, we wondered whether mlf interacted genetically with RUNX1-ETO in our model. Strikingly, we found that RUNX1-ETO leukemogenic activity was almost abolished in mlf mutant larvae; the number of circulating LZ-GFP+ cells was significantly decreased, and their differentiation was restored (Fig. 5 A and B). Furthermore, in the absence of mlf, RUNX1-ETO expression was barely detected, whereas robust staining was observed in control larvae (Fig. 5 C and D and Fig. S6B). Thus, we propose that the suppression of RUNX1-ETO–induced blood cell phenotypes observed in the absence of mlf reflects the role of MLF in RUNX1-ETO stabilization. In line with this hypothesis, Western blot analysis showed that the expression of RUNX1-ETO transfected in Kc cells was strongly down-regulated on dsRNA-mediated depletion of MLF (Fig. 5E).
Fig. 5.
MLF is required for RUNX1-ETO activity. (A and B) The phenotypes induced on expression of RUNX1-ETO in the LZ+ blood cell lineage are suppressed in the absence of mlf. (A) Absolute number of circulating LZ-GFP+ cells in third instar larvae. (B) Ratio of circulating progenitors (GFP+, PO45−) to differentiated (GFP+, PO45+) LZ+ cells in third instar larvae. (C–E) MLF is required for RUNX1-ETO expression in Drosophila. (C and D) Fluorescent immunostaining against RUNX1-ETO in lz-Gal4,UAS-GFP;UAS-RUNX1-ETO (C, control) and lz-Gal4,UAS-GFP;mlfΔ5–3/Δ5–3;UAS-RUNX1-ETO (D, mlfΔ5–3/Δ5–3) circulating larval blood cells. Nuclei were stained with DAPI. (E) Western blot analysis shows that RUNX1-ETO levels decrease after depletion of MLF by dsRNA in Kc cells. (F and G) In Kasumi-1 cells, MLF1 depletion by siRNA does not affect RUNX1-ETO mRNA levels (F; real-time qPCR), but does reduce RUNX1-ETO protein levels (G; Western blot analysis). The asterisk indicates a nonspecific band that provides a loading control. (H) Knockdown of MLF1 expression inhibits Kasumi-1 cell growth.
Finally, we asked whether MLF1 also regulates RUNX1-ETO expression in human. Kasumi-1 cells derived from a patient with AML carrying the t(8;21) translocation were found to constitutively express RUNX1-ETO, which is required for their proliferation (25). On transfection of an siRNA against MLF1, we found a significant reduction of MLF1 transcript levels, but no effect on RUNX1-ETO mRNA expression (Fig. 5F). However, consistent with our observations in Drosophila, Western blot analysis showed strongly decreased RUNX1-ETO protein levels (Fig. 5G), paralleled by reduced cell growth (Fig. 5H). Taken together, our results reveal a conserved functional relationship between MLF family members and the leukemogenic fusion protein RUNX1-ETO.
Discussion
Although deregulation of MLF1 has been linked to AML, the physiological role of MLF family members in hematopoiesis remains largely unknown. Focusing our analysis on Drosophila hematopoietic development, we have demonstrated that MLF controls blood cell homeostasis. In particular, we provide strong evidence that MLF is required to stabilize the RUNX factor LZ during crystal cell development. In addition, our findings suggest that the regulation of RUNX activity by MLF is conserved in humans, where it could play an important role in leukemogenesis.
Our findings reveal MLF's regulatory function in the control of crystal cell production. Actually mlf, which exhibits a rather ubiquitous expression pattern, is highly expressed in these blood cells, and mlf expression in this lineage is activated by SRP/LZ. We found that MLF controls crystal cell number in a cell-autonomous manner, chiefly by impinging on LZ levels. We propose that the induction of mlf expression by SRP/LZ contributes to crystal cell development by stabilizing LZ. As such, this gene regulatory network forms a two-component positive feedback loop that drives development forward by stabilizing the expression of lineage-specific regulators (26). Our findings also show that MLF controls lymph gland homeostasis, where it seems to promote hematopoietic progenitor maintenance. Although some of the factors controlling lymph gland development have been identified, no RUNX factor has been implicated in prohemocyte maintenance (1), suggesting that MLF has other partners in these cells. Of interest, MLF has been shown to bind Su(fu) and to possibly antagonize its function (27). Because Su(fu) negatively regulates both Hh and Wnt pathways (28, 29), which are required for prohemocyte maintenance (30), their premature differentiation in mlf mutants could result from increased Su(fu) activity. We anticipate that deciphering MLF's mode of action in the lymph gland will provide valuable insight into the regulation of blood cell progenitor fate.
Along with revealing the role of MLF in hematopoiesis, our findings shed light on the function and regulation of LZ. We found that decreased LZ levels in mlf mutant larvae resulted in an increased number of circulating LZ+ cells, but did not block these cells’ differentiation, suggesting that low levels of LZ are sufficient to induce crystal cell differentiation. In addition, expansion of the pool of LZ+ cells might reflect a slowdown in the cells’ rate of differentiation or a direct function of LZ in controlling blood cell proliferation or apoptosis. Along this line, LZ was shown to promote cell death in the eye, notably by regulating the expression of the Drosophila homolog of the Wilms’ tumor gene 1 (WT1) (31). Alternatively, decreased LZ levels may make crystal cell progenitors more susceptible to proliferative cues from the Notch pathway, which regulates larval crystal cell numbers (32, 33). In mammals, RUNX1 acts mostly as a brake on blood cell progenitor proliferation, and decreased RUNX1 dosage, as well as MDS/AML-associated mutations or translocations affecting RUNX1, tend to promote aberrant self-renewal (34). Interestingly, RUNX1-ETO also promotes hematopoietic progenitor cell expansion in Drosophila (14, 35), and both WT1 overexpression and activation of the Notch signaling pathway have been linked to RUNX1-ETO–induced AML (36, 37). Given these similarities, characterizing the function of LZ in the control of crystal cell number may have broader implications.
Notwithstanding the evolutionary distance between human and fly, the human MLF1 protein rescued mlf-associated crystal cell defects, including LZ down-regulation, whereas mlf and MLF1 were required for the stable expression of RUNX1-ETO in Drosophila and human leukemia cells, respectively. Thus, the regulation of RUNX turnover seems to be a conserved function of MLF family members. The proteasome was found to regulate RUNX1-ETO as well as other RUNX proteins in human cells (38, 39), and our data indicate that LZ is degraded in a proteasome-dependent manner in the absence of mlf. An important area of future inquiry will be to determine more precisely how RUNX stability is regulated by MLF. This is of particular interest given that altered RUNX levels are associated with several diseases in humans, including familial platelet disorders and AML for RUNX1 and cleidocranial displasia for RUNX2 (40). Actually, the slightly reduced LZ levels that we observed in mlf mutant eye discs suggests that the regulation of RUNX stability by MLF is not restricted to the hematopoietic system. Moreover, few genes required for RUNX1-ETO–induced AML have been identified so far, and our data suggest that MLF1 is a critical component for RUNX1-ETO leukemogenic activity. Indeed, along with the MDS/AML-associated t(3;5) translocation that generates the NPM-MLF1 fusion protein, MLF1 overexpression was correlated with malignant progression (17). The mechanisms underlying the oncogenic activity of NPM-MLF1 or MLF1 remain largely unknown, however. Similarly, the function of MLF1 in mammals remains poorly characterized. Exploring the relationship between MLF and RUNX factors could shed further light on MLF's role and mode of action.
Blood cell development is controlled by an intricate network of genes, the activity of which must be tightly controlled to ensure proper cell lineage choice, proliferation, and differentiation. Our present findings show that the dynamic and coordinated control of gene expression through positive feedback loops participates in the fine-tuning of hematopoiesis, and provides a framework for future investigations of the cross-regulatory interactions that control blood cell fate. Finally, our results open up new avenues of research into the mode of action of MLF family members as conserved regulators of RUNX protein stability, and we envision that Drosophila will provide a powerful model for deciphering MLF's function in hematopoiesis and leukemia.
Materials and Methods
Fly Strains.
We used the following Drosophila melanogaster strains: mlfΔ5–3, mlfΔC1 (23), UAS-mlfA (23), UAS-RUNX1-ETO (14), UAS-lz (P. Gergen, Stony Brook University, NY), lz-Gal4, UAS-GFP, and UAS-MLF1 (Bloomington Stock Center). To generate mlf-lacZ transgenic reporter lines, the mlf first intron was PCR-amplified from D. melanogaster genomic DNA and cloned into a pCasper-hsp43-lacZ vector. The resulting vector was used to produce transgenic lines by P element-mediated transformation into w1118 flies.
In Situ Hybridization, Immunostaining, and Hemocyte Counts.
Immunostaining and in situ hybridization were performed as described previously (9). For in situ hybridization, we used digoxigenin-UTP–labeled RNA probes and sheep anti-digoxigenin coupled to alkaline phosphatase (Roche). For immunostaining, we used mouse anti-LZ (Developmental Studies Hybridoma Bank), rabbit anti-MLF (27), rabbit anti–β-galactosidase (Cappel), mouse or rabbit anti-GFP (Torrey), mouse anti-P1/NimC1 (a kind gift from I. Ando, Biological Research Center, Szeged, Hungary), rabbit anti-AML1 (Calbiochem), and secondary antibodies coupled to Alexa Fluor 488 or 555 (Molecular Probes). Circulating larval hemocyte counts and LZ+ cell differentiation status were assessed as described previously (14). In brief, GFP expression from lz-Gal4 and UAS-GFP was used to determine LZ+ cell number, and DAPI staining was used to count the whole hemocyte population on individually bled third instar female larvae. To monitor LZ+ cell differentiation status, female third instar larvae were bled on polylysine-coated slides, and the proportion of LZ-GFP+ cells expressing PO45 was assessed after in situ hybridization coupled to immunostaining. Counts were performed on a minimum of 12 samples for each genotype.
Cell Culture, Transfection, Luciferase and Western Blot Assays, and qPCR.
For transfection, dsRNA treatment, luciferase assays, Western blot analyses, and real-time qPCR, Drosophila Kc cells were treated essentially as described previously (24). In brief, Kc cells were grown at 25 °C in Schneider's medium supplemented with 10% (vol/vol) FBS and 50 μg of penicillin/streptomycin. For dsRNA treatment and transfections, cells were plated on in vitro-transcribed and purified dsRNA at 24 h before transfection with Effectene (Qiagen). We used the following plasmids: pAc-MLFA (23), pAc-V5 (Invitrogen), pAc-LZ-V5, and p4xPO45-Firefly luciferase (24). pAc-RUNX1-ETO was generated by subcloning RUNX1-ETO ORF from pUAST-RUNX1-ETO (14) into pAc-V5. pAc-Renilla luciferase was used for transfection normalization. Three independent dsRNA targeting different regions of mlf were produced. The sequences of the T7-containing primers used to generate the dsRNA are available on request. Firefly and Renilla luciferase activity was measured using the Promega Dual Luciferase Reporter Assay System. PO45, Bc/PO54, and RP49 mRNA expression levels were measured by real-time qPCR (24). For proteasome inhibition, Kc cells were incubated for 8 h with MG132 (or DMSO) before being processed for analysis. Kasumi-1 cells were grown in RPMI-1640 medium (Sigma-Aldrich) supplemented with 2 mM l-glutamine and 10% FBS. siRNAs against MLF1 (5′-GGUGCUGGGUAAUAAGCAU-3′) or GFP (5′-GCAAGCUGACCCUGAAGUUCAU-3′) were transfected on day 0 and every 48 h thereafter by electroporation, as described previously (25). RNA and proteins were extracted on days 3 and 5. For real-time qPCR, MLF1 and RUNX1-ETO mRNA levels were normalized to GAPDH expression. Kasumi-1 cell numbers were counted every 3 d using the trypan blue exclusion assay. The following primary antibodies were used for Western blot analyses: mouse anti-V5 (Invitrogen), rabbit anti-MLF (27), mouse anti-ETO (Calbiochem), mouse anti-tubulin (Sigma-Aldrich), and rabbit anti-Renilla luciferase (MBL International). All experiments were reproduced at least three times. Means and SDs were calculated from independent triplicates.
Supplementary Material
Acknowledgments
We thank members of our teams for fruitful discussions and Y. Nakao and W. Sugano for their participation. We thank staff at the Toulouse (RIO) and Institut Jacques Monod (ImagoSeine) imaging platforms for assistance with confocal microscopy. S.B. was supported by fellowships from the Ministère de l'Enseignement Supérieur et de la Recherche and the Fondation pour la Recherche Médicale. S.M.-L. received fellowships from the Ministère de l'Enseignement Supérieur et de la Recherche and the Ligue Contre le Cancer. V.G. was supported by a postdoctoral fellowship from the Association pour la Recherche sur le Cancer. O.B. was supported by the Association for International Cancer Research. A.P. was supported by a grant from the Association pour la Recherche sur le Cancer. L.W. was supported by grants from the Association for International Cancer Research, Association pour la Recherche sur le Cancer, and Ligue Midi-Pyrénée Contre le Cancer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117317109/-/DCSupplemental.
References
- 1.Crozatier M, Vincent A. Drosophila: A model for studying genetic and molecular aspects of haematopoiesis and associated leukaemias. Dis Model Mech. 2011;4:439–445. doi: 10.1242/dmm.007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Odenike O, Rowley JD. Leukaemogenesis: More than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyoshi H, et al. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffman JA. Is Runx a linchpin for developmental signaling in metazoans? J Cell Biochem. 2009;107:194–202. doi: 10.1002/jcb.22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyama S, Mulloy JC. Molecular pathogenesis of core binding factor leukemia: Current knowledge and future prospects. Int J Hematol. 2011;94:126–133. doi: 10.1007/s12185-011-0858-z. [DOI] [PubMed] [Google Scholar]
- 6.Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science. 2000;288:146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- 7.Fossett N, Hyman K, Gajewski K, Orkin SH, Schulz RA. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc Natl Acad Sci USA. 2003;100:11451–11456. doi: 10.1073/pnas.1635050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waltzer L, Ferjoux G, Bataillé L, Haenlin M. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 2003;22:6516–6525. doi: 10.1093/emboj/cdg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferjoux G, Augé B, Boyer K, Haenlin M, Waltzer L. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev Biol. 2007;305:726–734. doi: 10.1016/j.ydbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Elagib KE, et al. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 11.Pencovich N, Jaschek R, Tanay A, Groner Y. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood. 2011;117:e1–e14. doi: 10.1182/blood-2010-07-295113. [DOI] [PubMed] [Google Scholar]
- 12.Tijssen MR, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Osman D, et al. A Drosophila model identifies calpains as modulators of the human leukemogenic fusion protein AML1-ETO. Proc Natl Acad Sci USA. 2009;106:12043–12048. doi: 10.1073/pnas.0902449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoneda-Kato N, et al. The t(3;5)(q25.1;q34) of myelodysplastic syndrome and acute myeloid leukemia produces a novel fusion gene, NPM-MLF1. Oncogene. 1996;12:265–275. [PubMed] [Google Scholar]
- 16.Rau R, Brown P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: Towards definition of a new leukaemia entity. Hematol Oncol. 2009;27:171–181. doi: 10.1002/hon.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto N, et al. Elevated MLF1 expression correlates with malignant progression from myelodysplastic syndrome. Leukemia. 2000;14:1757–1765. doi: 10.1038/sj.leu.2401897. [DOI] [PubMed] [Google Scholar]
- 18.Yoneda-Kato N, Tomoda K, Umehara M, Arata Y, Kato JY. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. EMBO J. 2005;24:1739–1749. doi: 10.1038/sj.emboj.7600656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winteringham LN, Kobelke S, Williams JH, Ingley E, Klinken SP. Myeloid Leukemia Factor 1 inhibits erythropoietin-induced differentiation, cell cycle exit and p27Kip1 accumulation. Oncogene. 2004;23:5105–5109. doi: 10.1038/sj.onc.1207661. [DOI] [PubMed] [Google Scholar]
- 20.Williams JH, et al. HLS7, a hemopoietic lineage switch gene homologous to the leukemia-inducing gene MLF1. EMBO J. 1999;18:5559–5566. doi: 10.1093/emboj/18.20.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno K, et al. Characterization of a Drosophila homologue of the human myelodysplasia/myeloid leukemia factor (MLF) Gene. 2000;260:133–143. doi: 10.1016/s0378-1119(00)00447-9. [DOI] [PubMed] [Google Scholar]
- 22.Sugano W, Yamaguchi M. Identification of novel nuclear localization signals of Drosophila myeloid leukemia factor. Cell Struct Funct. 2007;32:163–169. doi: 10.1247/csf.07039. [DOI] [PubMed] [Google Scholar]
- 23.Martin-Lannerée S, et al. Characterization of the Drosophila myeloid leukemia factor. Genes Cells. 2006;11:1317–1335. doi: 10.1111/j.1365-2443.2006.01023.x. [DOI] [PubMed] [Google Scholar]
- 24.Gobert V, et al. A genome-wide RNA interference screen identifies a differential role of the mediator CDK8 module subunits for GATA/ RUNX-activated transcription in Drosophila. Mol Cell Biol. 2010;30:2837–2848. doi: 10.1128/MCB.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez N, et al. The oncogenic fusion protein RUNX1-CBFA2T1 supports proliferation and inhibits senescence in t(8;21)-positive leukaemic cells. BMC Cancer. 2004;4:44. doi: 10.1186/1471-2407-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swiers G, Patient R, Loose M. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev Biol. 2006;294:525–540. doi: 10.1016/j.ydbio.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 27.Fouix S, et al. Over-expression of a novel nuclear interactor of Suppressor of fused, the Drosophila myelodysplasia/myeloid leukaemia factor, induces abnormal morphogenesis associated with increased apoptosis and DNA synthesis. Genes Cells. 2003;8:897–911. doi: 10.1046/j.1365-2443.2003.00685.x. [DOI] [PubMed] [Google Scholar]
- 28.Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol. 1998;8:583–586. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- 29.Meng X, et al. Suppressor of fused negatively regulates β-catenin signaling. J Biol Chem. 2001;276:40113–40119. doi: 10.1074/jbc.M105317200. [DOI] [PubMed] [Google Scholar]
- 30.Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wildonger J, Sosinsky A, Honig B, Mann RS. Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev. 2005;19:1034–1039. doi: 10.1101/gad.1298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-α in development and survival of Drosophila blood cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duvic B, Hoffmann JA, Meister M, Royet J. Notch signaling controls lineage specification during Drosophila larval hematopoiesis. Curr Biol. 2002;12:1923–1927. doi: 10.1016/s0960-9822(02)01297-6. [DOI] [PubMed] [Google Scholar]
- 34.Link KA, Chou FS, Mulloy JC. Core binding factor at the crossroads: Determining the fate of the HSC. J Cell Physiol. 2010;222:50–56. doi: 10.1002/jcp.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinenko SA, et al. Genetic manipulation of AML1-ETO–induced expansion of hematopoietic precursors in a Drosophila model. Blood. 2010;116:4612–4620. doi: 10.1182/blood-2010-03-276998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcalay M, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112:1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishida S, et al. AML1-ETO rapidly induces acute myeloblastic leukemia in cooperation with the Wilms tumor gene, WT1. Blood. 2006;107:3303–3312. doi: 10.1182/blood-2005-04-1656. [DOI] [PubMed] [Google Scholar]
- 38.Krämer OH, Müller S, Buchwald M, Reichardt S, Heinzel T. Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARα. FASEB J. 2008;22:1369–1379. doi: 10.1096/fj.06-8050com. [DOI] [PubMed] [Google Scholar]
- 39.Yang G, Thompson MA, Brandt SJ, Hiebert SW. Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene. 2007;26:91–101. doi: 10.1038/sj.onc.1209760. [DOI] [PubMed] [Google Scholar]
- 40.Cohen MM., Jr Perspectives on RUNX genes: An update. Am J Med Genet A. 2009;149A:2629–2646. doi: 10.1002/ajmg.a.33021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.