Abstract
Despite the importance of Salmonella infections in human and animal health, the target antigens of Salmonella-specific immunity remain poorly defined. We have previously shown evidence for antibody-mediating protection against invasive Salmonellosis in mice and African children. To generate an overview of antibody targeting in systemic Salmonellosis, a Salmonella proteomic array containing over 2,700 proteins was constructed and probed with immune sera from Salmonella-infected mice and humans. Analysis of multiple inbred mouse strains identified 117 antigens recognized by systemic antibody responses in murine Salmonellosis. Importantly, many of these antigens were independently identified as target antigens using sera from Malawian children with Salmonella bacteremia, validating the study of the murine model. Furthermore, vaccination with SseB, the most prominent antigenic target in Malawian children, provided mice with significant protection against Salmonella infection. Together, these data uncover an overlapping immune signature of disseminated Salmonellosis in mice and humans and provide a foundation for the generation of a protective subunit vaccine.
Keywords: antigen discovery, non-Typhi Salmonella bacteremia
Salmonella infections are responsible for gastroenteritis outbreaks in developed nations and typhoid fever and nontyphoidal Salmonella bacteremia in developing nations (1–4). Typhoid is caused by Salmonella enterica serovars (Typhi and Paratyphi) that are highly restricted to the human host and kill over 200,000 people every year (5–7). Most other Salmonella serovars can infect a variety of animals but only cause self-limiting gastroenteritis in humans (8). However, many of these same serovars cause serious disseminated infections in young children and patients with compromised host immunity (9, 10). These disseminated non-Typhi Salmonella (NTS) infections are particularly associated with HIV-infected individuals in Africa and Asia (11–14). Other groups susceptible to NTS infections include patients with immune deficiencies and young children in sub-Saharan Africa (15, 16). Although systemic bacterial infection can be treated with antimicrobials, Salmonella are increasingly resistant to antibiotics and the potential for new antibiotics is not encouraging (3, 17, 18). The development of an effective vaccine for systemic Salmonella infections remains an important global health priority (2, 3).
Although there are two licensed vaccines for typhoid, they provide only moderate protective efficacy (50–55% over 3 y) and are not widely used in endemic areas (2, 3, 19, 20). There is no current vaccine for NTS infection, but we have previously shown that Salmonella-specific antibody responses in young African children are associated with resistance to invasive NTS disease (16, 21) and that antibody is protective in the mouse model of Salmonellosis (22). Thus, there is an urgent need to improve the efficacy of currently licensed vaccines and/or develop new Salmonella vaccines that can protect against typhoid and NTS disease. The process of vaccine refinement and development would be aided by greater understanding of antigen targeting because maintaining or boosting expression of target antigens could improve the efficacy of live vaccines or these same antigens could be incorporated into new multivalent or conjugate subunit vaccines.
Immunity to systemic Salmonella infection is often studied using susceptible or resistant laboratory mouse strains (23). Many features of human Salmonellosis are reproduced in the murine model, including the route of intestinal entry, tissue and cellular tropism of the bacteria, the activation of innate and adaptive immunity, and development of chronic bacterial shedding and relapsing disease (24–29). However, as mice can only be infected with nontyphoidal strains of Salmonella, it is not clear whether this model is relevant for understanding typhoid or disseminated NTS infection. Furthermore, immune targets in both murine and human Salmonellosis are poorly characterized (30, 31), inhibiting detailed examination of immunity and preclinical vaccine development. However, proteomic microarrays can now be constructed containing complete or partial pathogen proteomes and can provide a comprehensive overview of antigenic targeting using sera from infected animals or patients (32–36).
Here we describe the construction of a large Salmonella proteomic array and the identification of Salmonella-specific immune targets using multiple laboratory mouse strains and natural human infection. Together, these data uncover the overlapping immune signature of murine and human Salmonellosis and demonstrate the potential for some of these antigens to be used as subunit vaccines against Salmonella infections.
Results
Vaccination with Attenuated Salmonella Generates a Systemic Antibody Response.
B cells are required for immunity to Salmonella infection (22, 37, 38), but the target antigens and precise role of antibody remain poorly defined. We infected C57BL/6 mice with the vaccine strain Salmonella enterica serovar Typhimurium BRD509 (39) and examined Salmonella-specific antibody production following a single or multiple doses. A single oral immunization induced serum Salmonella-specific IgM and IgG2c, and intestinal IgA in fecal pellets, but these responses were not elevated by subsequent immunizations (Fig. S1A). Intravenous immunization induced Salmonella-specific IgM and IgG2c responses that were boosted by additional immunizations (Fig. S1B). We previously reported that immunized B-cell–deficient mice are unable to resist secondary infection with virulent Salmonella (22). In contrast, immunized mice lacking IgA or the polymeric Ig receptor acquired robust protection against Salmonella infection (Fig. S1C). Thus, systemic, rather than mucosal, B-cell responses are important for protective immunity to Salmonella, but the antigenic targets of this response are largely unknown. Given these data, we examined antigen targeting of systemic IgG after multiple i.v. immunizations.
Construction and Validation of a Salmonella Protein Array.
The genome sequence of S. enterica serovar Typhi Ty2 was used to construct a protein array containing ∼60% of the Salmonella proteome. This genome was chosen due to the extensive homology between serovar Typhi and serovar Typhimurium genomes (40–42) and to facilitate screening with sera from typhoid and NTS patients. A set of 2,700 ORFs was chosen on the basis of whether they contained signal peptides; belonged to categories of “outer membrane,” “periplasmic,” “heat shock protein,” “chaperone,” “transport protein” “lipoprotein,” or “virulence associated protein”; or were homologous to serovar Typhimurium LT2. Proteins that were nonhomologous to Escherichia coli were also selected to minimize the potential for background cross-reactivity. Each selected ORF was amplified from S. enterica serovar Typhi Ty2 genomic DNA and cloned using high-throughput recombination. Proteins were expressed and printed onto nitrocellulose arrays, and 96% were confirmed by detecting HA- or HIS-tag expression.
Identification of Antigen Targets in Murine Salmonellosis.
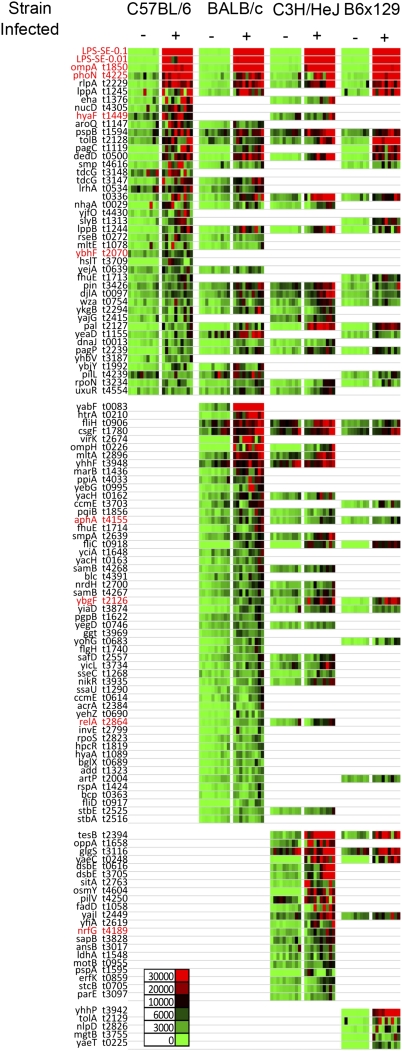
Different strains of genetically susceptible or resistant mice have been used to study the pathogenesis and immunology of Salmonella infection (23, 43–46). To perform a comprehensive analysis of antibody targeting in inbred mice, we examined immune responses in susceptible (C57BL/6, BALB/c, C3H/HeJ) and resistant (C57BL/6 × 129F1) mice infected with serovar Typhimurium BRD509. Salmonella protein arrays were probed using sera from 10 naive and 10 infected mice per strain, and these antigen profiles were collated onto heat maps showing all significant responses in infected mice (Fig. 1). Salmonella-specific IgG responses to 117 distinct antigens were detected among all mouse strains, 41 in C57BL/6 mice, 82 in BALB/c mice, 64 in C3H/HeJ mice, and 39 in C57BL/6 × 129 mice (Fig. 1). In each inbred mouse strain, a subset of antigenic targets was identified that were strain-specific whereas intrastrain variance between individual mice was low (Fig. S2). A subset of 29 antigens were recognized by Salmonella-specific IgG in at least three of four mouse strains, and 14 of these protein targets were common to all four inbred strains. These common antigens included ompA, phoN, pagC, csgF, and fliC, each of which has been identified during previous attempts at antibody profiling in murine and/or human Salmonellosis (Table S1) (30, 47–51). To our knowledge, none of the other proteins have been reported as target antigens in Salmonella-infected mice or humans and therefore represent potentially unique antigenic targets.
Fig. 1.
Identification of Salmonella antigenic targets in multiple inbred mouse strains. Groups of C57BL/6, BALB/c, C3H/HeJ, and B6 × 129 F1 mice were infected i.v. with 5 × 105 BRD509 and boosted 1 and 2 mo later. Blood was collected from naive mice, and 55 d after the last immunization, serum was tested for the ability to bind to Salmonella proteomic arrays. Bound IgG was detected using biotinylated anti-mouse IgG and SA-Surelight. Slides were scanned using a ProScanArray HT microarray scanner, and the signal intensity of each spot was quantified by ScanArray Express software (Perkin-Elmer). Signal intensity was normalized by using the R statistical environment (http://www.R-project.org). Normalized data were used for a Student's t test (SPSS software) in differential analyses of array data. Heat maps show signal intensity [red (strongest) to green (weakest)] of 10 naive and 10 immunized mice per group. Serodiagnostic antigens (P < 0.05) are presented in rows for easy comparison across all four mouse strains and are ranked by mean signal intensity within each strain. Antigens that were also antigenic targets in humans are highlighted in red.
Because mice were immunized with serovar Typhimurium and the protein arrays were generated using the S. Typhi Ty2 genome, we analyzed 20 publicly available Salmonella genome sequences for the presence of the 29 antigens and determined the percentage homology of amino acid sequences using the S. Typhi Ty2 genome as the reference (Dataset S1). Although there was 100% homology for each antigen between the two S. Typhi strains, one antigen (pilL) was not present in serovar Typhimurium SL1344 or D23580 and may represent aberrant detection of a cross-reactive response. A second antigen (pin) had lower homology (77.5%), and two additional antigens (lppA and lppB) are not annotated in the genome of the invasive African serovar Typhimurium isolate D23580 (41).
Antigenic Targeting During Primary Infection.
The antigens listed above were recognized by antibody from mice with a high titer of Salmonella-specific IgG following three immunizations with a vaccine strain of Salmonella (Fig. S1). It seemed possible that other unique antigens are transiently recognized during primary immunization. To examine this issue, we identified antigen targets at several different time points during primary immunization of C57BL/6 mice, when antibody titers were lower (Fig. S1). Interestingly, antibody responses to phoN peaked 42 d after immunization whereas responses to ompA were more prominent at day 80 (Fig. S3), suggesting that some antigen targeting follows distinct temporal kinetics. The majority of antigens were similar to those detected after tertiary immunization, including 10 of the top 12 antigens (Fig. S2). Thus, antigenic targeting after multiple immunizations is largely reflective of the primary B-cell response to Salmonella.
Antigenic Targeting During Salmonellosis in Malawian Children.
Although the murine model of Salmonella infection has been widely studied, its utility as a preclinical model of human Salmonellosis remains unclear (28, 52). We therefore examined antigen targeting using a well-characterized set of convalescent pediatric sera from 12 children admitted to hospital with Salmonella bacteremia or meningitis in Blantyre, Malawi, where NTS bacteremia is common in young children (16), and age- and sex-matched healthy controls. Ten children (aged 2–72 mo) had serovar Typhimurium in blood or cerebrospinal fluid cultures, whereas the eldest two children (90 and 151 mo) had fully sensitive S. Typhi, which affects older children in sub-Saharan Africa (53). Four children with serovar Typhimurium tested positively for HIV infection, and six had other recognized comorbidities including malaria, anemia, and malnutrition. All presented with fever, but only four had a history of diarrhea (Table S2).
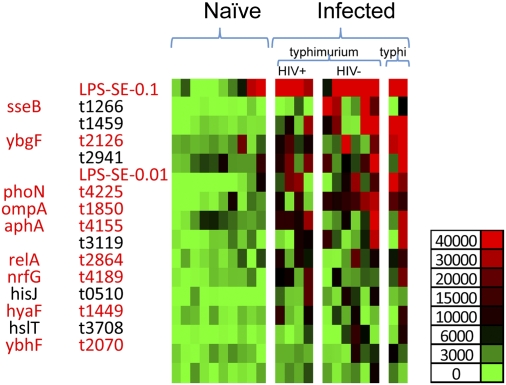
Although detection signals were lower than observed using murine sera, significant Salmonella-specific IgG responses to 14 antigens were detected in patients suffering from Salmonellosis compared with controls (Fig. 2). Genome comparison showed that all 14 antigens were encoded in all four publically available serovar Typhimurium genomes, including the sequenced invasive Malawian strain serovar Typhimurium D23580, with 95% or greater amino acid homology (Dataset S2). It was of considerable interest that 8/14 (57%) of the antigens detected by patient sera overlapped with antigens detected in Salmonella-infected mice (Figs. 1 and 2; Table S3). Furthermore, responses to two antigens, sseB and phoN, were higher in non-HIV versus HIV+ patients, suggesting that these antigens may be good vaccine candidates. In contrast, we did not detect differences in antigen targeting between patients infected with serovar Typhimurium and serovar Typhi, although there was a slightly higher antibody response in the two Typhi samples. Together, these data demonstrate extensive overlap in antigenic targeting during disseminated Salmonella infections of mice and humans.
Fig. 2.
Identification of Salmonella antigenic targets in Malawian children. Salmonella arrays were probed using 12 convalescent sera from Malawian children with blood culture-confirmed Salmonella bacteremia or 10 healthy HIV-uninfected controls. Bound IgG was detected using biotinylated anti-human IgG and SA-Surelight and microarray scanning. Normalized data were used for a Student's t test (SPSS software) in differential analyses of array data. Serodiagnostic antigens (P < 0.05) are in rows and are ranked by mean signal intensity in infected children. Antigens that were recognized in murine infection studies or used in subsequent vaccine experiments are highlighted in red. Individual samples are presented in columns, and infected patients have been sorted left to right on the basis of serovar Typhimurium or S. typhi infection and HIV infection status.
Bactericidal Activity of Human Convalescent and Control Sera.
Serum bacteridical assays were performed using Malawian child convalescent and control sera and the invasive Malawian serovar Typhimurium isolate D23580. Bactericidal titers were higher in the 12 convalescent sera (median 40,960; range 4,096–65,536) compared with age-matched control sera (range 1–16,384; Student's t test, P = 0.0013) (Fig. S4).
Subunit Vaccine Development in Murine Salmonellosis.
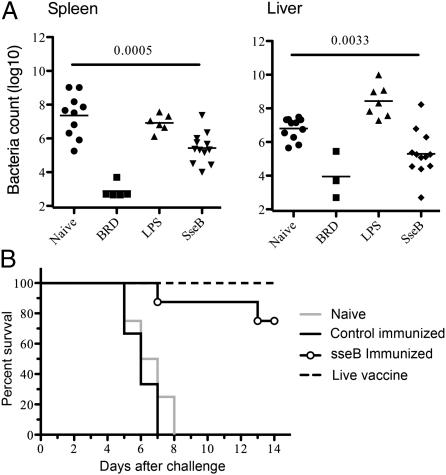
The extensive overlap in immune targeting between human and murine Salmonellosis indicates that the mouse may be a useful preclinical model for human Salmonellosis. Furthermore, the identification of numerous overlapping target antigens in human and murine Salmonellosis raises the possibility that these antigens could be used in diagnostics or as part of a subunit or conjugate vaccine. We examined the protective efficacy of three different antigens that broadly represented three different patterns of recognition, i.e., antigens detected in both mice and humans (e.g., phoN), in humans alone (e.g., sseB), or in mice alone (e.g., dedD). The recognition of phoN and sseB in HIV-uninfected individuals provided additional justification for examining these antigens. We produced large quantities of all three antigens and examined their protective capacity in the murine model. Only immunization of C57BL/6 mice with sseB provided significant protection against Salmonella infection, as measured by reduced bacterial burdens in both the spleen and the liver (Fig. 3A). Furthermore, although all control mice died 7–8 d after oral challenge with virulent Salmonella, around 70% of mice immunized with sseB survived until 14 d after challenge (Fig. 3B).
Fig. 3.
Immunization with SseB protects mice against Salmonella infection. (A) C57BL/6 mice were immunized twice i.v. with 100 μg of SseB and 10 μg of LPS at 4-wk intervals whereas control groups were left unimmunized (naive), injected with LPS alone, or immunized i.v. with 5 × 105 BRD509. Four weeks after boosting, mice were infected orally with 5 × 107 Salmonella SL1344, and bacterial burdens were determined in spleens and livers 7 d later. Data show the mean bacterial burden, and each individual mouse is also included on a scatter plot. Statistical significance was examined by Student's t test, and numbers indicate statistical significance between naive and SseB-immunized mice groups. (B) Groups of C57BL/6 mice were immunized twice i.v. with LPS alone, with SseB and LPS mixed in alum, or left unimmunized (naive). A positive control group was also vaccinated with 5 × 105 live attenuated Salmonella (BRD509). Four weeks after boosting, mice were infected orally with 1 × 106 Salmonella SL1344, and mice were examined every day for the development of a moribund state. Data show the percentage of surviving mice in each group and are representative of four to eight mice per group.
Discussion
Vaccine development for typhoid and NTS infections would benefit from a detailed understanding of antigenic targeting because the expression of these antigens could be modified in live attenuated vaccines or form the basis of subunit or conjugate vaccines (28, 31). Our approach was to identify antigen targets in mice immunized with a vaccine strain of Salmonella because this induces robust protective immunity to infection and requires Salmonella-specific B-cell responses (22, 37, 38, 54, 55). Our data demonstrate that immunized mice develop robust systemic IgG2c responses to Salmonella and that protective immunity develops in the absence of mucosal antibodies. These data confirm previous observations (56) and imply that serum IgG responses to Salmonella contribute to protection against infection, although it is not yet clear whether this is due to complement fixation (16), opsonization (21), or enhancement of antigen presentation (57, 58).
Current understanding of Salmonella-specific antibody targeting is based on empirical analysis of surface or secreted antigens such as ViCPS, lipopolysaccharide (LPS), flagellar components, and outer membrane proteins. A recent study examined antibody targeting of in vivo-expressed S. Typhi proteins and highlighted four proteins recognized in typhoid patients (pagC, tcfB, STY1648, and STY3683) (47). A subsequent study identified 57 S. Typhi proteins that bound to affinity columns containing antibodies from typhoid patients (30). Our proteomic approach extends these efforts and identifies additional antibody targets in four laboratory mouse strains used to study Salmonella infection. Antibody plays a prominent role in protective immunity in each of these strains after vaccination with attenuated Salmonella (22, 37, 38, 59). Importantly, our data uncover a common subset of target antigens between multiple strains, including several previously identified target antigens in mouse and human infection (30, 47–51). These include well-known surface and secreted antigens ompA and fliC (flagellin), each of which have been reported to be major immunogenic antigens (30, 47, 48, 50, 51), and flagellin alone can provide modest levels of protection against infection in the murine model (60, 61). Other antigens such as pagC, phoN, and csgF have been reported as targets of Salmonella-specific antibody, but their protective potential is unclear. The identification of these known antigens by our proteomic screen provides confidence that additional antigens are likely to be bona fide target antigens in murine and/or human Salmonella infection, although this will require further confirmation with larger numbers of sera. Interestingly, this set of antigens includes the outer membrane proteins lppA and lppB, involved in the induction of septic shock; fliH, which participates in flagellar assembly; and several other poorly characterized proteins. Fimbria-related proteins were not detected as major target antigens in infected mice, despite the fact that a group of these proteins were identified during analysis of human typhoid sera antigens (30). This may represent a difference between typhoid disease and the systemic Salmonellosis examined in our experiments, and future studies are planned to examine this issue directly. Other studies have reported an important protective effect of B-cell responses to OmpD (62); however, this protein is not present in the S. Typhi genome that was used to generate our proteomic array, but will be included in subsequent arrays.
Our analysis of sera from Malawian children with Salmonellosis identified eight antigens common to murine infection. Individual comparison with each mouse strain indicates that the profile of antibody responses in C3H mice is the closest to human infection (7/14), compared with C57BL/6 (5/14), BALB/c (5/14), and 129F1 (4/14), although this will require additional analysis of human sera. The extent of overlap was reassuring because natural infection might be more rigorously modeled by murine experiments that include a mucosal route of infection, a virulent strain of bacteria, and the presence of coinfections and other comorbidities. Each of these variables can be examined in laboratory experiments and may increase overlap between human and murine antigen targeting. The high titers of bactericidal activity in convalescent human sera compared with control sera suggest that antigens targeted may be key targets of bactericidal antibody. Furthermore, the presence of high bactericidal titers against serovar Typhimurium in the sera of the children who had had bacteremia with S. Typhi suggest that key protective antigens are common to both serovars. Few differences were observed between an S. Typhi and an serovar Typhimurium infection, but sseB and phoN were targeted more readily in HIV-uninfected patients. However, a larger study will be required to define antigen targeting in these subgroups with greater confidence. It will also be of interest to monitor changes in antigen targeting in patients over time and in response to antimicrobial treatment.
Given the lack of a vaccine against NTS, identification of target antigens in both mice and humans could facilitate the development of such a vaccine. Our data show that SseB immunization is protective in murine infection, reducing bacterial loads and increasing survival time after infection, which confirms and extends a previous report (63). It is not yet clear why a protein encoded by Salmonella pathogenicity island 2 would be recognized by antibody responses because these proteins would be expected to remain contained within infected cells. However, we have recently identified natural CD4 T-cell epitopes within SPI2 effector proteins (64), and it seems possible that promiscuous secretion of SPI-2 proteins may occur in vivo, as suggested by a recent report (65). Our ability to identify a protective antigen using proteomic screens provides some confidence that other protective antigens are likely to be found among the many target antigens detected in our study. However, the task of producing and testing each of these antigens in subunit vaccine trials remains a challenging objective.
In conclusion, we describe the construction of a large Salmonella proteomic array and identification of Salmonella-specific immune targets in the most commonly used laboratory mouse strains and in human infection. These data uncover a common immune signature of murine and human Salmonellosis and demonstrate the potential for these antigens to be used as subunit vaccines.
Materials and Methods
Mouse and Bacterial Strains.
C57BL/6, BALB/c, and C3H/HeJ mice were purchased from the National Cancer Institute or the Jackson Laboratory, and B6.129F1 mice were generated by intercrossing C57BL/6 with 129SvJ. All mice were cared for in accordance with University of Minnesota and University of California at Davis Research Animal Resource guidelines, and experiments were performed under Institutional Animal Care and Use Committee-approved protocols. Attenuated serovar Typhimurium strain BRD509 (aroA-aroD-), derived from SL1344 (59), was provided by D. Xu, University of Glasgow (Glasgow, United Kingdom).
Immunization and Infection of Inbred Mice.
Salmonella were cultured overnight and diluted in PBS after estimation of bacterial concentration using a spectrophotometer. For i.v. infections, 5 × 105 BRD509 were injected in the lateral tail vein. For oral infections, mice were administered 0.1 mL of 5% sodium bicarbonate before oral infection with 5 × 109 BRD509 or 5 × 107 SL1344. In some experiments, mice were infected with BRD509 on three occasions with 1 mo spacing between doses.
Antibody ELISAs.
Salmonella-specific IgM and IgG2c responses to Salmonella were measured using antibody ELISA. Briefly, 96-well microtiter plates were coated with heat-killed Salmonella and blocked using 10% FBS/PBS. Serum samples were added in serial dilutions before incubation with biotin-conjugated anti-mouse IgM or IgG2c (BD Bioscience and eBioscience) and HRP-conjugated streptavidin (Extravidin, Sigma-Aldrich). An HRP substrate (O-phenylenediamine dihydrochloride, Sigma-Aldrich) was used to develop the plates before analysis using an ELISA plate reader (SpectraMax M2, Molecular Devices).
Microarray Fabrication and Probing.
Microarrays were fabricated and probed as described previously (34–36). Plasmids were expressed using in vitro transcription/translation E. coli reactions (Expressway, Invitrogen), and reactions were printed onto nitrocellulose-coated glass FAST slides (Whatman) using an Omni Grid 100 microarray printer (Genomic Solutions). Each printed array contained positive and negative controls, including Salmonella, E. coli, and Francisella LPS, blank spots, PBS, no DNA, and human and mouse IgM and IgG. Before scanning, human serum samples were diluted in blocking buffer containing 10% E. coli lysate. Protein arrays were blocked and exposed to sera overnight before detection with biotinylated antibodies specific for mouse or human IgG. Streptavidin conjugated to Surelight was added before slides were scanned using a ProScanArray HT microarray scanner. Signal intensities were quantified using ScanArray Express software (Perkin-Elmer) and normalized using the R statistical environment (http://www.R-project.org). Normalized data were used for a Student's t test (SPSS software) in differential analyses of array data. Normalized data were retransformed into approximate raw values and presented in heat maps or plotted as graphs.
Mouse and Human Serum Samples.
Groups of C57BL/6, BALB/c, C3H/HeJ, and B6.129F1 mice were immunized i.v. with 5 × 105 BRD509 at monthly intervals, and blood was collected 55 d later. In other experiments, C57BL/6 mice were immunized i.v. with a single dose of 5 × 105 BRD509, and blood was collected 14, 28, 42, 57, 80, and 193 d later. Convalescent sera were obtained from African children with blood culture-confirmed Salmonella bacteremia who were admitted to Queen Elizabeth Central Hospital, Blantyre, Malawi, in 2006. Blood samples were taken 4–6 wk after acute presentation. Age- and sex-matched serum samples were obtained from healthy HIV-uninfected children in the Ndirande Township of Blantyre in 2006. All sera were separated after clotting at room temperature and were frozen in aliquots at −80 °C until required. Ethical approval for the collection of these sera and this work was granted by the Research and Ethics Committee, College of Medicine, University of Malawi.
Clinical Invasive Salmonella Isolates.
Blood and cerebrospinal fluid cultures were performed in the Microbiology Department of the Malawi-Liverpool-Wellcome Trust Clinical Research Program using a BacT/Alert System (BioMerieux), and antibiotic resistance was tested by standard disk diffusion as described previously (18). Nine Salmonella blood culture isolates and one cerebrospinal fluid isolate (from child 1) were serovar Typhimurium, and two were serovar Typhi. Molecular typing of NTS strains was by pulse-field gel electrophoresis (PFGE) of XbaI restriction endonuclease digests of genomic DNA using a CHEF-DR III PFGE instrument (Bio-Rad, Hercules) as previously described (66) and plasmid profiling using Plasmid Mini Purification Kits (Qiagen).
Serum Bactericidal Assays.
In vitro bactericidal assays to assess the sensitivity of NTS isolates to antibody-mediated killing were performed as previously described (16). Briefly, 106 cfu/mL of serovar Typhimurium D23580 (41) or the infecting Salmonella strain were added to a freshly thawed aliquot of control serum from healthy HIV-uninfected Malawian adults, and viable bacteria were determined by serial dilution and plating on LB agar. In vitro bactericidal assays using human serum used 75% baby rabbit serum (AbD Serotec) as a source of exogenous complement. The bactericidal titer was designated as the highest dilution of serum that could effect a 0.5 log 10 kill at 180 min.
Bioinformatic Analysis.
Sequence conservation of identified Salmonella antigens was obtained by multiple genome comparative analysis with a group of 20 publically available genomes. Sequenced strains and their National Center for Biotechnology Information accession numbers are listed in Table S1. Values are calculated as the percentage of identity at the amino acid level performing alignments using FASTA (fasta36.3.4; University of Virginia).
Production of Recombinant Proteins.
SseB, phoN, and dedD were cloned from S. enterica serovar Typhimurium genomic DNA, inserted into a His-tag vector, and expressed in E. coli BL21star DE3 (Invitrogen). Bacteria pellets were resuspended with guanidinium lysis buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate, 0.5 M NaCl) and disrupted on ice using probe sonication. Recombinant proteins were purified using the ProBond purification system (Invitrogen), dialyzed, and concentrated, and protein concentrations were determined using the Lowry method (Bio-Rad).
Immunization.
C57BL/6 mice were immunized i.v. with 100 μg of recombinant protein mixed with 10 μg LPS with or without alum and boosted 4 wk later. Control mice were injected with 10 μg of LPS alone or 5 × 105 BRD509. Four weeks after immunization, mice were infected orally with 5 × 107 SL1344, and infected mice were monitored daily for survival. Mice that were moribund were killed as required by our animal protocol. To determine bacterial colonization of spleens and livers, organs were recovered 7 d after infection and homogenized in PBS, and serial dilutions were plated onto MacConkey agar plates.
Supplementary Material
Acknowledgments
We thank Dr. Dennis Metzger for the kind gift of IgA- and polymeric Ig receptor-deficient mice. This work was supported by National Institutes of Health Grants AI073672 and AI55743 (to S.J.M.) and Grant AI091298 (to M.R.N.); by a Tropical Research Fellowship from the Wellcome Trust (067902/Z/02/Z) and a Clinical Research Fellowship from GlaxoSmithKline (both to C.A.M.); and by a Wellcome Trust Major Overseas Malawi-Liverpool-Wellcome Program award (award 084679/Z/08/Z to R.S.H.). M.A.K. was supported by National Institutes of Health/National Library of Medicine Biomedical Informatics Training Grant LM-07443-01 (to P. Baldi). This work was supported by a European Union FP7 Industry-Academia Partnerships and Pathways Grant ‘GENDRIVAX’ (Grant 251522) (to C.A.M.). P.H. was supported by a Medical Research Council PhD Studentship.
Footnotes
Conflict of interest statement: C.A.M. and F.N. are both employees of the Novartis Vaccines Institute for Global Health. C.A.M. has received grant support from GlaxoSmithKline.
This article is a PNAS Direct Submission.
See Commentary on page 47214721.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111413109/-/DCSupplemental.
References
- 1.Mead PS, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine MM. Enteric infections and the vaccines to counter them: Future directions. Vaccine. 2006;24:3865–3873. doi: 10.1016/j.vaccine.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 3.Crump JA, Mintz ED. Global trends in typhoid and paratyphoid fever. Clin Infect Dis. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: A systematic review and meta-analysis. Lancet Infect Dis. 2010;10:417–432. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 6.McClelland M, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 7.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos RL, et al. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–422. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Graham SM, et al. Nontyphoidal Salmonella infections of children in tropical Africa. Pediatr Infect Dis J. 2000;19:1189–1196. doi: 10.1097/00006454-200012000-00016. [DOI] [PubMed] [Google Scholar]
- 11.Gilks CF, et al. Life-threatening bacteraemia in HIV-1 seropositive adults admitted to hospital in Nairobi, Kenya. Lancet. 1990;336:545–549. doi: 10.1016/0140-6736(90)92096-z. [DOI] [PubMed] [Google Scholar]
- 12.Gordon MA, et al. Non-typhoidal Salmonella bacteraemia among HIV-infected Malawian adults: High mortality and frequent recrudescence. AIDS. 2002;16:1633–1641. doi: 10.1097/00002030-200208160-00009. [DOI] [PubMed] [Google Scholar]
- 13.Chierakul W, et al. The changing pattern of bloodstream infections associated with the rise in HIV prevalence in northeastern Thailand. Trans R Soc Trop Med Hyg. 2004;98:678–686. doi: 10.1016/j.trstmh.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Peters RP, et al. A prospective study of bloodstream infections as cause of fever in Malawi: Clinical predictors and implications for management. Trop Med Int Health. 2004;9:928–934. doi: 10.1111/j.1365-3156.2004.01288.x. [DOI] [PubMed] [Google Scholar]
- 15.van de Vosse E, Ottenhoff TH. Human host genetic factors in mycobacterial and Salmonella infection: Lessons from single gene disorders in IL-12/IL-23-dependent signaling that affect innate and adaptive immunity. Microbes Infect. 2006;8:1167–1173. doi: 10.1016/j.micinf.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 16.MacLennan CA, et al. The neglected role of antibody in protection against bacteremia caused by nontyphoidal strains of Salmonella in African children. J Clin Invest. 2008;118:1553–1562. doi: 10.1172/JCI33998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker D, et al. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 18.Gordon MA, et al. Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica Serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008;46(7):963–969. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 19.Guzman CA, et al. Vaccines against typhoid fever. Vaccine. 2006;24:3804–3811. doi: 10.1016/j.vaccine.2005.07.111. [DOI] [PubMed] [Google Scholar]
- 20.Fraser A, Paul M, Goldberg E, Acosta CJ, Leibovici L. Typhoid fever vaccines: Systematic review and meta-analysis of randomised controlled trials. Vaccine. 2007;25:7848–7857. doi: 10.1016/j.vaccine.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 21.Gondwe EN, et al. Importance of antibody and complement for oxidative burst and killing of invasive nontyphoidal Salmonella by blood cells in Africans. Proc Natl Acad Sci USA. 2010;107:3070–3075. doi: 10.1073/pnas.0910497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McSorley SJ, Jenkins MK. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:3344–3348. doi: 10.1128/iai.68.6.3344-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos RL, et al. Animal models of Salmonella infections: Enteritis versus typhoid fever. Microbes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 24.Jones BD, Falkow S. Salmonellosis: Host immune responses and bacterial virulence determinants. Annu Rev Immunol. 1996;14:533–561. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 25.Mittrücker HW, Kaufmann SH. Immune response to infection with Salmonella typhimurium in mice. J Leukoc Biol. 2000;67:457–463. doi: 10.1002/jlb.67.4.457. [DOI] [PubMed] [Google Scholar]
- 26.Mastroeni P, Sheppard M. Salmonella infections in the mouse model: Host resistance factors and in vivo dynamics of bacterial spread and distribution in the tissues. Microbes Infect. 2004;6:398–405. doi: 10.1016/j.micinf.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Crawford RW, et al. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci USA. 2010;107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffin AJ, McSorley SJ. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 2011;4:371–382. doi: 10.1038/mi.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffin AJ, Li LX, Voedisch S, Pabst O, McSorley SJ. Dissemination of persistent intestinal bacteria via the mesenteric lymph nodes causes typhoid relapse. Infect Immun. 2011;79:1479–1488. doi: 10.1128/IAI.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charles RC, et al. Characterization of anti-Salmonella enterica serotype Typhi antibody responses in bacteremic Bangladeshi patients by an immunoaffinity proteomics-based technology. Clin Vaccine Immunol. 2010;17:1188–1195. doi: 10.1128/CVI.00104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasan A, McSorley SJ. Visualizing the immune response to pathogens. Curr Opin Immunol. 2004;16:494–498. doi: 10.1016/j.coi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Prechl J, Papp K, Erdei A. Antigen microarrays: Descriptive chemistry or functional immunomics? Trends Immunol. 2010;31:133–137. doi: 10.1016/j.it.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Vigil A, Davies DH, Felgner PL. Defining the humoral immune response to infectious agents using high-density protein microarrays. Future Microbiol. 2010;5:241–251. doi: 10.2217/fmb.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies DH, et al. Profiling the humoral immune response to infection by using proteome microarrays: High-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giefing C, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. 2008;205:117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies DH, et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol. 2008;82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastroeni P, Simmons C, Fowler R, Hormaeche CE, Dougan G. Igh-6(-/-) (B-cell-deficient) mice fail to mount solid acquired resistance to oral challenge with virulent Salmonella enterica serovar typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect Immun. 2000;68:46–53. doi: 10.1128/iai.68.1.46-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittrücker HW, Raupach B, Köhler A, Kaufmann SH. Cutting edge: Role of B lymphocytes in protective immunity against Salmonella typhimurium infection. J Immunol. 2000;164:1648–1652. doi: 10.4049/jimmunol.164.4.1648. [DOI] [PubMed] [Google Scholar]
- 39.Strugnell R, et al. Characterization of a Salmonella typhimurium aro vaccine strain expressing the P.69 antigen of Bordetella pertussis. Infect Immun. 1992;60:3994–4002. doi: 10.1128/iai.60.10.3994-4002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng W, et al. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kingsley RA, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClelland M, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 43.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: The interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 44.Ravindran R, McSorley SJ. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology. 2005;114:450–458. doi: 10.1111/j.1365-2567.2005.02140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tam MA, Rydström A, Sundquist M, Wick MJ. Early cellular responses to Salmonella infection: Dendritic cells, monocytes, and more. Immunol Rev. 2008;225:140–162. doi: 10.1111/j.1600-065X.2008.00679.x. [DOI] [PubMed] [Google Scholar]
- 46.Valdez Y, Ferreira RB, Finlay BB. Molecular mechanisms of Salmonella virulence and host resistance. Curr Top Microbiol Immunol. 2009;337:93–127. doi: 10.1007/978-3-642-01846-6_4. [DOI] [PubMed] [Google Scholar]
- 47.Harris JB, et al. Identification of in vivo-induced bacterial protein antigens during human infection with Salmonella enterica serovar Typhi. Infect Immun. 2006;74:5161–5168. doi: 10.1128/IAI.00488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadallah F, et al. Production of specific monoclonal antibodies to Salmonella typhi flagellin and possible application to immunodiagnosis of typhoid fever. J Infect Dis. 1990;161:59–64. doi: 10.1093/infdis/161.1.59. [DOI] [PubMed] [Google Scholar]
- 49.Ortiz V, Isibasi A, García-Ortigoza E, Kumate J. Immunoblot detection of class-specific humoral immune response to outer membrane proteins isolated from Salmonella typhi in humans with typhoid fever. J Clin Microbiol. 1989;27:1640–1645. doi: 10.1128/jcm.27.7.1640-1645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown A, Hormaeche CE. The antibody response to Salmonellae in mice and humans studied by immunoblots and ELISA. Microb Pathog. 1989;6:445–454. doi: 10.1016/0882-4010(89)90086-7. [DOI] [PubMed] [Google Scholar]
- 51.Calderón I, et al. Antibodies to porin antigens of Salmonella typhi induced during typhoid infection in humans. Infect Immun. 1986;52:209–212. doi: 10.1128/iai.52.1.209-212.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siggins MK, et al. Absent bactericidal activity of mouse serum against invasive African nontyphoidal Salmonella results from impaired complement function but not a lack of antibody. J Immunol. 2011;186:2365–2371. doi: 10.4049/jimmunol.1000284. [DOI] [PubMed] [Google Scholar]
- 53.Feasey NA, et al. Typhoid fever and invasive nontyphoid salmonellosis, Malawi and South Africa. Emerg Infect Dis. 2010;16:1448–1451. doi: 10.3201/eid1609.100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 55.Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Adoptive transfer of immunity to oral challenge with virulent Salmonellae in innately susceptible BALB/c mice requires both immune serum and T cells. Infect Immun. 1993;61:3981–3984. doi: 10.1128/iai.61.9.3981-3984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uren TK, et al. Vaccine-induced protection against gastrointestinal bacterial infections in the absence of secretory antibodies. Eur J Immunol. 2005;35:180–188. doi: 10.1002/eji.200425492. [DOI] [PubMed] [Google Scholar]
- 57.Tobar JA, González PA, Kalergis AM. Salmonella escape from antigen presentation can be overcome by targeting bacteria to Fc gamma receptors on dendritic cells. J Immunol. 2004;173:4058–4065. doi: 10.4049/jimmunol.173.6.4058. [DOI] [PubMed] [Google Scholar]
- 58.Ugrinovic S, Ménager N, Goh N, Mastroeni P. Characterization and development of T-cell immune responses in B-cell-deficient (Igh-6(-/-)) mice with Salmonella enterica serovar Typhimurium infection. Infect Immun. 2003;71:6808–6819. doi: 10.1128/IAI.71.12.6808-6819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johanns TM, et al. Early eradication of persistent Salmonella infection primes antibody-mediated protective immunity to recurrent infection. Microbes Infect. 2011;13:322–330. doi: 10.1016/j.micinf.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J Immunol. 2000;164:986–993. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- 61.Strindelius L, Degling Wikingsson L, Sjöholm I. Extracellular antigens from Salmonella enteritidis induce effective immune response in mice after oral vaccination. Infect Immun. 2002;70:1434–1442. doi: 10.1128/IAI.70.3.1434-1442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gil-Cruz C, et al. The porin OmpD from nontyphoidal Salmonella is a key target for a protective B1b cell antibody response. Proc Natl Acad Sci USA. 2009;106:9803–9808. doi: 10.1073/pnas.0812431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rollenhagen C, Sörensen M, Rizos K, Hurvitz R, Bumann D. Antigen selection based on expression levels during infection facilitates vaccine development for an intracellular pathogen. Proc Natl Acad Sci USA. 2004;101:8739–8744. doi: 10.1073/pnas.0401283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SJ, et al. Temporal expression of bacterial proteins instructs host CD4 T cell expansion and Th17 development. PLoS Pathog. 2012;8:e1002499. doi: 10.1371/journal.ppat.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geddes K, Cruz F, Heffron F. Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog. 2007;3:e196. doi: 10.1371/journal.ppat.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kariuki S, et al. Characterisation of community acquired non-typhoidal Salmonella from bacteraemia and diarrhoeal infections in children admitted to hospital in Nairobi, Kenya. BMC Microbiol. 2006;6:101. doi: 10.1186/1471-2180-6-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.