Abstract
The motility of organisms is often directed in response to environmental stimuli. Rheotaxis is the directed movement resulting from fluid velocity gradients, long studied in fish, aquatic invertebrates, and spermatozoa. Using carefully controlled microfluidic flows, we show that rheotaxis also occurs in bacteria. Excellent quantitative agreement between experiments with Bacillus subtilis and a mathematical model reveals that bacterial rheotaxis is a purely physical phenomenon, in contrast to fish rheotaxis but in the same way as sperm rheotaxis. This previously unrecognized bacterial taxis results from a subtle interplay between velocity gradients and the helical shape of flagella, which together generate a torque that alters a bacterium's swimming direction. Because this torque is independent of the presence of a nearby surface, bacterial rheotaxis is not limited to the immediate neighborhood of liquid–solid interfaces, but also takes place in the bulk fluid. We predict that rheotaxis occurs in a wide range of bacterial habitats, from the natural environment to the human body, and can interfere with chemotaxis, suggesting that the fitness benefit conferred by bacterial motility may be sharply reduced in some hydrodynamic conditions.
Keywords: low Reynolds number, directional motion, chirality
The effectiveness and benefit of motility are largely determined by the dependence of movement behavior on environmental stimuli. For example, chemical stimuli may affect the spreading of tumor cells (1) and allow bacteria to increase uptake by swimming toward larger nutrient concentrations (2, 3), whereas hydrodynamic stimuli can stifle phytoplankton migration (4), allow protists to evade predators (5), and change sperm–egg encounter rates for external fertilizers (6). Microorganisms exhibit a broad range of directed movement responses, called “taxes”. Whereas some of these responses, such as chemotaxis (7) and thermotaxis (8), are active and require the ability to sense and respond to the stimulus, others, such as magnetotaxis (9) and gyrotaxis (4), are passive and do not imply sensing, instead resulting purely from external forces.
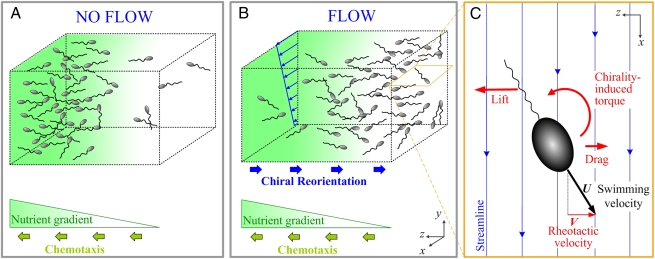
Chemotaxis is the best studied among these directional motions: Bacteria measure chemical concentrations and migrate along gradients (Fig. 1A). For instance, chemotaxis guides Escherichia coli to epithelial cells in the human gastrointestinal tract, favoring infection (10); Rhizobium bacteria to legume root hairs in soil, favoring nitrogen fixation (2); and marine bacteria to organic matter, favoring remineralization (3). Equally as pervasive as chemical gradients in microbial habitats are gradients in ambient fluid velocity or “shear” (Fig. 1B). Although nearly every fluid environment experiences velocity gradients—from laminar shear in bodily conduits and soil to turbulent shear in streams and oceans—the effect of velocity gradients on bacterial motility has received negligible attention compared with chemical gradients, partly due to the difficulty of studying motility under controlled flow conditions.
Fig. 1.
Bacterial chemotaxis and rheotaxis. (A) Many bacteria can climb chemical gradients by chemotaxis, for example to seek high nutrient concentrations. (B) Ambient velocity gradients can also affect motility, potentially hampering chemotaxis by orienting bacteria away from nutrient sources. The x–y plane is the flow gradient plane, defined by the direction of the flow (x) and the direction of the flow velocity gradient (y). (C) The mechanism responsible for bacterial rheotaxis, shown for a cell with a left-handed flagellum. The chirality of the flagellum causes a lift force along +z. This force is opposed by the drag on the cell body, producing a torque on the cell. This torque reorients the bacterium, which therefore has a component V of its swimming velocity U directed along –z. V is the rheotactic velocity.
Rheotaxis refers to changes in organism movement patterns due to shear. Rheotaxis is common in fish (11, 12), which actively sense shear and respond by either turning into the flow or fleeing high-flow regions. Certain insects use rheotaxis to escape droughts by swimming upstream in desert rivers (13). At smaller scales, spermatozoa exhibit rheotaxis, likely resulting from a passive hydrodynamic effect, whereby the combination of a gravitational torque and a shear-induced torque orients the swimming direction preferentially upstream (14–16). Responses to shear are also observed in copepods and dinoflagellates, which rely on shear detection to attack prey or escape predators (5, 17–19), orient in flow (20), and retain a preferential depth (21). Evidence of shear-driven motility in prokaryotes is limited to the upstream motion of mycoplasma (22), E. coli (23), and Xylella fastidiosa (24), all of which require the presence of a solid surface. In contrast, little is known about the effect of shear on bacteria freely swimming in the bulk fluid. Using Bacillus subtilis as a model organism, we here report that bacteria exhibit rheotaxis that is not conditional to the presence of a nearby surface and we demonstrate that this phenomenon results from the coupling of motility, shear, and cell morphology.
Results and Discussion
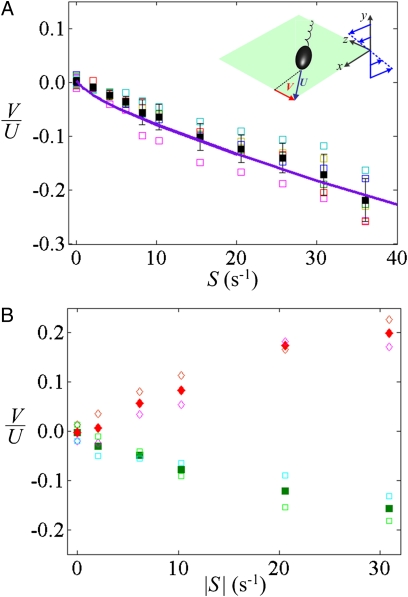
To expose bacteria to controlled shear flows, we injected a suspension of the smooth-swimming B. subtilis OI4139 (25) (Materials and Methods) in an H = 90-μm deep microfluidic channel. This strain has a 1 × 3-μm sausage-shaped body, multiple left-handed helical flagella, and swims at U = 40–65 μm⋅s−1. Bacteria were imaged a distance H/4 above the bottom of the microchannel, where the shear rate S was varied between 0 and 36 s−1. Cell-tracking software yielded the bacteria's drift velocity, V, perpendicular to the flow gradient plane (i.e., along z; Fig. 1C and Materials and Methods).
Experiments revealed a large and highly reproducible drift velocity along –z (Fig. 2A), whose magnitude increased with increasing shear rate. The drift velocity amounted to a considerable fraction of the swimming speed, with V/U = 6.5% ± 2.4% (mean ± SD) at a shear rate of S = 10 s−1 and 22% ± 4% at S = 36 s−1. Furthermore, bacteria imaged one-quarter depth below the top of the channel, where the shear rate had the same magnitude but opposite sign, exhibited the same drift velocity, only in the opposite direction (Fig. 2B). Taken together, these results indicate that the observed drift was caused by shear.
Fig. 2.
B. subtilis OI4139 exhibits rheotaxis. The rheotactic velocity, V, normalized by the mean swimming speed, U (55 μm⋅s−1), is shown as a function of the shear rate, S. (A) Data obtained one-quarter depth above the bottom of the microchannel. Open symbols of different colors refer to six replicate experiments (Materials and Methods). Black solid squares with bars denote the mean and SD of the six replicates. The solid line is the model prediction for a cell body radius of a = 0.5 μm and flagellar morphology corresponding to the flagellar bundle of B. subtilis (Materials and Methods). (Inset) Schematic of the coordinate system, flow, and bacterial motility. (B) Data obtained one-quarter depth below the top (red and pink) and one-quarter depth above the bottom (green and cyan) of the microchannel show an equal and opposite rheotactic velocity, V, because the shear rate S is equal in magnitude and opposite in sign at these two depths. Open symbols refer to two replicate experiments and solid symbols denote the mean of the two replicates.
This drift is surprising because many objects, such as spheres and ellipsoids, do not drift across streamlines at the low Reynolds numbers, Re = UFL/v < 0.02, characteristic of our experiments (26) (L is the object's length, UF the mean flow velocity, and v the kinematic viscosity of the fluid). However, this principle does not apply to chiral objects, like helices, which can drift in the direction perpendicular to the flow gradient plane (27, 28). This effect was recently demonstrated for nonmotile spirochetes (29) and originates from the hydrodynamic stresses on a helix in flow, which produce a net lift force on the helix that is normal to the flow gradient plane (Fig. 1C). This force—balanced by drag—causes the helix to drift across streamlines.
The helical shape of bacterial flagella suggests that the same phenomenon might be responsible for the observed drift. However, a model of the flagellar bundle of B. subtilis predicts a drift velocity that has the opposite direction (+z) to that observed (–z; Fig. 2A) and a magnitude of only 0.16 μm⋅s−1 (V/U = 0.29%) at S = 10 s−1, 22-fold smaller than measured (Fig. 2A). On the other hand, B. subtilis differs from a helix in two respects—motility and the presence of a cell body—suggesting a different drift mechanism. Because the body is not chiral, it is not subject to a lift force and the drag on it acts as an anchor for the drifting flagellar bundle (Fig. 1C). Together, the lift and drag forces produce a torque (Fig. 1C), which orients the cell away from the flow direction. This reorientation induces a component of swimming velocity along –z, in agreement with observations (Fig. 2A). According to this mechanism, then, drift results from a bias in the swimming direction mediated by shear and thus represents a case of rheotaxis.
To quantify the rheotactic velocity predicted from chirality-induced reorientation, we developed a mathematical model based on resistive force theory (RFT) (30). The model quantifies the forces and torques on a smooth-swimming B. subtilis cell in shear flow. For simplicity, we model B. subtilis as a sphere of radius a attached to a left-handed rigid helix that has the flagellar dimensions of B. subtilis: a three-turn left-handed helical bundle with axial length of 6 μm and diameter of 480 nm (31). RFT expresses the drag force per unit length as f = ζ// vr// + ζ⊥ vr⊥, where ζ// and ζ⊥ are the resistive force coefficients, and vr// and vr⊥ are the velocities relative to the fluid, parallel and perpendicular to a segment of the helix, respectively (30). Drag on each segment of the helix, assimilated to a thin rod, is anisotropic, with a greater resistance for motion perpendicular to the long axis compared with motion parallel to the long axis. In our model we use ζ⊥ /ζ// = 1.5, in accordance with the range of values proposed in the literature for low Reynolds number flow (ζ⊥/ζ// = 1.4–1.7) (30).
Integrating the local force and torque per unit length along the entire helix yields the total force and torque on the helix as a function of the linear and angular velocities of the helix. The drag forces and torques on a sphere of radius a moving with linear velocity v and angular velocity ω are given by 6πηav and 8πηa3ω, respectively (32), where η is the dynamic viscosity of the fluid. We use the fact that the translational drag in shear flow is the same as in uniform flow and that the effect of shear flow on rotational drag is to change the rotational rate only by an amount equal to half the vorticity.
For any given orientation (ψ,θ) of the cell axis, where ψ and θ are polar and azimuthal angles (Fig. S1), this condition yields the linear and angular velocities of the bacterium, including the rheotactic velocity vz(ψ,θ). The probability distribution of the cell orientation, c(ψ,θ), is determined by the competition between shear, which produces preferential orientation along streamlines and periodic orbits known as Jeffery orbits, and Brownian rotational diffusion, which randomizes the orientation. Specifically, c(ψ,θ) is governed by a Fokker–Planck equation 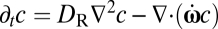 , where DR is the rotational diffusivity of B. subtilis (Materials and Methods) and
, where DR is the rotational diffusivity of B. subtilis (Materials and Methods) and 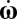 is the rate of change of the orientation angles due to shear. The steady-state solution for c(ψ,θ) was obtained numerically (using COMSOL Multiphysics) and used to quantify the mean rheotactic velocity
is the rate of change of the orientation angles due to shear. The steady-state solution for c(ψ,θ) was obtained numerically (using COMSOL Multiphysics) and used to quantify the mean rheotactic velocity 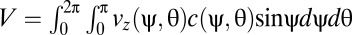 .
.
The model predictions (Fig. 2A, purple line) are in good agreement with observations (Fig. 2A, symbols), for both the direction of drift (–z) and the functional form of the drift velocity V, which increases sublinearly with S. For a model cell body of a = 0.5 μm radius, the agreement is also quantitative. Furthermore, the model correctly predicts that a change of sign in S reverses the direction of V, as found experimentally by contrasting data at one-quarter depth and three-quarters depth (Fig. 2B).
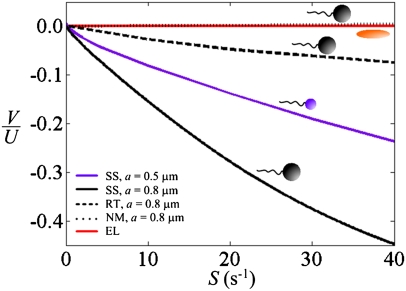
To demonstrate that rheotaxis results from the coupling of chirality, motility, and shear, we compare the predicted rheotactic velocity of motile bacteria, nonmotile bacteria, and hypothetical motile ellipsoids (i.e., nonchiral swimmers). For the latter, we apply the same model described above to quantify the orientational distribution (with a rotational diffusivity described in Materials and Methods) and prescribe the ellipsoids to move along their major axis at a constant speed relative to the fluid.
When S = 0, cell orientation is isotropic and thus V = 0 for all three cases. When |S| > 0, nonchiral swimmers undergo Jeffery orbits (26), swimming along +z and –z with equal probability, so that V = 0 for any shear rate (Fig. 3, red line). Nonmotile bacteria experience a lift force and drift slowly along +z (Fig. 3, dotted black line). For motile bacteria, this drift is overwhelmed by the reorientation of the swimming direction, resulting in a rapid drift along –z (Fig. 3, black and purple lines). Finally, we note that rheotaxis is reduced, but not eliminated, when one takes into account the tumbles characteristic of the movement pattern of many bacteria (Fig. 3, dashed black line, and Materials and Methods).
Fig. 3.
Predicted rheotactic velocity. A mathematical model reveals the mechanism behind bacterial rheotaxis. The rheotactic velocity, V, is plotted relative to the mean swimming speed, U (55 μm⋅s−1), as a function of the shear rate, S. SS refers to smooth-swimming bacteria, with a cell body of radius a. RT refers to bacteria that run and tumble. NM refers to nonmotile bacteria. EL refers to a hypothetical ellipsoid moving along its major axis at a constant speed relative to the fluid (i.e., a nonchiral swimmer). All model bacteria (SS, RT, and NM) have the same flagellar morphology, corresponding to the flagellar bundle of B. subtilis.
The rheotaxis described here results from the interaction between bacterial morphology, motility, and shear flow. As demonstrated by our model, this effect is independent of the presence of a nearby surface and can therefore occur also in the bulk fluid. This result is in contrast to previous observations of upstream swimming of E. coli cells in microfluidic shear flows, a process that hinges on the hydrodynamic interaction between the bacteria and the surface, which causes a torque that tilts the cell preferentially upstream (23). The upstream migration of X. fastidiosa, hypothesized to originate from a preferential upstream alignment due to a hydrodynamic torque (24), is also critically dependent on the presence of a surface: These bacteria have no flagella and can move only by migrating along surfaces through pili-mediated twitching motility. In our experiments, the channel top and bottom surfaces were responsible for the generation of shear, because they imposed no-slip conditions on the flow, but were sufficiently distant from the bacteria not to cause any hydrodynamic interaction. Therefore, shear from any origin—whether surface related or caused in the bulk fluid—can lead to the rheotactic movement pattern reported here.
Modeling revealed that bacterial rheotaxis is very sensitive to changes in morphology, suggesting that different species will exhibit different degrees of rheotaxis. For example, the rheotactic velocity increases with the size of the cell body, because a larger body yields a larger torque and, more subtly, reduces Brownian rotational diffusivity: Both effects increase the rheotactic velocity V. Using a spherical cell body with radius a = 0.5 μm, model predictions are in good quantitative agreement with the experimental data (Fig. 2A). An a = 0.8-μm cell body, obtained by matching the mean linear and torsional resistive coefficients of a sphere with those of B. subtilis (Materials and Methods), leads to a rheotactic velocity twice the observed one (Fig. 3). A separate mathematical model (SI Materials and Methods) indicates that this discrepancy is likely due to the wiggling in B. subtilis's trajectories (Figs. S2–S5).
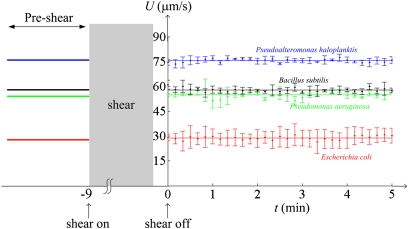
In larger organisms like fish (11, 12) and aquatic invertebrates (20) rheotaxis is active, as these organisms are able to measure shear and behaviorally respond to it. Shear-sensing mechanisms include the lateral line in fish (12), the setae on copepods’ antennae (18), and changes in the membrane's electrical potential in protists (33). In contrast, the agreement between observations and model results found here indicates that bacterial rheotaxis is a passive process. Rheotaxis in bacteria is thus analogous to rheotaxis in sperm and gyrotaxis in bottom-heavy dinoflagellates, which result from a passive hydrodynamic reorientation coupled with active motility (15, 16). To further verify that bacteria do not actively change their motility in response to shear, we performed experiments in which swimming speed was measured before and after exposure to shear (Materials and Methods). No change in speed was found for either B. subtilis or any of three other species tested (E. coli, Pseudoalteromonas haloplanktis, Pseudomonas aeruginosa) (Fig. 4). This result is consistent with the hypothesis that bacteria cannot actively sense and respond to shear.
Fig. 4.
Bacteria do not change swimming speed in response to shear. The mean speed of a population of bacteria, U, is shown before and after exposure to shear. Bars denote the SD among five replicate experiments and solid lines indicate speed before exposure to shear. The shear rate was S = 500 s−1.
By imposing a bias on the swimming direction that is outside the control of the cells, rheotaxis might reduce the effectiveness of bacterial motility. Because motile bacteria rely on chemotaxis to seek favorable microenvironments, the magnitude of this interference can be estimated by comparing the rheotactic velocity V with the chemotactic velocity VC. Typically, VC is a small fraction of the swimming speed U; for instance, VC/U = 5–15% for E. coli, with rare peaks of ∼30–35% for this and other species (34). The magnitude of rheotaxis is dependent on the hydrodynamic environment, because V increases as the shear rate S increases. For example, in oceans and lakes shear rates associated with large-scale flows are weak [S < 0.1 s−1 (35), and thus V/U < 0.1% (Fig. 2A) and V/VC << 1] and even turbulent shear is typically modest [S < 1 s−1 (36), and thus V/U < 1.3% (Fig. 2A) and V/VC < 25%], except for localized regions such as the outside edges of small-scale eddies, where it can reach S ∼10 s−1 (36). On the other hand, turbulent shear rates can be markedly higher in man-made turbulent flows, such as stirred tank bioreactors, where S ∼ 500–2,500 s−1 (37), corresponding to V/U > 67% and V/VC > 10. Therefore, rheotaxis will likely be weak in the water column of most natural water bodies, yet can be considerable in stronger turbulent flows, highlighting that the presence of a nearby surface is not essential for the process.
Still, the environment where bacteria will be most prone to undergo rheotaxis is the vicinity of a surface, where shear rates can be very large. High shear rates (S ∼ 1–10 s−1) can occur in flow through soil and very large shear rates arise in bodily flows, where S can reach 100–1,000 s−1 (38), leading to V/U ∼ 40–70%. In these environments, rheotactic velocities can be of the same magnitude or larger than chemotactic velocities and rheotaxis could either favor or hinder chemotaxis. We highlight that rheotaxis is expected to interfere with chemotaxis when the flow gradient plane is perpendicular to the chemoattractant gradient (Fig. 1B), whereas no interaction between chemotaxis and rheotaxis will take place when the flow gradient plane and the chemoattractant gradient are parallel to each other. Finally, we note that—coincidentally, it appears—upstream swimming of cells adjacent to surfaces is also triggered by shear rates above S ∼ 10 s−1, the same order-of-magnitude threshold found here for rheotaxis.
The overall fitness of free-swimming bacteria is largely shaped by their encounters with chemical and fluid dynamical gradients. Although the sophisticated swimming behaviors that bacteria use to exploit chemical gradients have been long known, our results show that gradients in flow can similarly affect motility and that this effect is not limited to the vicinity of a surface. Obtaining a deeper insight into the diversity of effects that environmental gradients—both chemical and physical—have on microbial movement behavior will ultimately provide a better understanding of the challenges that microbes face in real, complex microenvironments and the adaptive strategies they have evolved to overcome them.
Materials and Methods
Microfluidic Setup and Image Analysis.
We used a smooth-swimming strain of the bacterium B. subtilis, strain OI4139 (25), which is a mutant that almost never tumbles. The cells have an ellipsoidal body with average size of 1 × 3 μm. During normal swimming, the flagella form a three-turn left-handed helical bundle with axial length of 6 μm and diameter of 480 nm (31). The bacteria are grown using Cap assay minimal media (39), as described below.
To expose bacteria to a shear flow, we used a microfluidic channel with a rectangular cross section (for details on fabrication and materials, see ref. 29), having width W = 1,000 μm and depth H = 90 μm. Flow was driven by a syringe pump (PHD 2000 Programmable; Harvard Apparatus). The large aspect ratio W/H ensured a uniform flow across the channel's width, except within ∼100 μm from each sidewall. Observations were performed in the central 37.5 μm. The flow velocity in the channel, u, varies parabolically across the depth, y, as u(y) = (3UF/2)[1 – (2y/H)2], where UF is the mean flow speed and y = 0 denotes middepth (y positive upward). The shear rate, S = du/dy = –12yUF/H2, varies linearly over depth.
We imaged the bacteria at one-quarter depth above the bottom of the microchannel, y = −H/4, where the shear rate S was varied from 0 to 36 s−1. Imaging was performed using an inverted microscope (Eclipse TE2000-E; Nikon) with a 40× phase-contrast objective (NA = 0.6) and a CCD camera (PCO 1600; Cooke). For each shear rate, 25 sets of 1,000 images were recorded at 122 frames per second. Six independent replicate experiments were performed, each with a fresh bacterial culture (Fig. 2A). To further verify that the drift was due to chiral reorientation, a separate set of experiments (two replicates) was conducted at one-quarter depth below the top, y = H/4 (Fig. 2B, red), and one-quarter depth above the bottom, y = −H/4 (Fig. 2B, green). The two experiments at y = −H/4 were performed in addition to the ones shown in Fig. 2A.
Images were analyzed using BacTrack, an in-house cell-tracking Java software. BacTrack locates bacteria in each image and tracks them among two consecutive images, by applying a search radius (the maximum distance a bacterium can move between successive images). Because bacteria were also advected by the flow, different search radii were used for different shear rates. The search radius for each shear rate was determined by adding 100 μm⋅s−1 (an upper limit of swimming speed) to the flow speed, u, at the height y of the observations, and then multiplying by the time between two frames (8.2 ms). The drift velocity was obtained as the component of the velocity of bacteria perpendicular to the plane of the flow gradient (i.e., along z).
Growth Protocols.
B. subtilis OI4139 were prepared by streaking cells on a Tryptose blood agar plate [33 g Tryptose Blood Agar (Difco) and 3 g Agar in 1 L of distilled water] and growing overnight at 30 °C. Cells from a single colony were resuspended in 5 mL of Cap Assay Minimal media (50 mM KH2PO4, 50 mM K2HPO4, 1 mM MgCl2, 1 mM NH4SO4, 0.14 mM CaCl2, 0.01 mM MnCl2, 0.20 mM MgCl2), adding 15 μL HMT (5 mg/mL each of histidine, methionine, and tryptophan, filter sterilized), 1 mM IPTG, 50 μL Tryptone Broth [10 g Tryptone (Difco) and 5 g NaCl in 1 L of distilled water], and 50 μL 1 M Sorbitol (filter sterilized). The culture was incubated at 37 °C while shaking at 250 rpm until an optical density (OD)600 ∼ 0.2 was reached (∼5 h). Fifty microliters 5% (vol/vol) glycerol, 0.5 M Na-lactate (filter sterilized) was added to the culture, which was then shaken for an additional 15 min before the experiments.
P. haloplanktis (ATCC 700530) were grown overnight in 1% Tryptic Soy Broth [10 g of Tryptic Soy Broth (Difco) and 25 g NaCl in 1 L of distilled water] at room temperature on a shaker (175 rpm). Cultures were then diluted 1:20 in Artificial Sea Water (400 mM NaCl, 10 mM CaCl2·2H2O, 1.7 mM KBr, 10 mM KCl, 20 mM MgCl2·6H2O, 20 mM MgSO4·7H2O).
P. aeruginosa (ATCC 15962) were prepared by inoculating frozen stocks into 5 mL Luria Broth [25 g Luria Broth (Difco) into 1 L of distilled water] at 37 °C, while shaking at 180 rpm for 8 h.
E. coli HCB1 (courtesy of H. C. Berg, Harvard University, Cambridge, MA) were grown in Tryptone Broth at 37 °C on an orbital shaker (200 rpm) to midexponential phase (∼4 h) and then diluted 1:20 in Tryptone Broth before experiments.
Experiments to Test Whether Exposure to Shear Alters Swimming Behavior.
Experiments were performed by measuring the swimming speed U for a suspension of bacteria in the microchannel described above in the absence of flow, then subjecting the bacteria to a shear flow (mean value of the shear rate S = 500 s−1) for 9 min, and thereafter measuring U every 10 s for 5 min, again in the absence of flow. This procedure was done for four species (B. subtilis OI4139, P. haloplanktis, E. coli, and P. aeruginosa) with five replicate experiments per species. Due to the finite length of the microchannel, we programmed the syringe pump to alternate between the “infuse” and “refill” modes, so that the same bacteria imaged under quiescent conditions were retained in the microchannel for the duration of the experiment. Alternating between these two modes inverted the flow direction and thus the sign of the shear, but the magnitude of the shear was essentially constant (except for a few seconds during reversals). The mean shear rate of 500 s−1 was selected to expose bacteria to a large shear rate compared with that typically encountered in their natural habitats, with the intention of maximizing any change in behavior, while also keeping the Reynolds number of the bacteria below unity (Re = SL2/ν = 0.05, using L = 10 μm as the characteristic length of a bacterium including its flagella).
Brownian Rotational Diffusion.
The lateral and rotational drag coefficients of a bacterium can be obtained from RFT. The linear and angular velocities of the bacterium, v and ω, respectively, are given by requiring the total forces F and total moments M acting on the bacterium to vanish,
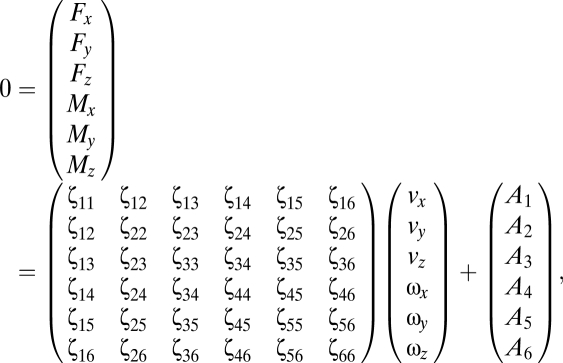 |
where A is the contribution due to shear and swimming. At (ψ = π/2, θ = 0), the bacterium is aligned with the x axis. At this orientation, ζ22 = ζ33 and ζ55 = ζ66 are the lateral and rotational drag coefficients, respectively. The Brownian rotational diffusivity DR can be estimated by using the Stokes–Einstein equation, DR = kT/ζ66.
Similarly, for an ellipsoid DR = kT/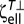 , where
, where 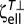 is the torsional resistive coefficient of an ellipsoid perpendicular to its major axis (40),
is the torsional resistive coefficient of an ellipsoid perpendicular to its major axis (40),
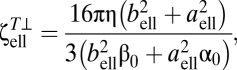 |
where aell and bell are the half-major and half-minor axes of the ellipsoid, respectively, α0 and β0 are given by
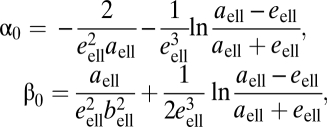 |
and 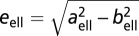 is the ellipsoid's eccentricity.
is the ellipsoid's eccentricity.
Equivalent Radius of the Spherical Cell Body Used in the Model.
B. subtilis has a prolate ellipsoidal body. Its size depends on the cell age and flagellar bundle formation and, on average, is 1 × 3 μm. In our model, we used a spherical cell body. To determine its radius a, one approach is to consider the sphere that has the same volume as B. subtilis, which yields a radius of a = 0.721 μm. A second approach is to match the average linear and torsional resistive coefficients between the ellipsoidal body and a sphere. The linear resistive coefficients parallel and perpendicular to the ellipsoid's major axis are given by (40)
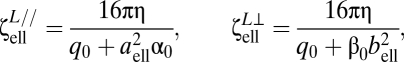 |
where
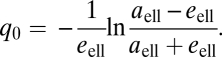 |
The torsional resistive coefficients are given by
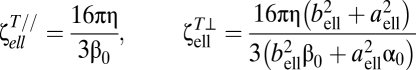 |
The average linear resistance 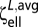 and torsional resistance
and torsional resistance 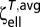 over all orientations can be computed as (41)
over all orientations can be computed as (41)
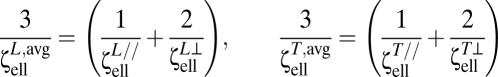 |
The linear and torsional resistances for a sphere are CF = 6πηa and CM = 8πηa3, respectively. For a 1 × 3 ellipsoid, by matching CF = 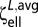 , we find an equivalent spherical radius of a = 0.807 μm. By matching CM =
, we find an equivalent spherical radius of a = 0.807 μm. By matching CM = 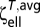 , we obtain a = 0.802 μm. Both conditions give an equivalent spherical radius close to a = 0.8 μm, which was therefore used in simulations (and is not too different from the value of 0.721 μm obtained on the basis of volume equivalence).
, we obtain a = 0.802 μm. Both conditions give an equivalent spherical radius close to a = 0.8 μm, which was therefore used in simulations (and is not too different from the value of 0.721 μm obtained on the basis of volume equivalence).
Effect of Tumbles on the Rheotactic Velocity.
Many bacteria, including wild-type B. subtilis and E. coli, alternate runs and tumbles (7). Because rheotaxis hinges on the preferential alignment of cells with streamlines, one might expect the large reorientation associated with tumbles to suppress rheotaxis. However, a simulation based on the tumbling statistics of E. coli (those of B. subtilis are unknown) shows that tumbles reduce, but do not eliminate rheotaxis (Fig. 3, dashed black line). In this simulation, the position x0 and Euler angles θ0 specifying orientation of each bacterium are updated as
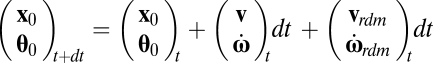 |
where dt is the time step of the simulation, v and 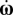 are the deterministic linear and angular velocities of the bacterium, and vrdm is the linear velocity and
are the deterministic linear and angular velocities of the bacterium, and vrdm is the linear velocity and 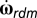 is the rate of change of Euler angles arising from the random motion due to Brownian translational and rotational diffusion. Note that
is the rate of change of Euler angles arising from the random motion due to Brownian translational and rotational diffusion. Note that 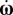 and the rotational velocities ω are related through Euler angle rotations. The deterministic velocities can be written as
and the rotational velocities ω are related through Euler angle rotations. The deterministic velocities can be written as
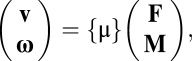 |
where
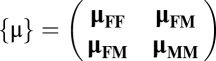 |
is the 6 × 6 symmetric mobility matrix. Brownian contributions obey (28)
where k is the Boltzmann constant, T is the temperature, 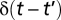 is a delta function, and the notation
is a delta function, and the notation 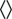 indicates an integral average over all time. These conditions state that thermal fluctuations lead to a Gaussian-distributed random velocity that is uncorrelated in time, has zero mean, and has a width related to the mobilities μ and kT via the Stokes–Einstein relation.
indicates an integral average over all time. These conditions state that thermal fluctuations lead to a Gaussian-distributed random velocity that is uncorrelated in time, has zero mean, and has a width related to the mobilities μ and kT via the Stokes–Einstein relation.
To model tumbling, we calculate the tumbling probability P = dt/τ (7), where τ is the mean run time (∼1 s for E. coli). We then generate a random number R uniformly distributed between 0 and 1 at every time step. If P > R, the cell tumbles; otherwise it continues along the current run. The tumble angle is taken from a Gaussian distribution with mean of 68° and SD of 39°, as appropriate for E. coli (42).
Supplementary Material
Acknowledgments
We thank G. Glekas and G. Ordal for providing B. subtilis and W. M. Durham, J. S. Guasto, and R. Rusconi for comments on the manuscript. This work was partially supported by National Science Foundation Grants CBET-0966000 (to R.S.), CBET-0967510 (to H.C.F. and T.R.P.), CBET-1067798 (to H.C.F.), and OCE-0744641-CAREER (to R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120955109/-/DCSupplemental.
References
- 1.Lam WC, et al. The chemotactic response of tumor cells. A model for cancer metastasis. Am J Pathol. 1981;104:69–76. [PMC free article] [PubMed] [Google Scholar]
- 2.Pandya S, Iyer P, Gaitonde V, Parekh T, Desai A. Chemotaxis of Rhizobium sp.S2 towards Cajanus cajan root exudate and its major components. Curr Microbiol. 1999;38:205–209. doi: 10.1007/pl00006788. [DOI] [PubMed] [Google Scholar]
- 3.Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA. 2008;105:4209–4214. doi: 10.1073/pnas.0709765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durham WM, Kessler JO, Stocker R. Disruption of vertical motility by shear triggers formation of thin phytoplankton layers. Science. 2009;323:1067–1070. doi: 10.1126/science.1167334. [DOI] [PubMed] [Google Scholar]
- 5.Jakobsen HH. Escape response of planktonic protists to fluid mechanical signals. Mar Ecol Prog Ser. 2001;214:67–78. [Google Scholar]
- 6.Riffell JA, Zimmer RK. Sex and flow: The consequences of fluid shear for sperm-egg interactions. J Exp Biol. 2007;210:3644–3660. doi: 10.1242/jeb.008516. [DOI] [PubMed] [Google Scholar]
- 7.Berg HC. E. coli in Motion. New York: Springer; 2004. [Google Scholar]
- 8.Paster E, Ryu WS. The thermal impulse response of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5373–5377. doi: 10.1073/pnas.0709903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. Magneto-aerotaxis in marine coccoid bacteria. Biophys J. 1997;73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Englert DL, Manson MD, Jayaraman A. Flow-based microfluidic device for quantifying bacterial chemotaxis in stable, competing gradients. Appl Environ Microbiol. 2009;75:4557–4564. doi: 10.1128/AEM.02952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold GP. Rheotropism in fishes. Biol Rev Camb Philos Soc. 1974;49:515–576. doi: 10.1111/j.1469-185x.1974.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 12.Montgomery JC, Baker CF, Carton AG. The lateral line can mediate rheotaxis in fish. Nature. 1997;389:960–963. [Google Scholar]
- 13.Lytle DA, Olden JD, McMullen LE. Drought-escape behaviors of aquatic insects may be adaptations to highly variable flow regimes characteristic of desert rivers. Southwest Nat. 2008;53:399–402. [Google Scholar]
- 14.Bretherton FP, Rothschild L. Rheotaxis of spermatozoa. Proc R Soc Lond B Biol Sci. 1961;153:490–502. [Google Scholar]
- 15.Roberts AM. Motion of spermatozoa in fluid streams. Nature. 1970;228:375–376. doi: 10.1038/228375a0. [DOI] [PubMed] [Google Scholar]
- 16.Pedley TJ, Kessler JO. Hydrodynamic phenomena in suspensions of swimming microorganisms. Annu Rev Fluid Mech. 1992;24:313–358. [Google Scholar]
- 17.Strickler JR, Bal AK. Setae of the first antennae of the copepod Cyclops scutifer (Sars): Their structure and importance. Proc Natl Acad Sci USA. 1973;70:2656–2659. doi: 10.1073/pnas.70.9.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visser AW. Hydromechanical signals in the plankton. Mar Ecol Prog Ser. 2001;222:1–24. [Google Scholar]
- 19.Maldonado EM, Latz MI. Shear-stress dependence of dinoflagellate bioluminescence. Biol Bull. 2007;212:242–249. doi: 10.2307/25066606. [DOI] [PubMed] [Google Scholar]
- 20.Richardson WB. Microcrustacea in flowing water: Experimental analysis of washout times and a field test. Freshw Biol. 1992;28:217–230. [Google Scholar]
- 21.Genin A, Jaffe JS, Reef R, Richter C, Franks PJS. Swimming against the flow: A mechanism of zooplankton aggregation. Science. 2005;308:860–862. doi: 10.1126/science.1107834. [DOI] [PubMed] [Google Scholar]
- 22.Rosengarten R, Klein-Struckmeier A, Kirchhoff H. Rheotactic behavior of a gliding mycoplasma. J Bacteriol. 1988;170:989–990. doi: 10.1128/jb.170.2.989-990.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill J, Kalkanci O, McMurry JL, Koser H. Hydrodynamic surface interactions enable Escherichia coli to seek efficient routes to swim upstream. Phys Rev Lett. 2007;98:068101-1–068101-4. doi: 10.1103/PhysRevLett.98.068101. [DOI] [PubMed] [Google Scholar]
- 24.Meng Y, et al. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol. 2005;187:5560–5567. doi: 10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szurmant H, Muff TJ, Ordal GW. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem. 2004;279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- 26.Jeffery GB. The motion of ellipsoidal particles in a viscous fluid. Proc R Soc Lond A Contain Pap Math Phys Character. 1922;102:161–179. [Google Scholar]
- 27.Brenner H. The Stokes resistance of an arbitrary particle–III. Chem Eng Sci. 1964;19:631–651. [Google Scholar]
- 28.Makino M, Doi M. Migration of twisted ribbon-like particles in simple shear flow. Phys Fluids. 2005;17:103605-1–103605-7. [Google Scholar]
- 29.Marcos, Fu HC, Powers TR, Stocker R. Separation of microscale chiral objects by shear flow. Phys Rev Lett. 2009;102:158103-1–158103-4. doi: 10.1103/PhysRevLett.102.158103. [DOI] [PubMed] [Google Scholar]
- 30.Childress S. Mechanics of Swimming and Flying. Cambridge, UK: Cambridge Univ Press; 1981. [Google Scholar]
- 31.Fujii M, Shibata S, Aizawa S-I. Polar, peritrichous, and lateral flagella belong to three distinguishable flagellar families. J Mol Biol. 2008;379:273–283. doi: 10.1016/j.jmb.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Happel J, Brenner H. Low Reynolds Number Hydrodynamics. Englewood Cliffs, NJ: Prentice-Hall; 1965. [Google Scholar]
- 33.Naitoh Y, Eckert R. Ionic mechanisms controlling behavioral responses of paramecium to mechanical stimulation. Science. 1969;164:963–965. doi: 10.1126/science.164.3882.963. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed T, Stocker R. Experimental verification of the behavioral foundation of bacterial transport parameters using microfluidics. Biophys J. 2008;95:4481–4493. doi: 10.1529/biophysj.108.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheriton OM, McManus MA, Stacey MT, Steinbuck JV. Physical and biological controls on the maintenance and dissipation of a thin phytoplankton layer. Mar Ecol Prog Ser. 2009;378:55–69. [Google Scholar]
- 36.Jimenez J. Oceanic turbulence at millimeter scales. Scientia Marina. 1997;61(Suppl 1):1–6. [Google Scholar]
- 37.Campesi A, Cerri MO, Hokka CO, Badino AC. Determination of the average shear rate in a stirred and aerated tank bioreactor. Bioprocess Biosyst Eng. 2009;32:241–248. doi: 10.1007/s00449-008-0242-4. [DOI] [PubMed] [Google Scholar]
- 38.Jadhav S, Bochner BS, Konstantopoulos K. Hydrodynamic shear regulates the kinetics and receptor specificity of polymorphonuclear leukocyte-colon carcinoma cell adhesive interactions. J Immunol. 2001;167:5986–5993. doi: 10.4049/jimmunol.167.10.5986. [DOI] [PubMed] [Google Scholar]
- 39.Zimmer MA, et al. The role of heterologous receptors in McpB-mediated signalling in Bacillus subtilis chemotaxis. Mol Microbiol. 2002;45:555–568. doi: 10.1046/j.1365-2958.2002.03035.x. [DOI] [PubMed] [Google Scholar]
- 40.Steinberger B, Petersen N, Petermann H, Weiss DG. Movement of magnetic bacteria in time-varying magnetic fields. J Fluid Mech. 1994;272:189–211. [Google Scholar]
- 41.Berg HC. Random Walks in Biology. Princeton: Princeton Univ Press; 1993. [Google Scholar]
- 42.Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.